Introduction
Humans have a homeostatic effect that maintains a
constant internal environment in response to various stimuli,
including changes in the external or internal environment. The
degradation, repair or stability of damaged proteins affects the
efficiency of cellular homeostasis and aging (1). The ubiquitin-proteasome system (UPS)
via ubiquitin and the 26S proteasome is essential for protein
homeostasis (2). Ubiquitin, a
small protein of 76 amino acids, binds to a target protein by
interacting with three enzymes: E1 (ubiquitin-activating enzyme),
E2 (ubiquitin-conjugating enzyme) and E3 (ubiquitin ligase). The E1
enzyme activates ubiquitin and generates a thioester intermediate
through ATP-dependent chemical bonding (3). The E2 enzyme receives the activated
ubiquitin and the E3 ligase delivers the ubiquitin to the target
protein. Ubiquitin forms a polyubiquitin chain such that the
c-terminal G76 (glycine) forms an isopeptide bond with the free
amino group K (lysine) or M (methionine). The binding sites of
ubiquitin are seven lysine residues (K6, K11, K27, K29, K33, K48
and K63) and one methionine residue (M1) (4). The role of ubiquitin varies,
depending on the position where it forms the polyubiquitin chain
(5). The K6-linked polyubiquitin
responds to mitochondrial homeostasis and DNA damage, whereas the
K11-linked polyubiquitin regulates the cell cycle (6). The K27-linked polyubiquitin chain
responds to DNA damage, K29-linked polyubiquitin regulates
lysosomal degradation and kinase modification, and K33-linked
polyubiquitin is involved in protein trafficking and kinase
modification (7,8). The K48-linked polyubiquitin and
K63-linked polyubiquitin are the most studied. The K48-linked
polyubiquitin regulates proteasomal degradation (9), whereas the K63-linked polyubiquitin
induces autophagic degradation (10,11). The M1-linked polyubiquitin is
involved in gene activation, DNA damage response and innate
immunity (12). The target
proteins tagged with polyubiquitin are directed to the 26S
proteasome and the protein separated from the polyubiquitin chain
passes through the 26S proteasome and is degraded. This process is
called the UPS and the ubiquitin dissociated from the substrates is
recycled in the UPS process (13). E3 ligase induces proteasomal
degradation by linking the polyubiquitin chain to the substrate
protein, but certain enzymes have a contrary role by breaking the
ubiquitin bond; these are the deubiquitinating enzymes (DUBs)
(14,15). DUBs are classified into 9
subfamilies: Jab1/Pab1/MPN metallo-enzyme motif protease, monocyte
chemotactic protein-induced protease, motif interacting with
ubiquitin-containing novel DUB family, Machado-Joseph disease
protein domain protease, ovarian tumor protease (OTU), permuted
papain fold peptidase of dsDNA viruses and eukaryotes, ubiquitin
C-terminal hydrolases protease, ubiquitin-specific protease (USP)
and zinc finger with UFM1-specific peptidase domain protein.
Ovarian tumor deubiquitinase 6A (also called OTU
domain-containing protein 6A; OTUD6A) is a member of the OTU
subfamily and is known to promote tumorigenesis in various
carcinomas. OTUD6A and dynamin-related protein 1 (Drp1), which are
upregulated in colorectal cancer tissues, interact with each other.
The OTUD6A stabilizes Drp1 by deubiquitination and promotes cancer
proliferation and mitochondrial fission (16). OTUD6A acts as a DUB of the cell
cycle regulator Aurora kinase-A (Aurora-A), which is overexpressed
in malignancies, and stabilizes Aurora-A (17). OTUD6A promotes prostate
tumorigenesis via deubiquitinating and stabilizing brahma-related
gene 1 androgen receptor (18),
and c-Myc (19). OTUD6A also
promotes breast cancer proliferation through deubiquitination and
stabilization of DNA topoisomerase II binding protein 1 (20). In addition, OTUD6A has a
differential expression depending on the presence or absence of p53
in the HCT116 cell line (21).
However, the mechanism and effect on cancer cells, which are
regulated by interaction between OTUD6A and p53, have not been
elucidated. Therefore, in the present study, substrates of OTUD6A
related to p53 signaling were discovered through liquid
chromatography-tandem mass spectrometry (LC-MS/MS) or
immunoprecipitation and it was demonstrated that the substrates
regulate cell proliferation in breast cancer cells. Therefore, it
is suggested that OTUD6A, which may regulate the p53 signaling
pathway, may be a therapeutic target for cancer treatment.
Materials and methods
Construction of expression vector
The full-length cDNA for OTUD6A (cat. no. 61416;
Addgene, Inc.) was subcloned into pcDNA3.1-6myc vector (cat. no.
V790-20; Invitrogen; Thermo Fisher Scientific, Inc.), pCS4-3XFlag
vector (cat. no. 55751; Addgene, Inc.) and pGEX-4T-1 vector (cat.
no. 27-4580-01; Pharmacia Biotech) and C152S, a catalytically
inactive mutant of OTUD6A, was mutated by PCR with the following
primer sequences: Forward, 5′-ACGGCCACAGCATGTAC-3′ and reverse,
5′-TACATGCTGTGGCCGTC-3′, Pfu polymerase (cat. no. CMT4002;
LaboPass; COSMOGENETECH, Inc) was used and the thermocycling
protocol was as follows: Denaturation was carried out at 95°C for 5
min. Subsequently, 15 cycles of denaturation at 95°C for 30 sec,
annealing at 54°C for 30 sec, and extension at 72°C for 5 min and
10 sec were performed. After extension at 72°C for 5 min, the
mixture was maintained at 4°C. The fragment was subcloned into
pcDNA3.1-6myc vector using BamHI (cat. no. R003S;
Enzynomics, Inc.) and EcoRI restriction enzymes (cat. no.
R002S; Enzynomics, Inc.). The cDNA for Flag-nucleolin was published
in a previous study by our group (22) and caspase-7-Flag (cat. no. 11815;
Addgene, Inc.) was used. Furthermore, hemagglutinin (HA)-tagged
ubiquitin (wild-type) and mutant constructs (K6, K11, K27, K29,
K33, K48 and K63) were used, the details of which were published in
a previous study by our group (23).
Cell culture and transfection
Human colon cancer cells [HCT116
(p53+/+); cat. no. CCL-247; American Type Culture
Collection (ATCC)] were grown in RPMI-1640 (cat. no. 31800-022;
Gibco; Thermo Fisher Scientific, Inc.) medium containing 10% fetal
bovine serum (FBS; cat. no. 12483-020; Gibco; Thermo Fisher
Scientific, Inc.) and 1% antibiotic-antimycotic (containing
penicillin, streptomycin and amphotericin B; cat. no. 15240-062;
Gibco; Thermo Fisher Scientific, Inc.). Human breast cancer cells
(MCF7; cat. no. HTB-22; ATCC) were grown in DMEM (cat. no.
12800-017; Gibco; Thermo Fisher Scientific, Inc.) containing 10%
FBS and 1% penicillin/streptomycin. The cells were grown in a 5%
CO2 incubator at 37°C. For transfection, 10 mM
polyethyleneimine reagent (Polysciences, Inc.), 3–6 µl/µg of DNA
and 150 mM NaCl (100 µl/ml of media) were used and incubated at
37°C. For western blot analysis, immunoprecipitation and
glutathione S-transferase (GST) pull-down assay, proteins were
purified from harvested cells 48 h after transfection.
Antibodies
Anti-Flag (1:5,000 dilution; cat. no. M185-3L; MBL
International Corporation), anti-Myc (1:100 dilution; 9E10
hybridoma cell media; cat. no. CRL-1729; ATCC) and anti-HA (1:1,000
dilution; cat. no. 11 583 816 001; Roche Diagnostics) antibodies
were used; furthermore, anti-β-actin (1:1,500 dilution; cat. no.
sc-47778), anti-nucleolin (1:1,500 dilution; cat. no. sc-8031),
anti-caspase-7 (1:200 dilution; cat. no. sc-56063), anti-poly(ADP
ribose) polymerase 1 (PARP1; 1:200 dilution; cat. no. sc-7150),
anti-X-linked inhibitor of apoptosis protein (XIAP; 1:200 dilution;
cat. no. sc-11426), anti-p53 (1:500 dilution; cat. no. sc-126),
anti-p27 (1:200 dilution; cat. no. sc-528) and anti-Bax (1:200
dilution; cat. no. sc-23959) antibodies were purchased from Santa
Cruz Biotechnology, Inc. and used for western blot and
immunoprecipitation assays.
Western blot analysis,
immunoprecipitation and LC-MS/MS
Cells were lysed using a lysis buffer [50 mM
Tris-HCl pH 7.5, 1 mM EDTA, 10% glycerol, 300 mM NaCl, 1% Triton
X-100, 10% SDS (1:100 dilution), a protease inhibitor cocktail
(PIC; 1:100 dilution; cat. no. 11697498001; Roche Diagnostics) and
phenylmethanesulfonyl fluoride (PMSF; 1:100 dilution;
MilliporeSigma)] and incubated on ice for 20 min after vortexing.
Centrifugation was performed at 16,200 × g for 20 min at 4°C and
the supernatants were used as samples. The total protein in the
samples was quantified using the Bradford assay, and samples were
boiled at 100°C for 7 min with 2X SDS and used for western blot
analysis. For each lane, 30 µg of protein was separated by size by
8% SDS-PAGE and transferred to polyvinylidene fluoride microporous
membranes (cat. no. IPVH00010; EMD Millipore) at 120 V for 90 min.
Proteins transferred to the membrane were blocked with 20 mM
Tris-HCl, 0.05% Tween 20 and 150 mM NaCl (TTBS) containing 5% BSA
(cat. no. BSAS 0.1; Bovogen Biologicals) or skimmed milk (cat. no.
232100; Becton, Dickinson and Company) at room temperature for 1 h
and then incubated at 4°C overnight with primary antibodies. The
membranes were then washed three times with TTBS for 7 min each and
then incubated at room temperature for 1 h with a secondary
antibody [mouse IgG (H+L) (1:30,000 dilution; cat. no. 074-1806;
LGC SeraCare)]. The membranes are then washed 3 times with TTBS for
7 min each and detected using enhanced chemiluminescence solution
(Young-In Frontier).
For immunoprecipitation, 2 mg of cell lysates were
incubated at 4°C on a rotator at 20 rpm overnight with an antibody
(1 µg per 500 µg of total protein) and incubated with 30 µl
resuspended volume of Protein A/G PLUS-Agarose Beads (cat. no.
sc-2003; Santa Cruz Biotechnology, Inc.) at 4°C on a rotator at 20
rpm for 2 h. The samples were centrifuged at 3,200 × g at 4°C for 5
min and subsequently, the beads were washed 2 times with wash
buffer [50 mM Tris-HCl pH 7.5, 1 mM EDTA, 10% glycerol, 300 mM
NaCl, 1% Triton X-100, PIC (1:100 dilution) and PMSF (1:100
dilution)] and then boiled in 2X SDS at 100°C for 7 min, followed
by the western blot procedure.
For LC-MS/MS, pCS4-3XFlag vector or Flag-OTUD6A was
transfected into HCT116 (p53+/+) cells. Cells were lysed and
immunoprecipitation was performed using anti-Flag antibody. Western
blot analysis was performed using 8% SDS-PAGE and gels were stained
with Coomassie Brilliant blue R 250 (cat. no. CI 42660;
MilliporeSigma) and G 250 (cat. no. CI 42655; MilliporeSigma)
solutions at room temperature for 15 min. LC-MS/MS analysis was
performed using the sample showing a higher expression in the
Flag-OTUD6A-transfected sample compared to the sample transfected
with the pCS4-3XFlag vector, which was used as a control.
GST pull-down assay
GST vector (pGEX-4T-1) and GST-OTUD6A were
transformed into the BL21 (DE3) bacterial strain (cat. no. C2527H;
New England BioLabs) and bacteria grown to an optical density value
at 600 nm of 0.6 at 37°C were incubated in Luria-Bertani broth
(cat. no. MB-L4488; KisanBio, Inc.) with 0.5 mM isopropyl
β-D-1-thiogalactopyranoside (Promega Corporation) at 18°C
overnight. The protein bound to GST-OTUD6A was boiled at 100°C with
2X SDS buffer and detected through western blot analysis. The
expression of GST and GST-OTUD6A was visualized with Coomassie
Brilliant Blue R 250 and G 250 solutions.
Protein stability assay
MCF7 cells were transfected with pCS4-Flag vector or
Flag-OTUD6A and incubated for 24 h after transfection. After 24 h,
cycloheximide (CHX; cat. no. 01810; MilliporeSigma) was treated at
a concentration of 50 µg/ml in a 5% CO2 incubator at
37°C. The cells were then harvested at different time-points (0, 6,
12 and 24 h) and cell lysates were used for western blot
analysis.
Ubiquitination and deubiquitination
assays
For the ubiquitination assay, Flag-nucleolin (or
caspase7-Flag) and HA-Ub were co-transfected into MCF7 cells. For
the deubiquitination assay, Myc-OTUD6A, Flag-nucleolin (or
caspase-7-Flag) and HA-Ub were co-transfected into MCF7 cells.
MG132 (cat. no. F1100; Ubiquitin Proteasome Biotechnologies, LLC)
was used to confirm the proteasomal-dependent degradation of the
substrate. Cells were treated with 10 mM concentration of MG132 for
4 h before harvest and cultured in a 5% CO2 incubator at
37°C. Both assays were performed using an ubiquitination assay kit
(cat. no. UBAK-100; D&P Biotech, Inc.) according to the
manufacturer's protocol. The cell lysates were immunoprecipitated
with an anti-Flag antibody. Western blot analysis was performed
using immunoprecipitated cell lysates according to the
immunoprecipitation procedure.
Colony-formation assay
pCS4-3XFlag vector or Flag-OTUD6A-transfected MCF7
cells (1.5×103 cells/100 mm culture dish) were seeded 24
h after transfection and cultured at 37°C in a 5% CO2
incubator for 14 days to form colonies. Next, the colonies were
stained using crystal violet (cat. no. 27210-0350; Junsei) diluted
with PBS at room temperature for 5 min. After washing with PBS, the
colonies were counted on each plate. Images were captured using a
DUALED Blue/White Transilluminator (cat. no. A6020; Bioneer
Corporation) and counting for colony formation analysis was
conducted using ImageJ software (version 1.4.3; National Institutes
of Health).
Cell cycle assay
MCF7 cells were synchronized in G1/S phase of the
cell cycle by double thymidine block. The thymidine block was
treated with thymidine (cat. no. T1895; MilliporeSigma) with the
final concentration of 2 mM. MCF7 cells were transfected with
pCS4-3XFlag vector or Flag-OTUD6A, or pCMV-Flag or Flag-nucleolin.
At 24 h after transfection, cells were incubated with thymidine at
a final concentration of 2 mM at 37°C for 18 h. The cells were then
washed with PBS and incubated in fresh media at 37°C for 9 h. For
double thymidine block, the second thymidine was applied at a final
concentration of 2 mM at 37°C for 18 h. The synchronized cells were
washed with PBS and released with fresh media. Cells were harvested
at 0, 3, 6, 12 and 24 h after release. A total of 1×106
cells were fixed with cold 70% ethanol for 30 min at −20°C and
washed with PBS to remove the ethanol. Fixed cells were stained
with 500 µl of FxCycle™ PI/RNase Staining Solution (cat. no.
F10797; Thermo Fisher Scientific, Inc.) at room temperature for 30
min and analyzed with a CytoFLEX flow cytometer (Beckman Coulter,
Inc.) and CytExpert software (version 2.4.0.28; Beckman Coulter,
Inc.).
Survival analysis
The survival analysis was performed by Kaplan-Meier
Plotter using the log-rank test (https://kmplot.com/analysis/). The Kaplan-Meier plot
of survival was based on the high or low OTUD6A expression in
patients with breast cancer from the Tang (2018, n=118) dataset
(P-value by log-rank test). Patients were divided by the median
value and the analysis was performed on 65 patients using the
selected dataset.
Statistical analysis
For all measured data, the values for all samples
obtained from at least three independent experiments were averaged
and the standard deviation or standard error was subsequently
calculated. Statistical analysis was performed using the unpaired
t-test and one-way analysis of variance followed by Tukey's
multiple-comparisons tests using GraphPad Prism version 5 (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Putative substrates that bind to
OTUD6A
Through multiplex reverse transcription-quantitative
PCR, our group previously confirmed that OTUD6A is highly expressed
in HCT116 (p53+/+) cells as compared to HCT116
(p53−/−) cells (21),
thereby indicating that OTUD6A is probably affected by p53.
However, the regulatory mechanisms of OTUD6A and p53 have not been
elucidated. p53 is a well-known tumor suppressor (24). It was expected that the
association of OTUD6A with p53 affects cell survival. Therefore,
the present study aimed to identify putative substrates for OTUD6A
related to p53 in HCT116 (p53+/+) cells via LC-MS/MS (Fig. 1A). LC-MS/MS studies determined
that nucleolin is a candidate substrate (Table I). Nucleolin is known to interact
with p53 (25,26) and regulates cell proliferation and
the DNA damage response by interacting with the herpes
virus-associated USP (HAUSP)-p53-MDM2 complex (22). Immunoprecipitation was performed
to determine whether nucleolin binds to OTUD6A. The band of
nucleolin was detected, indicating that OTUD6A and nucleolin
interact with each other (Fig.
1B). In addition, to investigate direct binding between OTUD6A
and nucleolin, the GST pull-down assay was performed using the
GST-OTUD6A fusion protein. The assay revealed that OTUD6A directly
binds to nucleolin (Fig. 1C). It
is known that p53 binds to nucleolin through the C-terminal domain
(25,26). Therefore, a GST pull-down assay
was performed to determine whether OTUD6A, nucleolin and p53 form a
complex (Fig. 1D). It was
demonstrated that the purified GST-OTUD6A protein binds to
nucleolin and p53 in a complex.
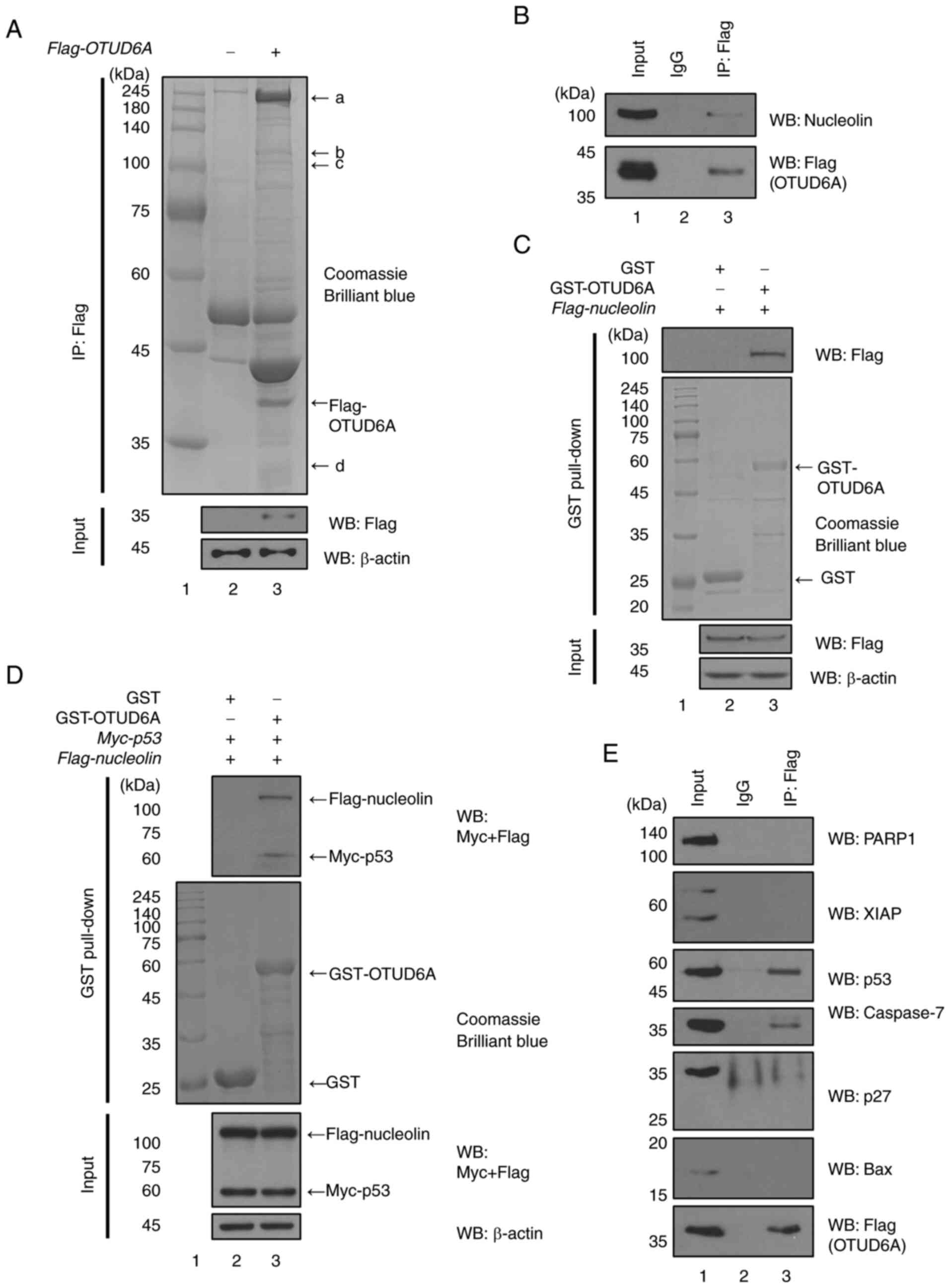 | Figure 1.Putative substrates that bind to
OTUD6A. (A) Flag-OTUD6A-transfected HCT116 (p53+/+) cell lysates
were immunoprecipitated with anti-Flag antibody. Proteins were
separated by size using 8% SDS-PAGE. The 1-dimensional
electrophoresis gels were used for the liquid chromatography tandem
mass spectrometry assay. (B) Flag-OTUD6A-transfected MCF7 cell
lysates were immunoprecipitated with an anti-Flag antibody. Bands
were detected with anti-Flag or anti-nucleolin antibody. (C) GST
and GST-OTUD6A fusion proteins were inducted with 0.5 mM
β-D-1-thiogalactopyranoside at 18°C for 16 h. GST and GST-OTUD6A
fusion proteins were incubated with lysates of
Flag-nucleolin-transfected MCF7 cells and WB was performed.
Flag-tagged nucleolin was blotted with anti-Flag antibody. (D) GST
and GST-OTUD6A fusion proteins were incubated with lysates of
Flag-nucleolin- and Myc-p53-transfected MCF7 cells and WB was
performed. (E) Flag-OTUD6A-transfected MCF7 cell lysates were
immunoprecipitated with anti-Flag antibody. Bands were detected
with anti-PARP1, anti-XIAP, anti-p53, anti-p27, anti-Bax,
anti-caspase-7 or anti-Flag antibody. OTUD6A, ovarian tumor
deubiquitinase 6A; PARP, poly (ADP ribose) polymerase; XIAP,
X-linked inhibitor of apoptosis protein; GST, glutathione
S-transferase; IP, immunoprecipitation; WB, western blot. |
 | Table I.List of proteins interacting with
ovarian tumor deubiquitinase 6A identified by liquid
chromatography-tandem mass spectrometry and proteomics
analysis. |
Table I.
List of proteins interacting with
ovarian tumor deubiquitinase 6A identified by liquid
chromatography-tandem mass spectrometry and proteomics
analysis.
| Sample ID | Protein name | Monoisotopic mass
(Da) | pI
value | Protein sequence
coverage, % | Number of matched
peptides |
|---|
| a | TSGA10 | 104514 | 5.51 | 1 | 2 |
| b | EPLIN-β | 85630 | 6.41 | 2 | 3 |
| c | Nucleolin | 76355 | 4.59 | 8 | 3 |
| d | BAP31 | 23621 | 9.57 | 9 | 1 |
| d | Porin | 38639 | 6.32 | 2 | 1 |
| d | RPS3A | 30154 | 9.75 | 6 | 1 |
In the present study, it was hypothesized that the
apoptotic proteins that function similarly to the tumor suppressor
p53 are also related to OTUD6A. Therefore, a binding assay between
OTUD6A and apoptotic proteins associated with p53 was performed in
MCF7 cells. The results revealed that Flag-OTUD6A and caspase-7 or
p53 bind to each other (Fig. 1E),
thereby demonstrating that OTUD6A binds to caspase-7.
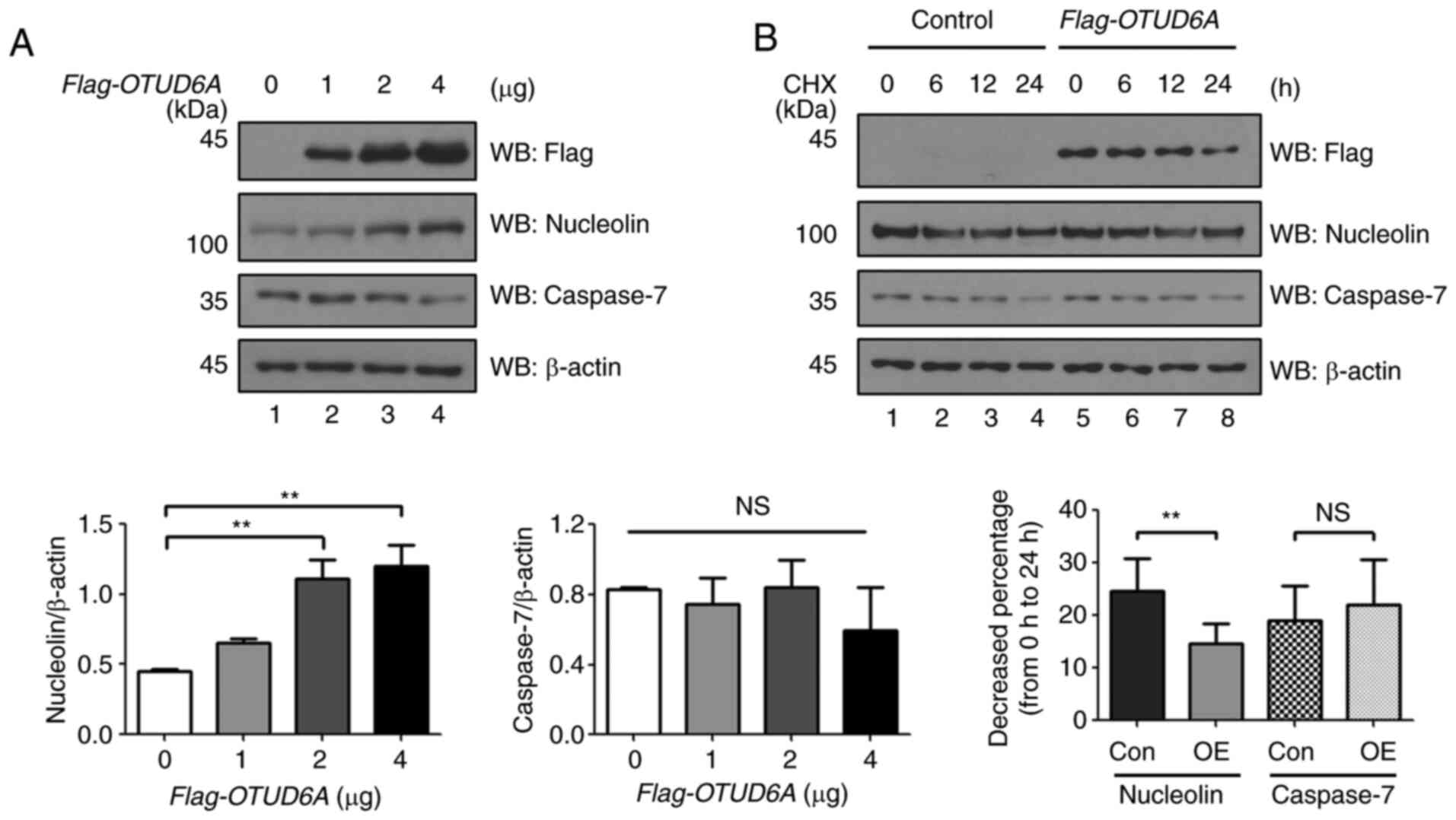
OTUD6A regulates the protein stability
of nucleolin
Next, the effect of OTUD6A on the protein stability
of nucleolin and caspase-7 was investigated. The expression level
of nucleolin was increased in a dose-dependent manner, but no
significant difference was obtained in the protein expression level
of caspase-7 in MCF7 cells transfected with varying concentrations
of Flag-OTUD6A (Fig. 2A). The
overexpression effect of Flag-OTUD6A on the protein half-life of
nucleolin and caspase-7 was further investigated using CHX, a
protein synthesis inhibitor. The expression level of nucleolin
decreased by 25% from 0 to 24 h in the mock control and decreased
by 15% from 0 to 24 h in Flag-OTUD6A-transfected cells. The
expression of caspase-7 protein was decreased by 19% from 0 to 24 h
in the mock control and decreased by 22% from 0 to 24 h in
Flag-OTUD6A-transfected cells, but there was no significant
difference. The half-life of nucleolin was significantly extended
in Flag-OTUD6A-transfected cells (Fig. 2B).
OTUD6A deubiquitinates nucleolin and
caspase-7 through the UPS
In order to investigate the function of OTUD6A as a
DUB, K mutants of ubiquitin remaining at only one of the K sites
were used for ubiquitination and deubiquitination assays (K6, K11,
K27, K29, K33, K48 and K63) (Fig.
3A). To investigate whether nucleolin is regulated by
proteasomal degradation, the ubiquitination assay was performed
using a proteasome inhibitor, MG132 (Fig. 3B). The ubiquitination level of
nucleolin was increased on the WT, K6-, K11-, K29-, K33-, K48 and
K63-linked polyubiquitin chains, but not on the K27-linked
polyubiquitin chain. The binding between OTUD6A and nucleolin was
confirmed by the GST pull-down assay (Fig. 1C). Therefore, the deubiquitination
assay was performed with Myc-OTUD6A (C152S), a catalytically
inactive form of OTUD6A, to examine whether OTUD6A functions as a
DUB for nucleolin. The expression level of the polyubiquitin chain
of nucleolin decreased in Myc-OTUD6A-transfected MCF7 cells, but no
alterations were observed in the Myc-OTUD6A (C152S)-transfected
MCF7 cells (Fig. 3C). The binding
between OTUD6A and caspase-7 was also demonstrated (Fig. 1D). Therefore, a deubiquitination
assay was performed to check the ubiquitination level of caspase-7
mediated by OTUD6A. When OTUD6A was overexpressed, the
ubiquitination level of caspase-7 decreased, whereas the
ubiquitination level of caspase-7-Flag remained unchanged in the
OTUD6A (C152S)-transfected MCF7 cells (Fig. 3D). These results suggest that
OTUD6A is a DUB for nucleolin and caspase-7. In addition, the
deubiquitination assay was performed using HA-Ub lysine mutants to
determine which lysine site-linked polyubiquitin chains were
removed by OTUD6A. The expression level of the K63-linked
polyubiquitin chain of caspase-7 was observed to be decreased, but
not the K48-linked polyubiquitin chain (Fig. 3E). These results indicate that the
binding of OTUD6A to caspase-7 affects cancer cells through the
removal of the K63-linked polyubiquitin chain, but not the
K48-linked polyubiquitin chain associated with proteasomal
degradation. Furthermore, the ubiquitination level of nucleolin was
found to be decreased on the K6-, K11-, K27-, K33- and K48-linked
polyubiquitin chains, but not on the K29- and K63-linked
polyubiquitin chains (Fig. 3F).
This indicates that OTUD6A regulates the protein stability of
nucleolin via deubiquitination of the K48-linked polyubiquitin
chain, which is well known to regulate proteasome-mediated protein
degradation (9).
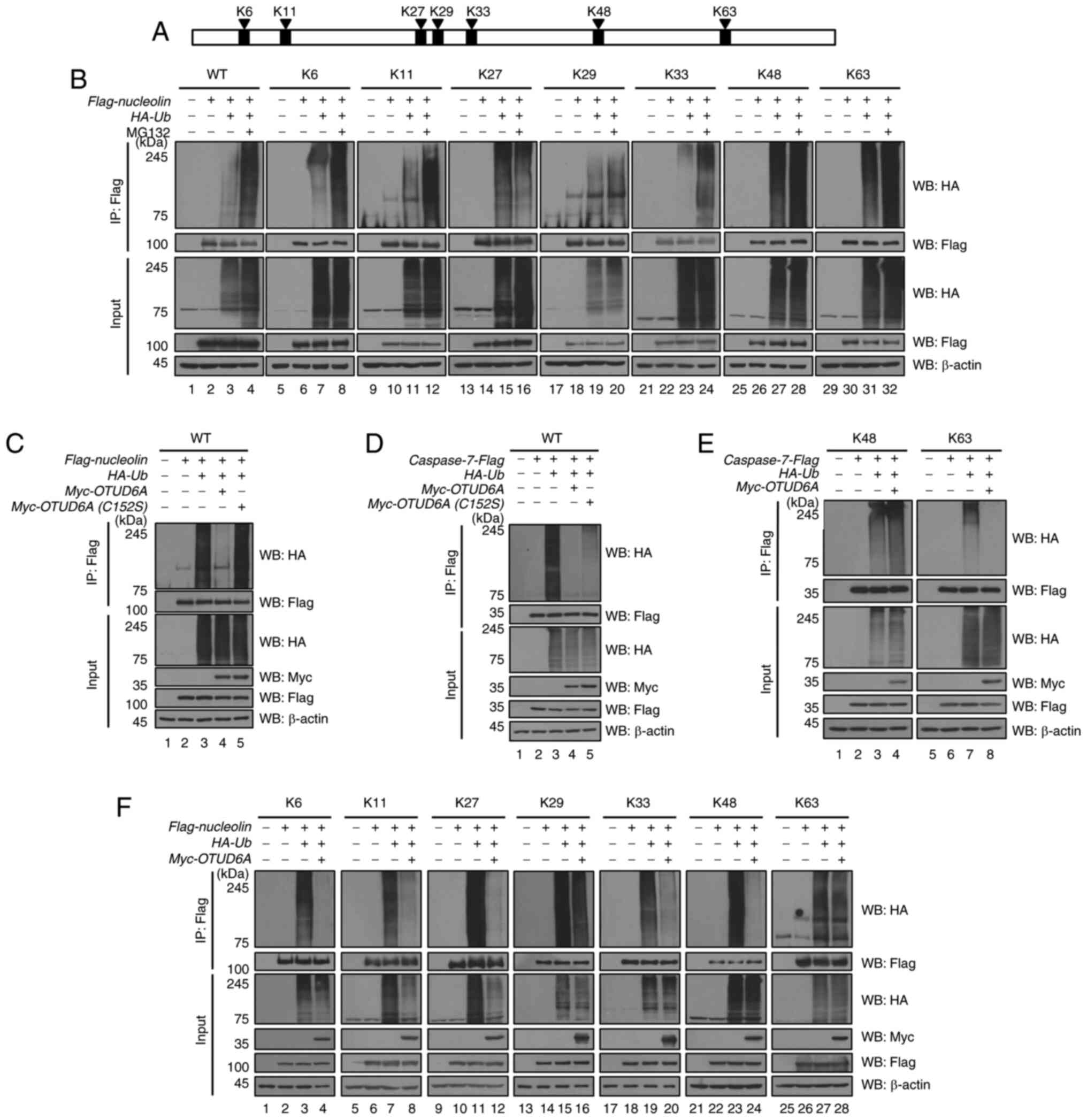 | Figure 3.OTUD6A deubiquitinates nucleolin and
caspase-7 through UPS. (A) HA-ubiquitin lysine mutants contain only
one lysine residue among the seven lysine sites of ubiquitin. (B)
The ubiquitination assay of Flag-nucleolin was performed in MCF7
cells with MG132. MCF7 cells were transfected with Flag-nucleolin
along with HA-Ub+K6, HA-Ub+K11,
HA-Ub+K27, HA-Ub+K29, HA-Ub+K33,
HA-Ub+48 or HA-Ub+K63 and treated with MG132.
(C) Flag-nucleolin was co-transfected with HA-Ub, Myc-OTUD6A or
Myc-OTUD6A (C152S) into MCF7 cells. (D) Caspase-7-Flag was
co-transfected with HA-Ub, Myc-OTUD6A or Myc-OTUD6A (C152S) into
MCF7 cells. (E) MCF7 cells were transfected with caspase-7-Flag or
Myc-OTUD6A along with HA-Ub+48 or HA-Ub+K63.
(F) MCF7 cells were transfected with Flag-nucleolin or Myc-OTUD6A
along with HA-Ub+K6, HA-Ub+K11,
HA-Ub+K27, HA-Ub+K29, HA-Ub+K33,
HA-Ub+48 or HA-Ub+K63. Cells were lysed and
WB was performed. Lysates were immunoprecipitated with an anti-Flag
antibody and polyubiquitin chains were detected with an anti-HA
antibody. OTUD6A, ovarian tumor deubiquitinase 6A; IP,
immunoprecipitation; WB, western blot; HA, hemagglutinin-tag
protein; Ub, ubiquitin. |
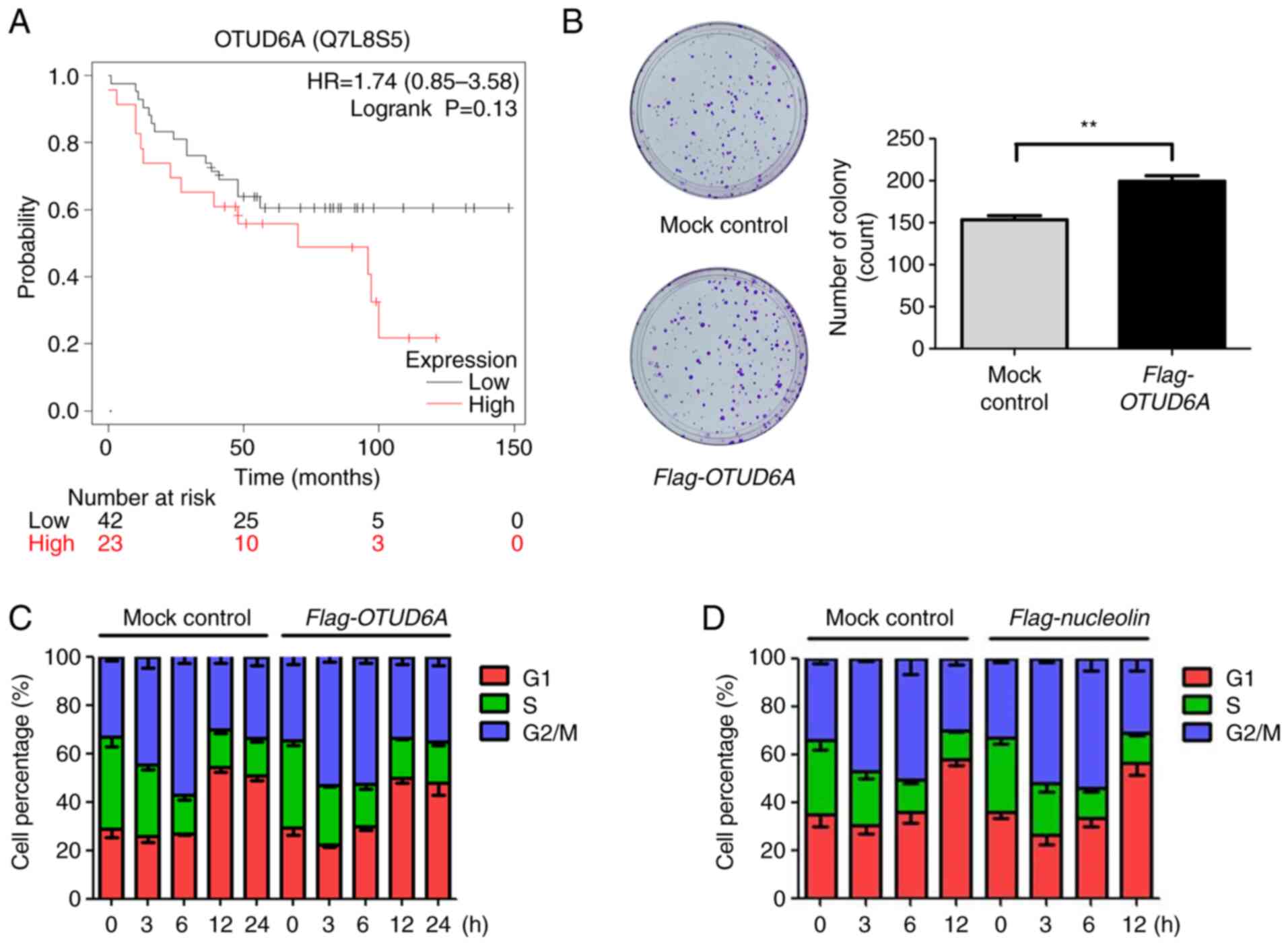
OTUD6A regulates cell
proliferation
It was investigated whether OTUD6A stabilizes the
protein stability of nucleolin through deubiquitination of the
K48-linked polyubiquitin chain (Fig.
3F). Nucleolin is known to regulate oncogenes in several
cancers (27). It has been
reported that nucleolin is upregulated by β-crystallin B2 (CRYβB2)
in breast cancer to induce tumorigenesis (28). In addition, caspase-7 is known as
an apoptotic protein (29).
Therefore, the effect of OTUD6A in cancer cells was investigated
through deubiquitination of nucleolin or caspase-7. First, a
survival analysis for patients with breast cancer was performed
using Kaplan-Meier Plotter. Higher expression of OTUD6A was
associated with an increased risk and poor survival (Fig. 4A). Next, a colony formation assay
was performed to investigate cell proliferation of cells with high
expression of OTUD6A. It was determined that the colony formation
ability was increased in Flag-OTUD6A-overexpressing cells, as
compared to the mock control (Fig.
4B). It was previously reported that downregulated nucleolin
protein is related to cell cycle arrest in ADP-induced cells
(30). Therefore, flow cytometric
analysis was performed to demonstrate cell cycle progression. The
results indicated that cell cycle progression was faster in MCF7
cells with overexpression of OTUD6A (Fig. 4C). In order to prove that the
promotion of cell cycle progression is due to upregulation of
nucleolin through overexpression of OTUD6A, cell cycle analysis was
performed with overexpression of nucleolin. Similar to the effect
of overexpression of OTUD6A in Fig.
4C, the cell cycle progression of the MCF7 cells transfected
with Flag-nucleolin was promoted (Fig. 4D). These data suggest that OTUD6A
regulates cell proliferation by promoting cell cycle progression in
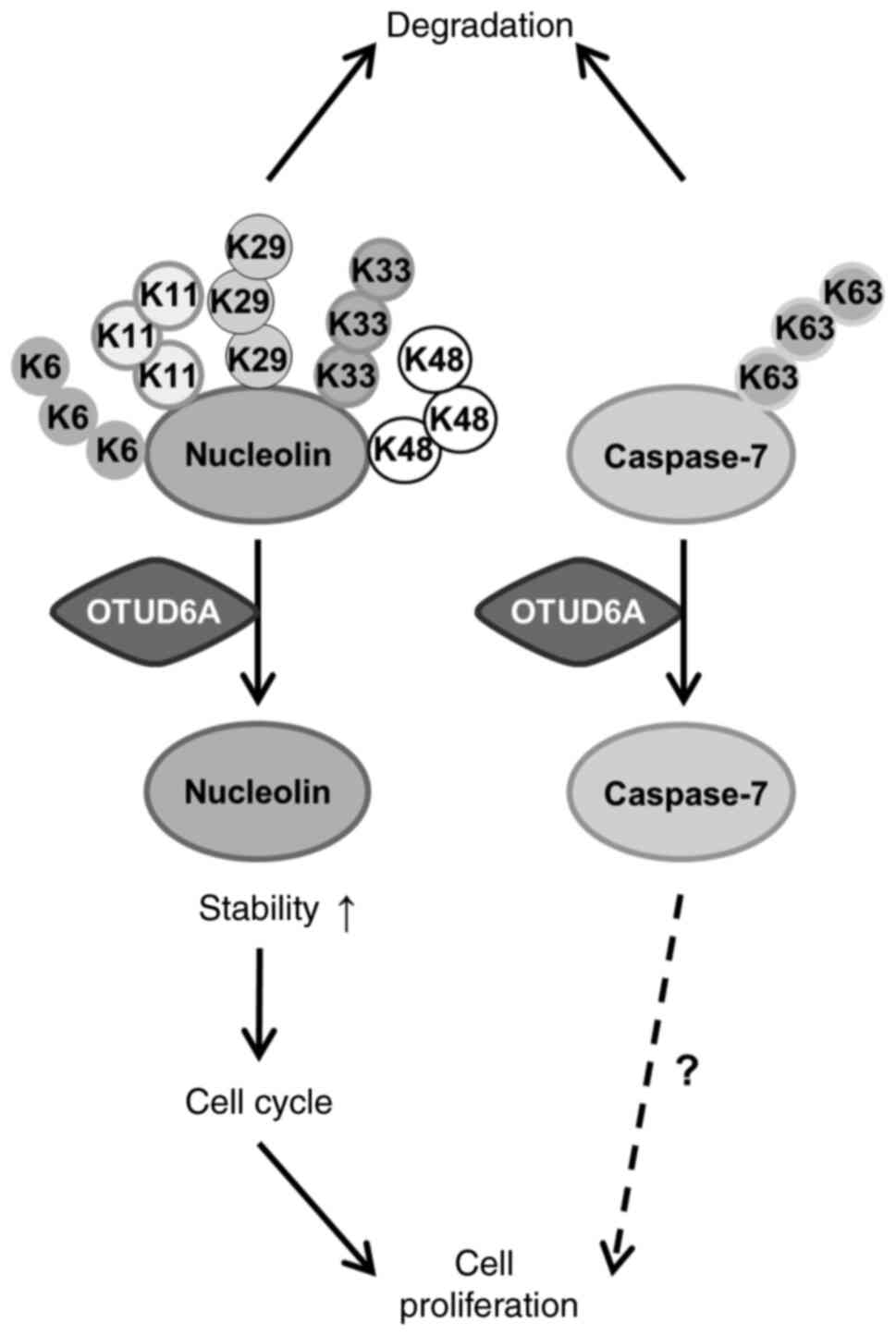
breast cancer (Fig. 5).
Discussion
It has been reported that DUBs function to stabilize
the protein by dissociating the ubiquitin bond of the polyubiquitin
chain and inducing protein degradation (31). Another important function of DUB
family members is to regulate cancer progression by inhibiting the
degradation and stabilizing cancer-related proteins (32). Certain DUBs have been identified
to be involved in cancer chemoresistance (33) and metastasis (34). Therefore, identifying
cancer-related DUBs would be beneficial for anticancer therapy. p53
is a well-known tumor suppressor (35). However, p53 is frequently mutated
in human cancers and the frequency varies depending on the tissue
and type of cancer (36).
Identifying the DUBs for the p53 signaling pathway may be helpful
as a potential target for anticancer therapy. A previous study by
our group reported on the changes in the expression level of OTUD6A
in the presence and absence of p53 (21). OTUD6A is known to promote cell
proliferation in several carcinomas, such as cervical cancer,
colorectal carcinoma (16) and
prostate cancer cell lines (18,19). However, the mechanism of the
association between OTUD6A and p53 is yet to be elucidated. In the
present study, it was hypothesized that OTUD6A controls cell
proliferation through substrates associated with p53.
In order to identify the putative substrates,
LC-MS/MS was first performed using overexpression of Flag-OTUD6A in
HCT116 (p53+/+) cells. Among the potential candidates
identified, nucleolin that binds to HAUSP and p53 was selected
(22). MicroRNA (miRNA/miR)-577
regulates testis-specific gene 10 protein isoform (TSGA10), and
TSGA10 restores the expression of p53 reduced by miR-577 (37). The LIM domain and actin-binding 1
(LIMA1) gene encoding epithelial protein lost in neoplasm (EPLIN)
is transcribed by p53 and knockdown of LIMA1 partially suppressed
the cell invasion inhibitory effect of p53 (38). In addition, EPLIN has different
roles depending on the isoform (39). However, it is not known whether
TSGA10, EPLIN-β, B-cell receptor associated protein 31, porin and
v-fos transformation effector protein directly bind to p53. On the
other hand, the interaction between nucleolin and p53 is known. It
has been demonstrated that nucleolin binds to p53 through the
C-terminal GAR domain of nucleolin (645–707 aa) (25) and p53 binds to nucleolin through
the C-terminal domain of p53 (320–393 aa) (26). Therefore, p53-binding nucleolin
was selected as a putative target of OTUD6A. Immunoprecipitation
and GST pull-down assays revealed the direct binding between OTUD6A
and nucleolin, and interaction in the OTUD6A-nucleolin-p53 complex.
In addition, it was hypothesized that OTUD6A would interact with
apoptotic proteins associated with p53, which would function as a
tumor suppressor. Therefore, immunoprecipitation was performed with
Flag-OTUD6A-transfected cell lysate and western blot analysis was
performed by applying antibodies of apoptotic proteins. The results
confirmed that both caspase-7 and p53 bind to OTUD6A. Caspase-7 is
a protease that regulates inflammation and apoptosis (40). The p53-caspase-7-PARP signaling
pathway is known to be associated with cordycepin-induced apoptosis
(41). In addition, the first
intron of human and mouse caspase-7 is involved in binding to p53
(42). Therefore, nucleolin and
caspase-7 were set as new target candidates for OTUD6A.
The present study therefore investigated whether
OTUD6A regulates the protein stability of nucleolin or caspase-7. A
dose-dependent increase in the protein expression levels of
nucleolin was obtained with increasing expression of Flag-OTUD6A.
In addition, examination of the half-lives of nucleolin and
caspase-7 after exposure to CHX, a protein synthesis inhibitor,
revealed that the half-life of nucleolin was increased in
Flag-OTUD6A-overexpressing MCF7 cells. However, caspase-7 exhibited
no significant change in both protein expression level and
half-life.
Ubiquitin has a different role depending on the
lysine site at which it forms the polyubiquitin chain. Typically,
the K48-linked polyubiquitin chain induces proteasomal degradation
through the 26S proteasome and the K63-linked polyubiquitin chain
responds to lysosomal targeting, DNA damage and stress response
(43,44). OTUD6A regulates the K48-linked
polyubiquitin chain of nucleolin, whereas OTUD6A regulates the
K63-linked polyubiquitin chain of caspase-7. Based on these
results, it is expected that OTUD6A increases the stabilization of
the nucleolin by deubiquitination on the K48-linked polyubiquitin
chain that induces proteasomal degradation. However, the K63-linked
polyubiquitin chain of nucleolin was regulated by MG132. This is a
result different from the known proteasomal degradation-independent
function of the K63-linked polyubiquitin chain. It is possible that
other K sites formed a branch from the K63-linked polyubiquitin
chain, leading to an association with proteasomal degradation
(45). In addition, no
alterations were obtained in the protein expression level of
caspase-7, since there is no difference in the level of the
K48-linked polyubiquitin chain. Although there was no change in the
protein level of caspase-7, it may be expected that it will respond
to DNA repair or stress response, which is one of the cellular
roles for the K63-linked polyubiquitin chain. In addition, it is
suggested that OTUD6A may regulate caspase-7 by responding to
lysosomal targets and not by regulating the protein expression
level by OTUD6A.
The present study confirmed that OTUD6A acts as a
DUB for nucleolin and caspase-7. Nucleolin is known to act as an
oncogene in various cancers. Overexpression of nucleolin is a
predictor of poor prognosis of lung cancer (46) and prostate cancer (47). Upregulation of nucleolin by CRYβB2
promotes tumorigenesis of triple-negative breast cancer (28), and activation of the nucleolin-AKT
pathway by CBP/p300-interacting transactivator with E/D-rich
carboxy-terminal domain-2 promotes tumorigenesis of prostate cancer
(48). XIAP induces bladder
cancer invasion and lung cancer metastasis by enhancing the
nucleolin-mediated Rho GDP-dissociation inhibitor 2 stability
(49), and nucleolin promotes
cervical cancer by activating EGFR signaling involved in tumor
growth and invasion (50). The
interaction of nucleolin and ErbB2 induced tumorigenesis in
ErbB2-positive breast cancer (51), whereas inhibition of nucleolin and
ErbB2 inhibited tumorigenesis in ErbB2-positive breast cancer
(52). In addition, inhibition of
the nucleolin protein by nucleolin antagonist N6L promoted the
normalization of tumor vasculature and impaired the progression of
pancreatic cancer (53).
Caspase-7 is a known apoptotic protein (54). Therefore, flow cytometric analysis
was performed to investigate the effect of OTUD6A on cell
proliferation and cell cycle progression. Overexpression of OTUD6A
induced cell proliferation through the promotion of cell cycle
progression. In addition, flow cytometric analysis was performed
following overexpression of nucleolin to prove that the promotion
of the cell cycle by OTUD6A is due to the upregulation of nucleolin
induced by OTUD6A. It was indicated that overexpression of
nucleolin promoted cell cycle progression in MCF7 cells. Therefore,
the present results suggest that OTUD6A stabilizes nucleolin by
deubiquitination and that upregulated nucleolin increases cell
proliferation by promoting cell cycle progression.
Taken together, the present results indicate that
nucleolin and caspase-7, the substrates of OTUD6A, are potential
anticancer therapy targets. However, further studies are required
to discover the synergistic effect and regulatory mechanism in the
p53 signaling pathway of OTUD6A and nucleolin or caspase-7.
Acknowledgements
The authors would like to thank Ms. Hae-Seul Choi,
Ms. Hyeon-Ah Do, Mr. Hong-Beom Park and Mr. Sang-Soo Park (all from
the Department of Biomedical Science, CHA University, Seongnam,
Korea) for proofreading the manuscript.
Funding
This research was supported by the Basic Science Program
Research Program through the National Research Foundation of Korea
funded by the Ministry of Education (grant no.
2019R1A6A1A03032888).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SHK and KHB designed the study and confirm the
authenticity of all the raw data. SHK performed most of the
experiments and wrote the manuscript. Both authors read and agreed
to the published version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chondrogianni N and Gonos ES: Proteasome
function determines cellular homeostasis and the rate of aging. Adv
Exp Med Biol. 694:38–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bard JAM, Goodall EA, Greene ER, Jonsson
E, Dong KC and Martin A: Structure and function of the 26S
proteasome. Annu Rev Biochem. 87:697–724. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bachiller S, Alonso-Bellido IM, Real LM,
Pérez-Villegas EM, Venero JL, Deierborg T, Armengol JA and Ruiz R:
The ubiquitin proteasome system in neuromuscular disorders: Moving
beyond movement. Int J Mol Sci. 21:64292020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park HB, Kim JW and Baek KH: Regulation of
Wnt signaling through ubiquitination and deubiquitination in
cancers. Int J Mol Sci. 21:39042020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park HB and Baek KH: E3 ligases and
deubiquitinating enzymes regulating the MAPK signaling pathway in
cancers. Biochim Biophys Acta Rev Cancer. 1877:1887362022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Caputi FF, Rullo L, Stamatakos S,
Candeletti S and Romualdi P: Interplay between the endogenous
opioid system and proteasome complex: Beyond signaling. Int J Mol
Sci. 20:14412019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suresh B, Lee J, Kim H and Ramakrishna S:
Regulation of pluripotency and differentiation by deubiquitinating
enzymes. Cell Death Differ. 23:1257–1264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suresh B, Lee J, Kim KS and Ramakrishna S:
The importance of ubiquitination and deubiquitination in cellular
reprogramming. Stem Cells Int. 2016:67059272016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grice GL and Nathan JA: The recognition of
ubiquitinated proteins by the proteasome. Cell Mol Life Sci.
73:3497–3506. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kwon YT and Ciechanover A: The ubiquitin
code in the ubiquitin-proteasome system and autophagy. Trends
Biochem Sci. 42:873–886. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dosa A and Csizmadia T: The role of
K63-linked polyubiquitin in several types of autophagy. Biol Futur.
73:137–148. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lafont E, Hartwig T and Walczak H: Paving
TRAIL's path with ubiquitin. Trends Biochem Sci. 43:44–60. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan T, Yan F, Ying M, Cao J, He Q, Zhu H
and Yang B: Inhibition of ubiquitin-specific proteases as a novel
anticancer therapeutic strategy. Front Pharmacol. 9:10802018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lange SM, Armstrong LA and Kulathu Y:
Deubiquitinases: From mechanisms to their inhibition by small
molecules. Mol Cell. 82:15–29. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fhu CW and Ali A: Dysregulation of the
ubiquitin proteasome system in human malignancies: A window for
therapeutic intervention. Cancers (Basel). 13:15132021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi L, Liu J, Peng Y, Zhang J, Dai X,
Zhang S, Wang Y, Liu J and Long J: Deubiquitinase OTUD6A promotes
proliferation of cancer cells via regulating Drp1 stability and
mitochondrial fission. Mol Oncol. 14:3169–3183. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim HJ and Kim J: OTUD6A Is an aurora
kinase a-specific deubiquitinase. Int J Mol Sci. 22:19362021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu X, Zhao J, Yu G, Zhang X, Sun J, Li L,
Yin J, Niu Y, Ren S, Zhu Y, et al: OTUD6A promotes prostate
tumorigenesis via deubiquitinating Brg1 and AR. Commun Biol.
5:1822022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peng Y, Liu J, Wang Z, Cui C, Zhang T,
Zhang S, Gao P, Hou Z, Liu H, Guo J, et al: Prostate-specific
oncogene OTUD6A promotes prostatic tumorigenesis via
deubiquitinating and stabilizing c-Myc. Cell Death Differ.
29:1730–1743. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Y, Huang X, Zhu D, Wei M, Luo J, Yu
S, Tian Y and Zheng X: Deubiquitinase OTUD6A promotes breast cancer
progression by increasing TopBP1 stability and rendering tumor
cells resistant to DNA-damaging therapy. Cell Death Differ. Jun
29–2022.(Epub ahead of print). View Article : Google Scholar
|
|
21
|
Kim SY, Kwon SK, Lee SY and Baek KH:
Ubiquitin-specific peptidase 5 and ovarian tumor deubiquitinase 6A
are differentially expressed in p53+/+ and
p53−/− HCT116 cells. Int J Oncol. 52:1705–1714.
2018.PubMed/NCBI
|
|
22
|
Lim KH, Park JJ, Gu BH, Kim JO, Park SG
and Baek KH: HAUSP-nucleolin interaction is regulated by p53-Mdm2
complex in response to DNA damage response. Sci Rep. 5:127932015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park JH, Kim SY, Cho HJ, Lee SY and Baek
KH: YOD1 deubiquitinates NEDD4 involved in the hippo signaling
pathway. Cell Physiol Biochem. 54:1–14. 2020.PubMed/NCBI
|
|
24
|
Roth JA, Swisher SG and Meyn RE: p53 tumor
suppressor gene therapy for cancer. Oncology (Williston Park).
13:148–154. 1999.PubMed/NCBI
|
|
25
|
Bhatt P, d'Avout C, Kane NS, Borowiec JA
and Saxena A: Specific domains of nucleolin interact with Hdm2 and
antagonize Hdm2-mediated p53 ubiquitination. FEBS J. 279:370–383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Daniely Y, Dimitrova DD and Borowiec JA:
Stress-dependent nucleolin mobilization mediated by p53-nucleolin
complex formation. Mol Cell Biol. 22:6014–6022. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jia W, Yao Z, Zhao J, Guan Q and Gao L:
New perspectives of physiological and pathological functions of
nucleolin (NCL). Life Sci. 186:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan Y, Narayan A, Cho S, Cheng Z, Liu JO,
Zhu H, Wang G, Wharram B, Lisok A, Brummet M, et al: CRYbetaB2
enhances tumorigenesis through upregulation of nucleolin in triple
negative breast cancer. Oncogene. 40:5752–5763. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mahib MR, Hosojima S, Kushiyama H,
Kinoshita T, Shiroishi T, Suda T and Tsuchiya K: Caspase-7 mediates
caspase-1-induced apoptosis independently of Bid. Microbiol
Immunol. 64:143–152. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang W, Luo J, Xiang F, Liu X, Jiang M,
Liao L and Hu J: Nucleolin down-regulation is involved in
ADP-induced cell cycle arrest in S phase and cell apoptosis in
vascular endothelial cells. PLoS One. 9:e1101012014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McClurg UL and Robson CN: Deubiquitinating
enzymes as oncotargets. Oncotarget. 6:9657–9668. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Poondla N, Chandrasekaran AP, Kim KS and
Ramakrishna S: Deubiquitinating enzymes as cancer biomarkers: New
therapeutic opportunities? BMB Rep. 52:181–189. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu X, Zhang Y, Luo Q, Wu X, Huang F, Shu
T, Wan Y, Chen H and Liu Z: The deubiquitinase USP11 promotes
ovarian cancer chemoresistance by stabilizing BIP. Signal Transduct
Target Ther. 6:2642021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Z, Li J, Ou Y, Yang G, Deng K, Wang
Q, Wang Z, Wang W, Zhang Q, Wang H, et al: CDK4/6 inhibition blocks
cancer metastasis through a USP51-ZEB1-dependent deubiquitination
mechanism. Signal Transduct Target Ther. 5:252020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Machado-Silva A, Perrier S and Bourdon JC:
p53 family members in cancer diagnosis and treatment. Semin Cancer
Biol. 20:57–62. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutations in human cancers. Science. 253:49–53.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yuan X, He J, Sun F and Gu J: Effects and
interactions of MiR-577 and TSGA10 in regulating esophageal
squamous cell carcinoma. Int J Clin Exp Pathol. 6:2651–2667.
2013.PubMed/NCBI
|
|
38
|
Ohashi T, Idogawa M, Sasaki Y and Tokino
T: p53 mediates the suppression of cancer cell invasion by inducing
LIMA1/EPLIN. Cancer Lett. 390:58–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Taha M, Aldirawi M, Marz S, Seebach J,
Odenthal-Schnittler M, Bondareva O, Bojovic V, Schmandra T, Wirth
B, Mietkowska M, et al: EPLIN-alpha and -beta Isoforms modulate
endothelial cell dynamics through a spatiotemporally differentiated
interaction with actin. Cell Rep. 29:1010–1026.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lamkanfi M and Kanneganti TD: Caspase-7: A
protease involved in apoptosis and inflammation. Int J Biochem Cell
Biol. 42:21–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Y, Yang SH, Hueng DY, Syu JP, Liao CC
and Wu YC: Cordycepin induces apoptosis of C6 glioma cells through
the adenosine 2A receptor-p53-caspase-7-PARP pathway. Chem Biol
Interact. 216:17–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang C, Kaushal V, Haun RS, Seth R, Shah
SV and Kaushal GP: Transcriptional activation of caspase-6 and −7
genes by cisplatin-induced p53 and its functional significance in
cisplatin nephrotoxicity. Cell Death Differ. 15:530–544. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou B and Zeng L: Conventional and
unconventional ubiquitination in plant immunity. Mol Plant Pathol.
18:1313–1330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Akutsu M, Dikic I and Bremm A: Ubiquitin
chain diversity at a glance. J Cell Sci. 129:875–880.
2016.PubMed/NCBI
|
|
45
|
Ohtake F, Tsuchiya H, Saeki Y and Tanaka
K: K63 ubiquitylation triggers proteasomal degradation by seeding
branched ubiquitin chains. Proc Natl Acad Sci USA. 115:E1401–E1408.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Valerio-Fernandes A, Fonseca NA, Goncalves
N, Cruz AF, Pereira MI, Gregório AC, Moura V, Ladeirinha AF,
Alarcão A, Gonçalves J, et al: Nucleolin overexpression predicts
patient prognosis while providing a framework for targeted
therapeutic intervention in lung cancer. Cancers (Basel).
14:22172022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Firlej V, Soyeux P, Nourieh M, Huet E,
Semprez F, Allory Y, Londono-Vallejo A, de la Taille A, Vacherot F
and Destouches D: Overexpression of nucleolin and associated genes
in prostate cancer. Int J Mol Sci. 23:44912022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shin SH, Lee GY, Lee M, Lee M, Kang J,
Shin HW, Chun YS and Park JW: Aberrant expression of CITED2
promotes prostate cancer metastasis by activating the nucleolin-AKT
pathway. Nat Commun. 9:41132018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu Y, Jin H, Xu J, Gu J, Li X, Xie Q,
Huang H, Li J, Tian Z, Jiang G, et al: XIAP overexpression promotes
bladder cancer invasion in vitro and lung metastasis in vivo via
enhancing nucleolin-mediated Rho-GDIbeta mRNA stability. Int J
Cancer. 142:2040–2055. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yan J, Zhang Y, Ren C, Shi W and Chen L:
Involvement of nuclear protein C23 in activation of EGFR signaling
in cervical cancer. Tumour Biol. 37:905–910. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wolfson E, Goldenberg M, Solomon S,
Frishberg A and Pinkas-Kramarski R: Nucleolin-binding by ErbB2
enhances tumorigenicity of ErbB2-positive breast cancer.
Oncotarget. 7:65320–65334. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wolfson E, Solomon S, Schmukler E,
Goldshmit Y and Pinkas-Kramarski R: Nucleolin and ErbB2 inhibition
reduces tumorigenicity of ErbB2-positive breast cancer. Cell Death
Dis. 9:472018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gilles ME, Maione F, Cossutta M,
Carpentier G, Caruana L, Maria SD, Houppe C, Destouches D, Shchors
K, Prochasson C, et al: Nucleolin targeting impairs the progression
of pancreatic cancer and promotes the normalization of tumor
vasculature. Cancer Res. 76:7181–7193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Brentnall M, Rodriguez-Menocal L, De
Guevara RL, Cepero E and Boise LH: Caspase-9, caspase-3 and
caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell
Biol. 14:322013. View Article : Google Scholar : PubMed/NCBI
|