In parallel to DNA and protein, there are >170
types of biochemical modifications that occur in RNA (1,2). The
most remarkable RNA modification is methylation. Numerous studies
have indicated that methylation modifications are able to alter RNA
structure, stability and function. These modifications include
7-methylguanine, 5-methylcytosine,
N6,2'-O-dimethyladenosine (m6Am),
N1-methyladenosine (m1A), 5-hydroxymethylcytosine and
N6-methyladenosine (m6A) (3-5).
Since 1974, m6A has been gradually identified as the most abundant
modification in human RNAs (6) and
has been proven to affect numerous functional processes of RNAs,
such as alternative splicing (7-9),
decay (10), subcellular
distribution (11) and translation
(12).
Mostly transcribed from DNAs, long noncoding RNAs
(lncRNAs) are transcripts of >200 nt in length (13). To date, numerous studies have
indicated that lncRNAs exert various functions in epigenetic
regulation (14), mRNA
transcription or degradation, alternative splicing and translation
(15). Undoubtedly, lncRNAs have
roles in the pathogenesis, development, diagnosis and treatment of
various disorders, including numerous cancers (16), neurological diseases (17) and autoimmune dysregulation
(18).
The interplay between lncRNAs with m6A modification
has been reported to have important roles in cancers (19). With the development of
next-generation sequencing, studies have indicated that lncRNAs are
modified in this way using methylated RNA immunoprecipitation and
sequencing analysis (20). Given
the important roles of m6A-modified ncRNAs in carcinogenesis,
accumulating evidence has focused on m6A-modified lncRNAs (21,22).
The present review summarized how m6A affects lncRNAs and the
functions of m6A-modified lncRNAs in cancers.
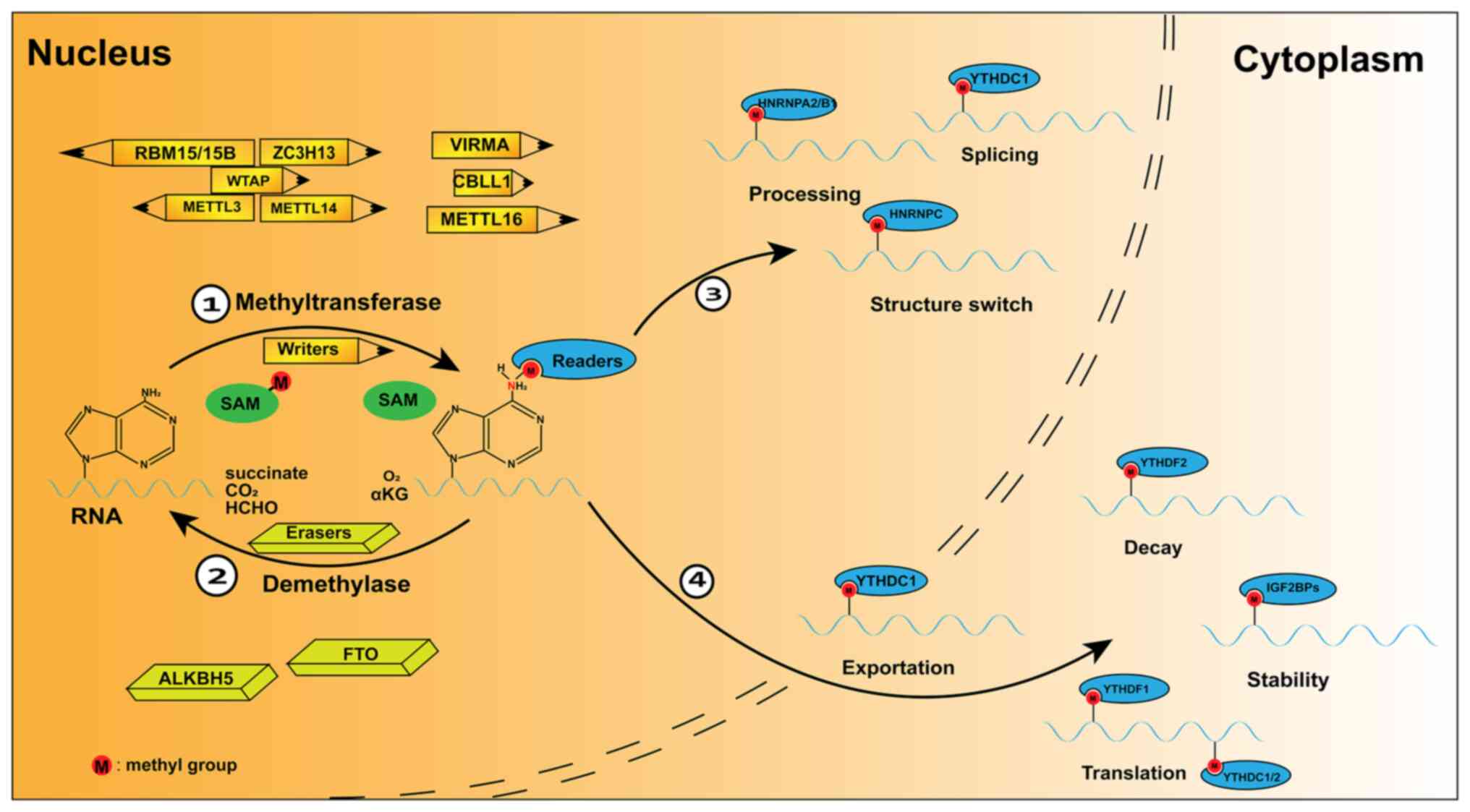
M6A modification is defined as methylation or
demethylation and recognition of the methyl group on the sixth
nitrogen atom of adenine (A) in RNAs (6), which is reversible due to the
interplay among three m6A regulators, writers, erasers and readers
(23). The writers attach the
methyl group to the m6A sites, whereas the erasers remove it.
Readers recognize m6A marks and link RNAs with diverse regulatory
mechanisms (Fig. 1) (24,25).
Writers are methyltransferases that utilize
S-adenosylmethionine (SAM) to provide methyl groups for the
methylation of RNAs (26). The
first m6A writer was identified as a complex, which contains the
following proteinic subunits: methyltransferase-like 3 (METTL3),
METTL14 and their cofactors: Wilms' tumour 1-associating protein
(WTAP), vir-like m6A methyltransferase associated (VIRMA), zinc
finger CCCH-type containing 13, casitas B-lineage
lymphoma-transforming sequence-like protein 1 and RNA-binding motif
protein 15/15B (22,27-29).
METTL3 is highly conserved and is able to transfer the methyl group
of SAM to the targeted A (30).
METTL14 acts as a platform for RNA binding (31) and may be used to construct a steady
heterodimer with METTL3 to effect m6A modification (32). In addition, WTAP is another subunit
of the m6A methyltransferase complex (MTC). It was reported that
the interplay between WTAP and METTL3/METTL14 is required for the
chemical activity of m6A writers in vivo (33). METTL16 is a newly identified writer
that catalyses the m6A methylation of ncRNAs (34). Similar to METTL3, METTL16 contains
the Rossmann-like fold of class I methyltransferases and uses SAM
as the methyl donor but appears to have additional regulatory and
RNA binding domains (35).
Consistent with studies that focus on dysregulated m6A writers,
abnormal lncRNAs interacting with writers have been identified in
various cancers (36,37).
M6A modification is reversable and erasers have
important roles in its reversion. Two essential erasers, namely fat
mass and obesity-associated protein (FTO) and AlkB homologue 5
(ALKBH5) (23,38), belong to the α-ketoglutarate
(α-KG)-dependent ALKB dioxygenase family and clear m6A methylation
in an Fe(II)- and α-KG-dependent manner (39). First, they oxidize m6A to form
N6-hydroxymethyladenosine (hm6A). Next, they catalyse
the conversion of hm6A to N6-formyladenosine (f6A).
Finally, f6A is converted to A and demethylation is completed
(40).
ALKBH5 is another RNA demethylase. It has been
indicated that demethylation induced by ALKBH5 contributes to the
dysregulation of certain oncogenic lncRNAs in tumours. Numerous
clinical trials based on novel drugs targeting ALKBH5 are in
progress. Of note, ALKBH5 may become a considerable target for
cancer diagnosis, therapy and prognosis (23).
M6A readers, including the YT521-B homology domain
(YTHD) family, heterogeneous nuclear ribonucleoprotein (HNRNP)
family and insulin-like growth factor 2 mRNA binding protein
(IGF2BP) family, may function as binding proteins to recognize m6A
on RNAs (42). Different readers
have distinct functions on methylated RNA.
Among diverse readers, the YTH domain family,
including YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2, has been
indicated to interact with m6A modification on RNAs (10). YTHDF1 promotes the initiation of
translation by interacting with eukaryotic initiation factor 3
(eIF3, which interacts with mRNA in 48S complexes and participates
in the progression of translation) (43). YTHDF2 recognizes m6A sites on mRNAs
and promotes their decay (44).
YTHDF3 is associated with lncRNA decay (45) and recruits effectors involved in
translation (46). YTHDC1
participates in m6A-modified lncRNA-mediated gene silencing
(22) and modulates the nuclear
transportation of mRNA after being m6A modified (46). YTHDC2 binds to circular RNAs and
interferes with their sponging of targeted microRNAs (miRNAs/miRs)
(47), preferentially functioning
as an m6A reader to enhance translation efficiency or decrease mRNA
abundance (48).
HNRNPs are a type of RNA binding proteins (RBPs),
some of which may act as m6A readers (41). HNRNPA2/B1 binds to m6A sites to
enhance the progression of primary miRNAs in an m6A-dependent
manner. Furthermore, HNRNPA2/B1 may combine with certain binding
sites indirectly via a mechanism called the 'm6A switch' (49), which means that reversible m6A
modification within the RNA duplex changes the local RNA structures
and accessibility for binding proteins such as HNRNPA2/B1 and
HNRNPC. The function of HNRNPA2/B1 in m6A-dependent biological
processes still requires further study. HNRNPC also binds to
m6A-modified RNAs through an 'm6A switch' mechanism (21). Liu et al (50) reported HNRNPG as a novel m6A
reader, as it utilizes the low-complexity region for the
recognition of m6A modification and participates in the alteration
of splicing diversity.
IGF2BPs recognize and bind RNAs with m6A
modifications. Subsequently, the stability and translation of
targeted RNAs may be promoted by IGF2BPs in an m6A-dependent
manner, which may strongly affect gene expression levels.
IGF2BP1/2/3 are newly discovered m6A readers that may protect mRNAs
with m6A modifications from degradation (51). In addition, IGF2BPs stabilize the
mRNAs of certain oncogenes with m6A modifications, promoting the
progression of certain cancers (52).
To date, attempts have been made to illustrate the
regulatory mechanisms of lncRNAs in cancers. In addition, m6A
modification appears to be an upstream mechanism that regulates
lncRNAs (36,37). Through the joint efforts of
numerous researchers, additional m6A writers, erasers, readers and
lncRNAs regulated by m6A will be discovered, which will certainly
broaden our horizon regarding the functional features of
m6A-modified lncRNAs in cancers.
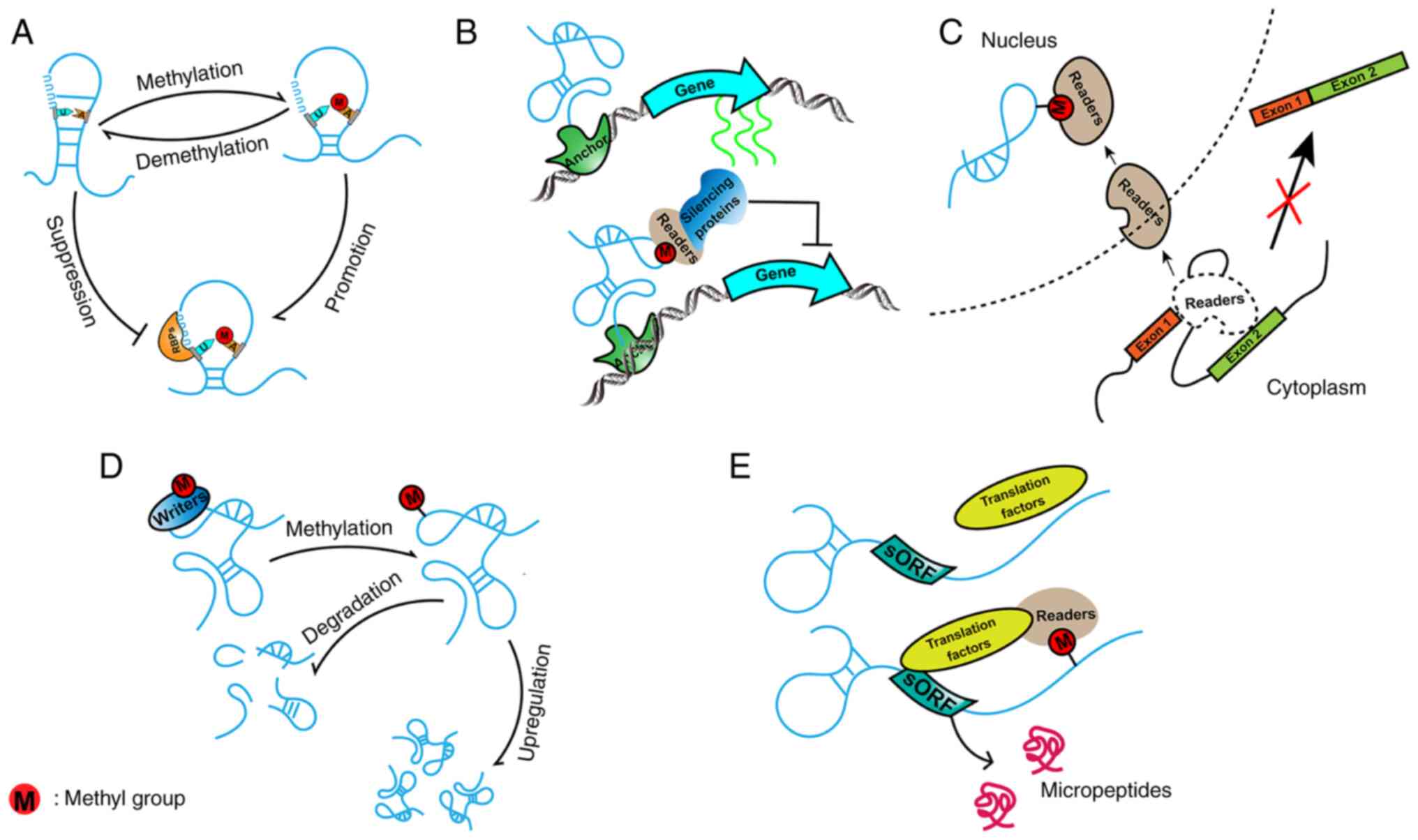
Parallel analysis of RNA structure indicated that
the structure of RRACH (R=A or G; H=A, U or C) motifs that contain
m6A differs from those that lack m6A modification. In vitro
studies indicated that whether m6A leads to RNA secondary structure
destabilization or stabilization relies upon its position within or
at the edge of duplexes (53). Due
to disrupting base complementary pairing between A and U, m6A
modification within duplexes makes the opposite strand more
single-stranded and more accessible (21). While m6A sites on lncRNAs may bind
readers, localized complementary chains may also expose a
U-containing binding motif for RBPs (54). In addition, reversible m6A
modification changes the local structure of lncRNAs, altering the
RBPs attached to lncRNAs, referred to as the 'm6A switch' (Fig. 2A). For instance, m6A methylation of
lncRNA metastasis associated lung adenocarcinoma transcript 1
(MALAT1) affects its duplex structure and regulates the
inter-action with HNRNPC/HNRNPG, which suggests that m6A
modification participates in structural switching of lncRNAs and
contributes to the binding strength of proteins (21,50,54).
M6A-modified lncRNAs are recognized by readers.
Readers may also bind to partners that have distinct roles in
transcriptional regulation, resulting in the assembly of
transcriptional ribonucleoprotein complexes. Targeting activated
transcriptional regions, m6A-modified lncRNAs with transcriptional
ribonucleoprotein complexes may lead to alterations in gene
transcription (Fig. 2B). This
mechanism requires that m6A-modified lncRNAs may be recognized by
readers and assist readers to recruit transcriptional
ribonucleoprotein complexes. It is different from epigenetic
regulation via methylation at promoters, whose regulation depends
on methyltransferases directly binding to promoters other than
m6A-modified lncRNAs.
Splicing has a vital role in regulating gene
expression by removing introns and joining exons together. During
the process of splicing, diverse exons from the same encoded gene
may be alternatively selected. This mechanism leads to the
diversity of mRNA variants and protein products (55). It has been reported that m6A
modifications affect alternative splicing (56). Furthermore, Ninomiya et al
(57) demonstrated that during the
process of thermal stress recovery, m6A-modified lncRNA highly
repetitive satellite III (HSATIII) is recognized by YTHDC1,
resulting in decreased YTHDC1-induced mRNA alternative splicing in
the cytoplasm (Fig. 2C). In
addition, m6A-modified HSATIII binding to YTHDC1 was surrounded by
nuclear stress bodies, providing a reaction crucible for CDC-like
kinase 1 to phosphorylate Ser/Arg-rich splicing factors, which were
previously indicated to have an impact on pre-mRNA splicing during
thermal stress (57).
Several studies have indicated that m6A-modified
lncRNAs exhibit differential stability and abundance compared to
those without m6A modification in different types of cancer
(Fig. 2D). For instance, Xue et
al (58) determined that
METTL3-mediated methylation stabilizes lncRNA abhydrolase domain
containing 11-antisense RNA 1, exerting its function in non-small
cell lung cancer (NSCLC), while Guo et al (59) investigated whether ALKBH5 was able
to increase the abundance of lncRNA nuclear enriched abundant
transcript 1 (NEAT1) in colon cancer tissues and associated with
poor prognosis.
However, the upstream mechanisms by which m6A
modification changes lncRNA stability and abundance are still
unclear. On the one hand, m6A modification itself may change the
intermolecular forces or spatial structures of local lncRNAs to
affect their stability or degradation. Furthermore, m6A regulators
may interfere with RNA degradosome recognition or binding to
special sites on lncRNAs, thereby regulating the process of lncRNA
degradation. Specifically, writers may interfere with degrading
factors binding to key sites on lncRNAs, erasers may eliminate the
methyl group on lncRNAs and remove special signals recognized by
degrading complexes for starting the process of RNA decomposition,
and m6A readers likely recruit other partners to form complexes
that interfere with the degradation of m6A-modified lncRNAs.
Due to the lack of canonical open reading frames
(ORFs >100 aa), lncRNAs are considered untranslatable (60,61).
Of note, a study identified hundreds of short or small ORFs (sORFs
or smORFs) in the sequence of lncRNAs that actually encode peptides
(62). Mechanistically, certain
m6A motifs on lncRNAs may be close to crucial sequences of sORFs or
smORFs. M6A readers not only recognize m6A modification on lncRNAs
but also recruit translation factors at the same time. As a result,
micropeptides encoded by lncRNAs are produced in an m6A-dependent
manner (Fig. 2E). Wu et al
(63) indicated that the YTHDF1
reader, which interacts with the translation machinery, promotes
protein translation and recognizes m6A modification on lncRNA
LINC00278. This m6A motif is near the stop codon of a
LINC00278-encoded micropeptide, which is termed yin-yang 1
(YY1)-blocking micropeptide (YY1BM). Subsequently, YY1BM is
translated in an m6A-dependent manner, ultimately resulting in
apoptosis of esophageal squamous cell carcinoma (ESCC).
Despite these findings, numerous points remain to be
elucidated, e.g. how m6A influences lncRNA translation. Recently, a
genome-wide study identified ~100 lncRNA-encoded micropeptides that
regulate induced pluripotent stem cell growth (64). However, whether m6A modification
participates in lncRNA translation and micropeptide roles in
cancerous cells still requires extensive validation. In addition,
m6A modification has been indicated to promote mRNA export from the
nucleus and it has been suggested that m6A modification may affect
the subcellular diffusion of lncRNAs. For instance, Wu et al
(65) reported that upregulated
METTL3 subsequently localizes lncRNA RP11 primarily in the nucleus.
However, how m6A modification alters the subcellular distribution
of lncRNA remains elusive and requires further study.
In recent decades, emerging evidence has emphasized
the essential role of m6A-modified lncRNAs in cancers, which has
laid a solid foundation for future clinical applications targeting
m6A-modified lncRNAs, even if their mechanisms of pathogenesis
differ. Below, a brief review of relevant research in this area is
provided (Fig. 3).
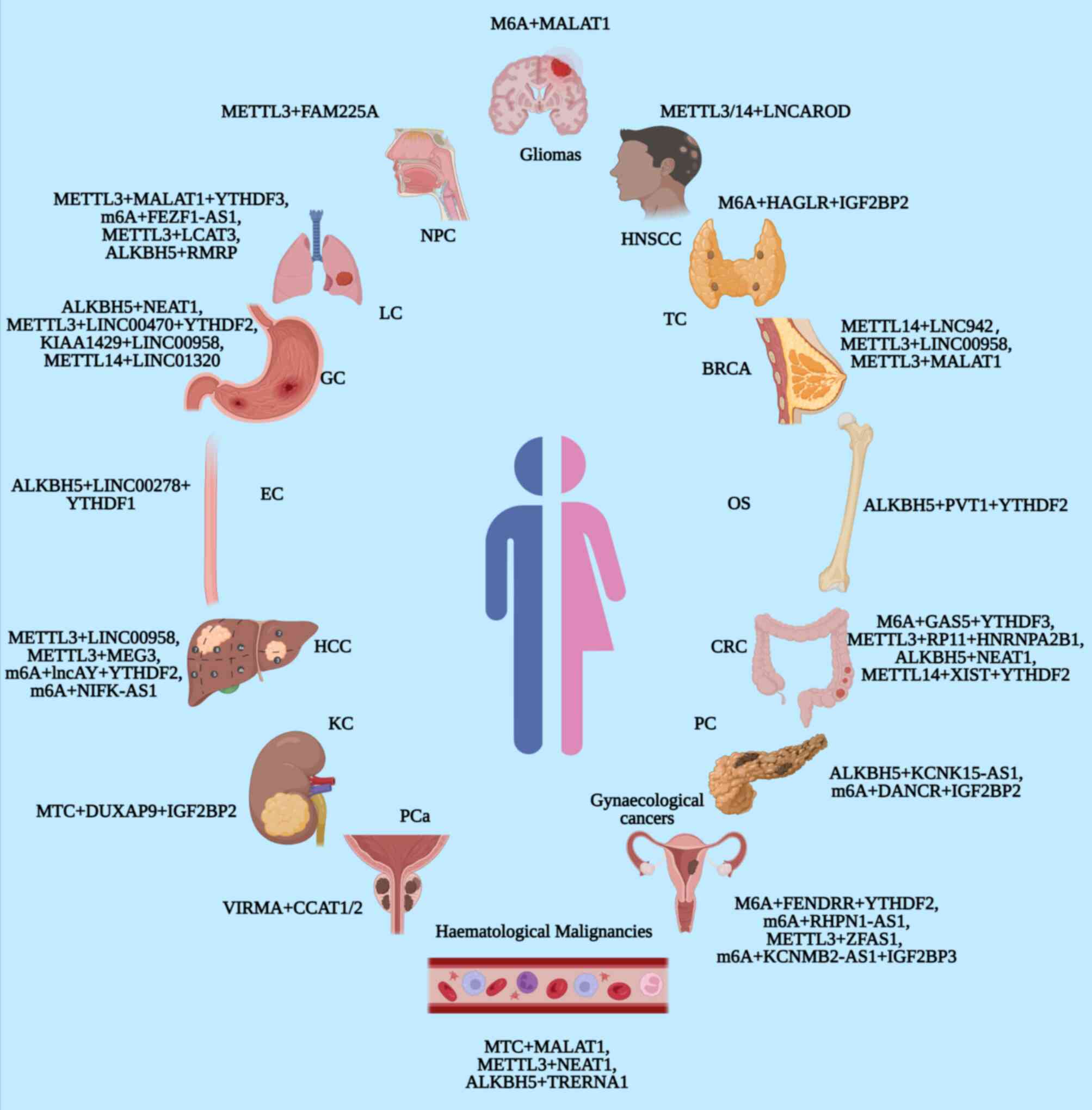
Originating from mesenchymal cells, OS is one of the
most aggressive tumour types in the musculoskeletal system
(66). Multiple studies have
revealed that dysregulated m6A-modified lncRNAs may participate in
multiple phenotypes in OS. Chen et al (67) indicated that lncRNA PVT1, an
oncogenic lncRNA verified in different tumour types, is upregulated
in patients with OS. The m6A demethylase ALKBH5 'erased' the m6A
mark of PVT1, subsequently decreasing the binding of PVT1 and
YTHDF2, which is a common reader that promotes the decay of
m6A-modified RNAs. ALKBH5-induced PVT1 upregulation promoted OS
development both in vitro and in vivo. The study's
conclusion suggests that ALKBH5-PVT1 may be a potential biomarker
for OS (67). Evidence is emerging
that m6A abundance and lncRNA abnormalities are involved in OS
(68,69), indicating that m6A-modified lncRNAs
may be better biomarkers for cancer. This provides a future
direction for OS therapy that warrants further investigation.
EC is regarded as a globally common invasive tumour
type in the digestive tract with a 5-year overall survival rate of
~20-30% (70). With the
development of detection techniques, the role of m6A-modified
lncRNAs in EC has been gradually revealed. Wu et al
(63) indicated that smoking leads
to ALKBH5 upregulation and removes m6A marks on the Y-linked lncRNA
LINC00278. The classical m6A modification motif of LINC00278 that
interacts with YTHDF1 is near the stop codon of YY1BM and the
neighbouring YTHDF1 facilitates translation of the micropeptide
YY1BM. YY1BM suppresses the transcription of eukaryotic elongation
factor 2 kinase by blocking contact between YY1 and androgen
receptor, resulting in apoptosis of ESCC. Mechanistically, smoking
inhibits YY1BM translation, correspondingly inducing male ESCC
progression (63). It is
foreseeable that a deeper understanding of the complexity of
m6A-modified lncRNAs involved in EC will offer novel and promising
applications for EC management in the future.
CRC remains one of the most common neoplasms in the
lower digestive tract globally. In China, CRC is also a major cause
of tumour-associated mortality (75). Researchers have identified numerous
m6A-modified lncRNAs related to the tumorigenesis and metastasis of
CRC. Ni et al (76)
revealed that lncRNA GAS5 binds to the WW domain of yes-associated
protein (YAP) directly and subsequently leads to phosphorylation
and ubiquitin-mediated decomposition of YAP, inhibiting CRC
progression. However, the m6A reader YTHDF3 facilitates
m6A-modified GAS5 degradation and reverses YAP-mediated inhibition
of tumour progression. Wu et al (65) discovered that METTL3-mediated
methylation of lncRNA RP11 is recognized by HNRNPA2B1. After
binding with HNRNPA2B1, m6A-modified RP11 facilitates the decay of
seven in absentia homolog 1 (SIAH1) and F-box only protein 45
(FBXO45) mRNA, possibly through HNRNPA2B1 recruiting factors that
participate in the mRNA degradation process, such as P bodies. The
mRNA of SIAH1 and FBXO45 encodes proteins that function as
ubiquitin E3 ligases and further mediate zinc finger E-box binding
homeobox 1 protein degradation via the ubiquitin-proteasome
pathway. In this way, m6A-modified RP11 and accelerated mRNA
degradation trigger the dissemination of CRC. Guo et al
(59) indicated that ALKBH5
increased lncRNA NEAT1 expression in colon cancer tissues and was
associated with poor prognosis. Yang et al (77) reported that m6A-modified XIST, an
oncogenic lncRNA, is degraded after recognition by the m6A reader
YTHDF2. Furthermore, METTL14 suppresses the proliferation and
invasion of CRC cells by degrading XIST in an m6A-dependent manner.
Further exploration of m6A-related lncRNAs in CRC is necessary to
lay a solid foundation for their future use in CRC diagnosis and
treatment.
PC is one of the most malignant tumour types and
lacks specific symptoms and effective treatment (81). In recent decades, the morbidity of
PC has increased 6-fold in China (82). Recent studies indicated that
m6A-modified lncRNAs have critical roles in PC. He et al
(83) found that lncRNA KCNK15-AS1
levels were decreased by m6A methylation and functioned as an
antitumor lncRNA by inhibiting pancreatic cancer cell migration,
invasion and epithelial to mesenchymal transition (EMT). As an m6A
eraser, ALKBH5 mediates the increase in KCNK15-AS1 via
demethylation. Owing to the higher levels of KCNK15-AS1, the
antitumor effects were recovered in PC. Hu et al (84) demonstrated that the stability of
lncRNA DANCR, which was reported to promote cancer stemness-like
properties, was strengthened by the interaction of IGF2BP2 with
DANCR at the A664 site in PC. As a result, m6A-mediated
upregulation of DANCR promoted cell proliferation, stemness-like
properties and tumorigenesis of PC cells. In general, research in
this field is progressing rapidly. With continued research on PC
diagnosis, treatment and prognosis, profound changes will occur in
the near future.
Along with changes in the environment and lifestyle,
lung cancer (LC) has gradually become the most frequently occurring
cancer type (11.6% of total cases) and the major cause of
cancer-associated mortality (18.4% of all cancer-associated deaths)
(71,85). Studies have indicated that
m6A-modified lncRNAs may serve as biomarkers for the dual purpose
of timely screening and targeted therapy of lung cancer. Jin et
al (37) demonstrated that
METTL3-induced methylation upregulated the levels of lncRNA MALAT1
and the stability of MALAT1 was increased by the METTL3/YTHDF3
complex in NSCLC. Upon binding to METTL3-modified MALAT1, YTHDF1/3
recruits eIF3b to the translation initiation complex, enhancing
translation of YAP, which is associated with the development,
progression and poor prognosis of NSCLC. Furthermore, MALAT1
functions as a ceRNA to sponge miR-1914-3p and inhibit its binding
to the 3'-untranslated region (3'-UTR) of YAP mRNA to decrease YAP
expression. Finally, m6A-modified MALAT1 facilitates malignant
phenotypes of NSCLC cells. Similarly, Song et al (86) discovered that the m6A-modified
lncRNA FEZF1-AS1 facilitates NSCLC progression via the
miR-516b-5p/ITGA11 axis. Qian et al (87) reported that m6A modification
mediated by METTL3 contributes to the increasing level and enhanced
stability of lncRNA LCAT3 in lung adenocarcinoma (LUAD). LCAT3
interacts with far upstream element binding protein 1 (FUBP1),
resulting in activation of c-Myc expression, which is a typical
oncogenic transcriptional factor. Thus, LCAT3 promotes malignant
phenotypes of LUAD cells via the FUBP1/c-Myc axis. Yu and Zhang
(88) observed that ALKBH5
upregulated lncRNA RMRP expression via demethylation in both
patients with LUAD and cell lines, conveying a poorer prognosis of
LUAD. Despite numerous challenges, it is thought that m6A-modified
lncRNAs have the potential to be applied in the diagnosis and
therapy of LC.
PCa is the most commonly new diagnosed cancer in
males and the 5-year survival rate of patients with PCa with
distant metastasis is as low as ~30% (91). Currently, a comprehensive analysis
of how m6A-modified lncRNAs function in the clinicopathological
characteristics, malignant progression, and prognosis of PCa is
lacking. Barros-Silva et al (92) demonstrated that VIRMA, a subunit of
MTC, is the most common writer in PCa, and m6A methylation may
stabilize lncRNA CCAT1 and lncRNA CCAT2, which participate in the
alteration of the proto-oncogene MYC. Regarding clinical relevance,
VIRMA and upregulation of both lncRNAs (CCAT1 and CCAT2) predicts
worse disease-free survival. Although their roles are not entirely
understood, m6A-modified lncRNAs in PCa are thought to be very
attractive biomarkers for early diagnosis, targeted therapy and
improved prognosis in the future.
According to GLOBOCAN 2020, BRCA is the most
commonly diagnosed cancer and the leading cause of
cancer-associated death in females (71). Recently, solid evidence has led to
research on the role of m6A-modified lncRNAs in BRCA. Sun et
al (93) found that METTL14
recognizes and binds lncRNA LNC942 at a specific motif. Thereafter,
METTL14-methylated LNC942 promotes the mRNA stability and
expression of CXCR4 and CYP1B1, which induce EMT and drug
resistance, respectively. Finally, m6A-modified LNC942 mediates
BRCA cell vitality and EMT. Rong et al (94) revealed that METTL3-mediated m6A
modification promotes the stability of LINC00958. Moreover,
m6A-modified LINC00958 acts as a ceRNA to sponge miR-378a-3p and
positively regulate YY1, which is involved specifically in
metastasis, chemoimmunoresistance and EMT. Thus, m6A-modified
LINC00958 promotes the tumour progression of BRCA cells via the
miR-378a-3p/YY1 axis. Likewise, Zhao et al (95) found that the METTL3-modified lncRNA
MALAT1 facilitates the pathogenesis of BRCA through the
miR-26b/high-mobility-group A2 (HMGA2) axis. HMGA2 is a
transcriptional regulator that is known to promote stemness,
aggression and tumour heterogenicity. The differential expression
of m6A-related lncRNAs in BRCA makes them exciting candidates for
diagnostic markers or therapeutic targets for BRCA.
Gynaecological cancers originate from and affect
women's reproductive organs, while endometrial, ovarian and
cervical cancers are the most frequent and major gynaecological
cancers in females (96). Studies
have indicated that m6A-modified lncRNAs have pivotal roles in the
tumorigenesis and metastasis of gynaecological cancers. Shen et
al (97) observed that
abundant m6A modification increases the degradation of lncRNA
FENDRR by recruiting the m6A reader YTHDF2 in endometrioid
endometrial carcinoma (EEC). Subsequently, downregulation of lncRNA
FENDRR results in the accumulation of SOX4 protein, which is
reported to promote the proliferation of EEC cell lines via
epigenetic repression of miR-129-2. Thus, m6A-modified FENDRR
boosts the proliferation of EEC cells and further aggravates the
pathological process of EEC. Wang et al (98) discovered that m6A upregulates the
levels of lncRNA RHPN1-AS1 by decreasing RNA decay in epithelial
ovarian cancer (EOC). Subsequently, RHPN1-AS1 acts as a ceRNA to
sponge miR-596, consequently increasing leucine zipper/EF
hand-containing transmembrane-1 (LETM1) expression, which has an
oncogenic role as an activator of the focal adhesion kinase
(FAK)/PI3K/Akt pathway in EOC. Therefore, RHPN1-AS1 facilitates EOC
cell growth and metathesis via the miR-596/LETM1 axis. Likewise,
Yang et al (99) revealed
that METTL3 methylation is required for lncRNA ZFAS1 to sequester
miR-647 by acting as a ceRNA, which was previously reported to be
involved in tumour progression, ultimately resulting in enhanced
cervical cancer (CC) proliferation and metastasis. Sun et al
(100) also revealed that lncRNA
KCNMB2-AS1 is stabilized by m6A modification and promotes CC growth
by acting as a ceRNA to sponge miR-130b-5p and miR-4294. Despite
these studies having contributed to the understanding of the
disease pathophysiology, challenges in gynaecological cancers still
require to be addressed by comprehensive research to accelerate the
advancement of prognostic, diagnostic and therapeutic
approaches.
Neoplasms of the haematopoietic and lymphoid tissues
are closely linked to the circulatory and immune systems,
accounting for ~9% of all cancers (101). Due to their roles in the
regulation of blood cell turnover, m6A-modified lncRNAs are likely
to participate in the tumorigenesis of haematopoietic malignancies.
Focusing on tumorigenesis fusion proteins produced by aberrant
chromosomal translocations, Chen et al (102) reported that lncRNA MALAT1 binds
to these fusion proteins and is localized in nuclear speckles after
m6A modification. Then, m6A-modified MALAT1 and fusion proteins
serve as a functional platform for METTL14 to methylate chimeric
mRNA. After binding YTHDC1, m6A-modified chimeric mRNA is exported
to the cytoplasm, leading to upregulation of the fusion protein,
which is involved in the tumorigenesis of leukaemia. Yao et
al (103) discovered that
METTL3-mediated methylation increases the stability of lncRNA
NEAT1. Subsequently, m6A-modified NEAT1 acts as a ceRNA to sponge
miR-766-5p and subsequently increases levels of cyclin-dependent
kinase inhibitor 1A (CDKN1A), which is the target gene of
miR-766-5p and serves as a negative cell cycle regulator. Finally,
m6A-modified NEAT1 suppresses the progression of chronic myelocytic
leukaemia by regulating the miR-766-5p/CDKN1A axis. Song et
al (104) reported that
ALKBH5 decreases the m6A modification of lncRNA TRERNA1, resulting
in increased TRERNA1 in diffuse large B-cell lymphoma (DLBCL).
TRERNA1 then recruits EZH2 to the p21 promoter region, leading to
the repression of this cyclin-dependent inhibitor. Finally, TRERNA1
regulates cell proliferation and is associated with poor prognosis
in patients with DLBCL. The development of sensitive detection
methods and targeted inhibitors for m6A-modified lncRNAs may have
potential in the clinical management of haematological
malignancies, particularly when combined with existing options.
HNSCC is the sixth most frequently occurring
neoplasm globally, claiming ~450,000 lives every year (91). To date, the complex molecular
mechanisms of m6A-modified lncRNAs and their influence on the
tumorigenesis and development of HNSCC still require to be
explored. Ban et al (105)
discovered that MTC-induced m6A modification increases the
stability of the lncRNA LNCAROD. As a scaffold for the interaction
between Y-box binding protein 1 (YBX1) and heat-shock 70-kDa
protein 1A (HSPA1A), m6A-modified LNCAROD shields YBX1 from
proteasomal degradation by promoting the YBX1-HSPA1A
protein-protein interaction, leading to HNSCC cell growth by
stabilizing the YBX1 protein. It is still necessary to verify the
vital role of m6A-modified lncRNAs in HNSCC, understand the
mechanisms of tumorigenesis and progression in depth and identify
more effective therapeutic approaches.
Due to early invasion and distant metastasis, the
5-year survival rate of NPC remains <20% (106). Accumulating studies suggest that
dysregulation of m6A-modified lncRNAs has a pivotal role in the
occurrence and metastasis of NPC. Zheng et al (107) identified that m6A modification
improves the stability of lncRNA FAM225A. Consequently,
upregulation of FAM225A facilitates its function as a ceRNA to
sponge miR-590-3p and miR-1275, resulting in increased integrin β3
(ITGB3) expression, which is one of the activators in the
FAK/PI3K/Akt pathway. Functionally, FAM225A has been confirmed to
promote NPC tumorigenesis.
Characterized by high tumour heterogeneity and poor
survival, gliomas appear to be one of the deadliest intracranial
cancer types (108). Recently,
scientists demonstrated that abnormal m6A-modified lncRNAs are
associated with the occurrence and growth of gliomas. Chang et
al (109) indicated that
METTL3 was elevated in isocitrate dehydrogenase (IDH) wild-type
glioma, and with the assistance of HuR, upregulated levels of m6A
methylation increase the stability of lncRNA MALAT1. M6A-methylated
MALAT1 then activates the NF-κB signalling pathway to promote the
malignant progression of IDH wild-type gliomas (109). The association between
m6A-modified lncRNAs and oncology is being increasingly explored,
while studies in this field have the potential to lay a solid
foundation for elucidating the mechanisms of brain tumours.
Thyroid cancer (TC) is the most frequently occurring
cancer type arising from the endocrine system and has exhibited an
increasing trend in recent decades (91). However, its morbidity is
significantly heterogeneous in terms of different regional factors,
ethnic backgrounds and genders (110). To date, various m6A-modified
lncRNAs have been studied. Dong et al (111) reported that the m6A reader
IGF2BP2 recognizes m6A modification of lncRNA HAGLR and upregulates
the expression and stability of HAGLR. Subsequently, elevated HAGLR
markedly promotes TC cell proliferation, migration and invasion
(111). The features of
m6A-modified lncRNAs may be applied to the clinical management of
TC and have the potential to expand the current knowledge regarding
TC tumorigenesis.
Emerging evidence has revealed that m6A-modified
lncRNAs participate in tumour cell proliferation, invasion and
migration primarily through two different mechanisms: regulating
lncRNA stability (58,59) and translation (63). However, fewer studies on cancers
are relevant to the other three functions of m6A-modified lncRNAs,
such as changing accessibility to binding proteins (50,54),
mediating gene transcriptional silencing (22) and affecting mRNA precursor
alternative splicing (57), which
still require to be explored in depth. Furthermore, novel
bioinformatics analyses based on sequencing databases focused on
m6A-modified lncRNA roles in the tumour immune microenvironment,
such as immunocyte infiltration and responses to immune checkpoint
inhibitors (112,113), require in-depth studies to
resolve challenges that are highly relevant to clinical
application.
To explore the biological functions of m6A
modification, it is urgent to clarify where m6A modifications occur
and how many of these modifications exist in RNA molecules. To
date, various novel detection technologies for m6A modification
have been developed that benefit from mass spectrometric techniques
and high-throughput sequencing (Table
I). These detection technologies include antibody-based
methods, such as methylated RNA immunoprecipitation sequencing
(meRIP-seq), photo-cross-linking-assisted m6A sequencing strategy,
m6A individual-nucleotide resolution UV crosslinking and
immunoprecipitation, m6A-level isoform-characterization sequencing,
digestion-based methods named m6A-sensitive
RNA-endoribonuclease-facilitated sequencing or MAZTER-seq, a
gene-editing based method named deamination adjacent to RNA
modification targets-sequencing, and metabolism-based methods
including m6A-label-seq and FTO-assisted m6A selective chemical
labeling method sequencing. Among of them, meRIP-seq adopts
specific antibodies of methylated RNAs to capture methylated RNA
segments for sequencing (20),
which is widely used in investigating methylated RNAs (114,115).
In addition, bioinformatics is a powerful tool that
is able to identify likely RNA methylation sites based on
statistical evidence (Table II).
Among these public databases for prediction, SRAMP and m6A2Target
appear to be applicated more widely (116,117). Of note, LITHOPHONE is a
computational framework specifically designed for predicting m6A
sites of lncRNAs (118). In
addition, the use of public databases may allow researchers to
reduce the cost of preliminary investigations and facilitate
experimental design.
Although m6A detection and prediction methods are
continuously improving, it is urgent to develop more accurate
methods to identify or predict m6A sites on lncRNAs. In addition,
public databases to explore m6A-modified lncRNAs must be
established to broaden the horizon regarding their association with
biological features of human cancers.
In conclusion, m6A modification of lncRNAs has
pivotal roles in gene expression through several mechanisms,
including the adaptation in structure and binding proteins of
lncRNAs, direct and/or indirect regulation of transcription,
alteration of mRNA precursor splicing and change of lncRNA
stability and translation. Undoubtedly, lncRNAs modified by m6A
writers, erasers or readers are involved in the development and
progression of various human malignancies. However, further
investigations are required in the following aspects.
The tumour microenvironment remains a hot spot in
cancer research and a number of studies have identified lncRNAs as
key regulators in the tumour microenvironment (121). However, only a small number of
investigations have focused on the function of m6A-modified lncRNAs
in the tumour microenvironment. It is possible that single-cell
sequencing technology is used to scrutinize the m6A-modified
lncRNAs in different cell types of the tumour microenvironment.
Current detection procedures of m6A-modified
lncRNAs are complex and lack single molecule resolution. The
technologies to quantify m6A-modified lncRNAs are also limited
(122). For instance,
transcriptome-wide m6A-seq methods frequently require a large
amount of RNA, so it is difficult to apply in clinical practice due
to the limitation of patient-derived samples. It is necessary to
establish new detection approaches, which allow the detection of
m6A-modified lncRNAs at a low concentration such as in tumour
tissues, circulating tumour cells, exosomes and faeces, and a more
comprehensive public database to collect information of lncRNAs
modified by m6A, which will facilitate the investigation of
m6A-modified lncRNAs as regulators, biomarkers and potential
therapeutic targets in human cancer.
Not applicable.
YH and XD drafted the manuscript and completed the
visualization. MC performed the literature search and selection,
and participated in reviewing the paper. LH and JS performed
revision of the manuscript, and conducted project administration
and funding acquisition. All authors read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The project was supported by the National Natural Science
Foundation of China (grant no. 81773187), Tianjin High School
Program for Young and Middle-aged Talents Backbone and Tianjin
Young Medical Talents Program and the Tianjin Commission Scientific
Research Plan Project (grant no. 2018KJ064).
|
1
|
Liu J and Jia G: Methylation modifications
in eukaryotic messenger RNA. J Genet Genomics. 41:21–33. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boccaletto P, Machnicka MA, Purta E,
Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R,
Limbach PA, Kotter A, et al: MODOMICS: A database of RNA
modification pathways. 2017 update. Nucleic Acids Res.
46:D303–D307. 2018. View Article : Google Scholar :
|
|
3
|
Li J, Yang X, Qi Z, Sang Y, Liu Y, Xu B,
Liu W, Xu Z and Deng Y: The role of mRNA m6A methylation
in the nervous system. Cell Biosci. 9:662019. View Article : Google Scholar
|
|
4
|
Dominissini D, Nachtergaele S,
Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni
A, Salmon-Divon M, Clark WC, et al: The dynamic
N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature.
530:441–446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
You C, Dai X and Wang Y:
Position-dependent effects of regioisomeric methylated adenine and
guanine ribonucleosides on translation. Nucleic Acids Res.
45:9059–9067. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartosovic M, Molares HC, Gregorova P,
Hrossova D, Kudla G and Vanacova S: N6-methyladenosine demethylase
FTO targets pre-mRNAs and regulates alternative splicing and 3'-end
processing. Nucleic Acids Res. 45:11356–11370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roundtree IA and He C: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Trends Genet. 32:320–321.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar
|
|
11
|
Fustin JM, Doi M, Yamaguchi Y, Hida H,
Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I
and Okamura H: RNA-methylation-dependent RNA processing controls
the speed of the circadian clock. Cell. 155:793–806. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen J, Zhang YC, Huang C, Shen H, Sun B,
Cheng X, Zhang YJ, Yang YG, Shu Q, Yang Y and Li X: m6A
regulates neurogenesis and neuronal development by modulating
histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics.
17:154–168. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu J, Bai J, Zhang X, Lv Y, Gong Y, Liu L,
Zhao H, Yu F, Ping Y, Zhang G, et al: A comprehensive overview of
lncRNA annotation resources. Brief Bioinform. 18:236–249. 2017.
|
|
14
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bhat SA, Ahmad SM, Mumtaz PT, Malik AA,
Dar MA, Urwat U, Shah RA and Ganai NA: Long non-coding RNAs:
Mechanism of action and functional utility. Noncoding RNA Res.
1:43–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen F, Li Z, Deng C and Yan H:
Integration analysis for novel lncRNA markers predicting tumor
recurrence in human colon adenocarcinoma. J Transl Med. 17:2992019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zimmer-Bensch G: Emerging roles of long
non-coding RNAs as drivers of brain evolution. Cells. 8:13992019.
View Article : Google Scholar
|
|
18
|
Robinson EK, Covarrubias S and Carpenter
S: The how and why of lncRNA function: An innate immune
perspective. Biochim Biophys Acta Gene Regul Mech. 1863:1944192020.
View Article : Google Scholar :
|
|
19
|
Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G,
Yuan W, Kan Q and Sun Z: The interplay between m6A RNA methylation
and noncoding RNA in cancer. J Hematol Oncol. 12:1212019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3'UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu N, Dai Q, Zheng G, He C, Parisien M
and Pan T: N(6)-methyladenosine-dependent RNA structural switches
regulate RNA-protein interactions. Nature. 518:560–564. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m(6)A RNA methylation promotes
XIST-mediated transcriptional repression. Nature. 537:369–373.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar :
|
|
24
|
Lan Q, Liu PY, Haase J, Bell JL,
Huttelmaier S and Liu T: The critical role of RNA m6A
methylation in cancer. Cancer Res. 79:1285–1292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Y, Hsu PJ, Chen YS and Yang YG:
Dynamic transcriptomic m6A decoration: Writers, erasers,
readers and functions in RNA metabolism. Cell Res. 28:616–624.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yao L, Man CF, He R, He L, Huang JB, Xiang
SY, Dai Z, Wang XY and Fan Y: The Interaction between
N6-Methyladenosine modification and non-coding RNAs in
gastrointestinal tract cancers. Front Oncol. 11:7841272021.
View Article : Google Scholar
|
|
27
|
Deng X, Su R, Weng H, Huang H, Li Z and
Chen J: RNA N6-methyladenosine modification in cancers:
Current status and perspectives. Cell Res. 28:507–517. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5'sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wen J, Lv R, Ma H, Shen H, He C, Wang J,
Jiao F, Liu H, Yang P, Tan L, et al: Zc3h13 regulates nuclear RNA
m6A methylation and mouse embryonic stem cell
self-renewal. Mol Cell. 69:1028–1038.e6. 2018. View Article : Google Scholar
|
|
30
|
Bokar JA, Shambaugh ME, Polayes D, Matera
AG and Rottman FM: Purification and cDNA cloning of the
AdoMet-binding subunit of the human mRNA
(N6-adenosine)-methyltransferase. RNA. 3:1233–1247. 1997.PubMed/NCBI
|
|
31
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Structural basis of
N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature.
534:575–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
33
|
Horiuchi K, Kawamura T, Iwanari H, Ohashi
R, Naito M, Kodama T and Hamakubo T: Identification of Wilms' tumor
1-associating protein complex and its role in alternative splicing
and the cell cycle. J Biol Chem. 288:33292–33302. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Warda AS, Kretschmer J, Hackert P, Lenz C,
Urlaub H, Höbartner C, Sloan KE and Bohnsack MT: Human METTL16 is a
N6-methyladenosine (m6A) methyltransferase
that targets pre-mRNAs and various non-coding RNAs. EMBO Rep.
18:2004–2014. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ruszkowska A: METTL16,
Methyltransferase-like protein 16: Current insights into structure
and function. Int J Mol Sci. 22:21762021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zuo X, Chen Z, Gao W, Zhang Y, Wang J,
Wang J, Cao M, Cai J, Wu J and Wang X: M6A-mediated upregulation of
LINC00958 increases lipogenesis and acts as a nanotherapeutic
target in hepatocellular carcinoma. J Hematol Oncol. 13:52020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jin D, Guo J, Wu Y, Du J, Yang L, Wang X,
Di W, Hu B, An J, Kong L, et al: m6A mRNA methylation
initiated by METTL3 directly promotes YAP translation and increases
YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to
induce NSCLC drug resistance and metastasis. J Hematol Oncol.
14:322021. View Article : Google Scholar
|
|
38
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun T, Wu R and Ming L: The role of m6A
RNA methylation in cancer. Biomed Pharmacother. 112:1086132019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang T, Kong S, Tao M and Ju S: The
potential role of RNA N6-methyladenosine in Cancer progression. Mol
Cancer. 19:882020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shi H, Wei J and He C: Where, When, and
How: Context-dependent functions of RNA methylation writers,
readers, and eras. Mol Cell. 74:640–650. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Frye M, Harada BT, Behm M and He C: RNA
modifications modulate gene expression during development. Science.
361:1346–1349. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yan J, Huang X, Zhang X, Chen Z, Ye C,
Xiang W and Huang Z: LncRNA LINC00470 promotes the degradation of
PTEN mRNA to facilitate malignant behavior in gastric cancer cells.
Biochem Biophys Res Commun. 521:887–893. 2020. View Article : Google Scholar
|
|
45
|
Hu Y, Tang J, Xu F, Chen J, Zeng Z, Han S,
Wang F, Wang D, Huang M, Zhao Y, et al: A reciprocal feedback
between N6-methyladenosine reader YTHDF3 and lncRNA DICER1-AS1
promotes glycolysis of pancreatic cancer through inhibiting
maturation of miR-5586-5p. J Exp Clin Cancer Res. 41:692022.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N6-methyladenosine-modified RNA. Cell Res. 27:315–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Luxton HJ, Simpson BS, Mills IG, Brindle
NR, Ahmed Z, Stavrinides V, Heavey S, Stamm S and Whitaker HC: The
oncogene metadherin interacts with the known splicing proteins
YTHDC1, Sam68 and T-STAR and plays a novel role in alternative mRNA
splicing. Cancers (Basel). 11:12332019. View Article : Google Scholar
|
|
48
|
Ding Y, Wang M and Yang J: Circular RNA
midline-1 (circMID1) promotes proliferation, migration, invasion
and glycolysis in prostate cancer. Bioengineered. 13:6293–6308.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Alarcón CR, Goodarzi H, Lee H, Liu X,
Tavazoie S and Tavazoie SF: HNRNPA2B1 Is a mediator of
m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu N, Zhou KI, Parisien M, Dai Q,
Diatchenko L and Pan T: N6-methyladenosine alters RNA structure to
regulate binding of a low-complexity protein. Nucleic Acids Res.
45:6051–6063. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhu P, He F, Hou Y, Tu G, Li Q, Jin T,
Zeng H, Qin Y, Wan X, Qiao Y, et al: A novel hypoxic long noncoding
RNA KB-1980E6.3 maintains breast cancer stem cell stemness via
interacting with IGF2BP1 to facilitate c-Myc mRNA stability.
Oncogene. 40:1609–1627. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Roost C, Lynch SR, Batista PJ, Qu K, Chang
HY and Kool ET: Structure and thermodynamics of N6-methyladenosine
in RNA: A spring-loaded base modification. J Am Chem Soc.
137:2107–2115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhou KI, Parisien M, Dai Q, Liu N,
Diatchenko L, Sachleben JR and Pan T: N(6)-Methyladenosine
modification in a long noncoding RNA hairpin predisposes its
conformation to protein binding. J Mol Biol. 428:822–833. 2016.
View Article : Google Scholar
|
|
55
|
Dong S, Wu Y, Liu Y, Weng H and Huang H:
N6-methyladenosine steers RNA metabolism and regulation
in cancer. Cancer Commun (Lond). 41:538–559. 2021. View Article : Google Scholar
|
|
56
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ninomiya K, Iwakiri J, Aly MK, Sakaguchi
Y, Adachi S, Natsume T, Terai G, Asai K, Suzuki T and Hirose T:
m6 A modification of HSATIII lncRNAs regulates
temperature-dependent splicing. EMBO J. 40:e1079762021. View Article : Google Scholar
|
|
58
|
Xue L, Li J, Lin Y, Liu D, Yang Q, Jian J
and Peng J: m6 A transferase METTL3-induced lncRNA
ABHD11-AS1 promotes the Warburg effect of non-small-cell lung
cancer. J Cell Physiol. 236:2649–2658. 2021. View Article : Google Scholar
|
|
59
|
Guo T, Liu DF, Peng SH and Xu AM: ALKBH5
promotes colon cancer progression by decreasing methylation of the
lncRNA NEAT1. Am J Transl Res. 12:4542–4549. 2020.PubMed/NCBI
|
|
60
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Okazaki Y, Furuno M, Kasukawa T, Adachi J,
Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al:
Analysis of the mouse transcriptome based on functional annotation
of 60,770 full-length cDNAs. Nature. 420:563–573. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wu Q, Wright M, Gogol MM, Bradford WD,
Zhang N and Bazzini AA: Translation of small downstream ORFs
enhances translation of canonical main open reading frames. EMBO J.
39. pp. e1047632020, View Article : Google Scholar
|
|
63
|
Wu S, Zhang L, Deng J, Guo B, Li F, Wang
Y, Wu R, Zhang S, Lu J and Zhou Y: A novel micropeptide encoded by
Y-linked LINC00278 links cigarette smoking and AR signaling in male
esophageal squamous cell carcinoma. Cancer Res. 80:2790–2803. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chen J, Brunner AD, Cogan JZ, Nuñez JK,
Fields AP, Adamson B, Itzhak DN, Li JY, Mann M, Leonetti MD and
Weissman JS: Pervasive functional translation of noncanonical human
open reading frames. Science. 367:1140–1146. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wu Y, Yang X, Chen Z, Tian L, Jiang G,
Chen F, Li J, An P, Lu L, Luo N, et al: m6A-induced
lncRNA RP11 triggers the dissemination of colorectal cancer cells
via upregulation of Zeb1. Mol Cancer. 18:872019. View Article : Google Scholar
|
|
66
|
Biazzo A and De Paolis M:
Multidisciplinary approach to osteosarcoma. Acta Orthop Belg.
82:690–698. 2016.
|
|
67
|
Chen S, Zhou L and Wang Y: ALKBH5-mediated
m6A demethylation of lncRNA PVT1 plays an oncogenic role
in osteosarcoma. Cancer Cell Int. 20:342020. View Article : Google Scholar
|
|
68
|
Chen J, Tian Y, Zhang Q, Ren D, Zhang Q,
Yan X, Wang L, He Z, Zhang W, Zhang T and Yuan X: Novel insights
into the role of N6-Methyladenosine RNA modification in bone
pathophysiology. Stem Cells Dev. 30:17–28. 2021. View Article : Google Scholar
|
|
69
|
Li D, Yang C, Yin C, Zhao F, Chen Z, Tian
Y, Dang K, Jiang S, Zhang W, Zhang G and Qian A: LncRNA, important
player in bone development and disease. Endocr Metab Immune Disord
Drug Targets. 20:50–66. 2020. View Article : Google Scholar
|
|
70
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar
|
|
71
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang J, Guo S, Piao HY, Wang Y, Wu Y,
Meng XY, Yang D, Zheng ZC and Zhao Y: ALKBH5 promotes invasion and
metastasis of gastric cancer by decreasing methylation of the
lncRNA NEAT1. J Physiol Biochem. 75:379–389. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yang D, Chang S, Li F, Ma M, Yang J, Lv X,
Huangfu L and Jia C: m6 A transferase
KIAA1429-stabilized LINC00958 accelerates gastric cancer aerobic
glycolysis through targeting GLUT1. IUBMB Life. 73:1325–1333. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Hu N and Ji H: N6-methyladenosine
(m6A)-mediated up-regulation of long noncoding RNA LINC01320
promotes the proliferation, migration, and invasion of gastric
cancer via miR495-5p/RAB19 axis. Bioengineered. 12:4081–4091. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou
A, Liu J, Che L and Li J: Long noncoding RNA GAS5 inhibits
progression of colorectal cancer by interacting with and triggering
YAP phosphorylation and degradation and is negatively regulated by
the m6A reader YTHDF3. Mol Cancer. 18:1432019.
View Article : Google Scholar
|
|
77
|
Yang X, Zhang S, He C, Xue P, Zhang L, He
Z, Zang L, Feng B, Sun J and Zheng M: METTL14 suppresses
proliferation and metastasis of colorectal cancer by
down-regulating oncogenic long non-coding RNA XIST. Mol Cancer.
19:462020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wu J, Pang R, Li M, Chen B, Huang J and
Zhu Y: m6A-induced LncRNA MEG3 suppresses the proliferation,
migration and invasion of hepatocellular carcinoma cell through
miR-544b/BTG2 signaling. Onco Targets Ther. 14:3745–3755. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chen MH, Fu LS, Zhang F, Yang Y and Wu XZ:
LncAY controls BMI1 expression and activates BMI1/Wnt/beta-catenin
signaling axis in hepatocellular carcinoma. Life Sci.
280:1197482021. View Article : Google Scholar
|
|
80
|
Chen YT, Xiang D, Zhao XY and Chu XY:
Upregulation of lncRNA NIFK-AS1 in hepatocellular carcinoma by
m6A methylation promotes disease progression and
sorafenib resistance. Hum Cell. 34:1800–1811. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Mizrahi JD, Surana R, Valle JW and Shroff
RT: Pancreatic cancer. Lancet. 395:2008–2020. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Yin MY, Xi LT, Liu L, Zhu JZ, Qian LJ and
Xu CF: Pancreatic cancer incidence and mortality patterns in
2006-2015 and prediction of the epidemiological trend to 2025 in
China. World J Clin Cases. 10:4404–4413. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P,
Liu D, Tian L, Yin J, Jiang K and Miao Y: ALKBH5 inhibits
pancreatic cancer motility by decreasing long non-coding RNA
KCNK15-AS1 methylation. Cell Physiol Biochem. 48:838–846. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Hu X, Peng WX, Zhou H, Jiang J, Zhou X,
Huang D, Mo YY and Yang L: IGF2BP2 regulates DANCR by serving as an
N6-methyladenosine reader. Cell Death Differ. 27:1782–1794. 2020.
View Article : Google Scholar :
|
|
85
|
Chen Y, Zitello E, Guo R and Deng Y: The
function of LncRNAs and their role in the prediction, diagnosis,
and prognosis of lung cancer. Clin Transl Med. 11:e3672021.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Song H, Li H, Ding X, Li M, Shen H, Li Y,
Zhang X and Xing L: Long noncoding RNA FEZF1AS1 facilitates
nonsmall cell lung cancer progression via the ITGA11/miR516b5p
axis. Int J Oncol. 57:1333–1347. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Qian X, Yang J, Qiu Q, Li X, Jiang C, Li
J, Dong L, Ying K, Lu B, Chen E, et al: LCAT3, a novel
m6A-regulated long non-coding RNA, plays an oncogenic role in lung
cancer via binding with FUBP1 to activate c-MYC. J Hematol Oncol.
14:1122021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yu H and Zhang Z: ALKBH5-mediated m6A
demethylation of lncRNA RMRP plays an oncogenic role in lung
adenocarcinoma. Mamm Genome. 32:195–203. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Tan L, Tang Y, Li H, Li P, Ye Y, Cen J,
Gui C, Luo J, Cao J and Wei J: N6-Methyladenosine modification of
LncRNA DUXAP9 promotes renal cancer cells proliferation and
motility by activating the PI3K/AKT signaling pathway. Front Oncol.
11:6418332021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer Statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Barros-Silva D, Lobo J, Guimarães-Teixeira
C, Carneiro I, Oliveira J, Martens-Uzunova ES, Henrique R and
Jerónimo C: VIRMA-dependent N6-Methyladenosine modifications
regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in
prostate cancer. Cancers (Basel). 12:7712020. View Article : Google Scholar
|
|
93
|
Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W,
Lu S, Xu D, Wu Y, Chen Q, et al: LNC942 promoting METTL14-mediated
m6A methylation in breast cancer cell proliferation and
progression. Oncogene. 39:5358–5372. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Rong D, Dong Q, Qu H, Deng X, Gao F, Li Q
and Sun P: m6A-induced LINC00958 promotes breast cancer
tumorigenesis via the miR-378a-3p/YY1 axis. Cell Death Dis.
7:272021. View Article : Google Scholar
|
|
95
|
Zhao C, Ling X, Xia Y, Yan B and Guan Q:
The m6A methyltransferase METTL3 controls epithelial-mesenchymal
transition, migration and invasion of breast cancer through the
MALAT1/miR-26b/HMGA2 axis. Cancer Cell Int. 21:4412021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Di Fiore R, Suleiman S, Drago-Ferrante R,
Felix A, O'Toole SA, O'Leary JJ, Ward MP, Beirne J, Yordanov A,
Vasileva-Slaveva M, et al: LncRNA MORT (ZNF667-AS1) in cancer-is
there a possible role in gynecological malignancies? Int J Mol Sci.
22:78292021. View Article : Google Scholar
|
|
97
|
Shen J, Feng XP, Hu RB, Wang H, Wang YL,
Qian JH and Zhou YX: N-methyladenosine reader YTHDF2-mediated long
noncoding RNA FENDRR degradation promotes cell proliferation in
endometrioid endometrial carcinoma. Lab Invest. 101:775–784. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wang J, Ding W, Xu Y, Tao E, Mo M, Xu W,
Cai X, Chen X, Yuan J and Wu X: Long non-coding RNA RHPN1-AS1
promotes tumorigenesis and metastasis of ovarian cancer by acting
as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany
NY). 12:4558–4572. 2020. View Article : Google Scholar
|
|
99
|
Yang Z, Ma J, Han S, Li X, Guo H and Liu
D: ZFAS1 exerts an oncogenic role via suppressing miR-647 in an
m6A-dependent manner in cervical cancer. Onco Targets
Ther. 13:11795–11806. 2020. View Article : Google Scholar :
|
|
100
|
Zhang Y, Wang D, Wu D, Zhang D and Sun M:
Long noncoding RNA KCNMB2-AS1 stabilized by
N6-Methyladenosine modification promotes cervical cancer
growth through acting as a competing endogenous RNA. Cell
Transplant. 29:9636897209643822020.
|
|
101
|
Ghafouri-Fard S, Esmaeili M and Taheri M:
Expression of non-coding RNAs in hematological malignancies. Eur J
Pharmacol. 875:1729762020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Chen ZH, Chen TQ, Zeng ZC, Wang D, Han C,
Sun YM, Huang W, Sun LY, Fang K, Chen YQ, et al: Nuclear export of
chimeric mRNAs depends on an lncRNA-triggered autoregulatory loop
in blood malignancies. Cell Death Dis. 11:5662020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Yao FY, Zhao C, Zhong FM, Qin TY, Wen F,
Li MY, Liu J, Huang B and Wang XZ: m6A Modification of
lncRNA NEAT1 regulates chronic myelocytic leukemia progression via
miR-766-5p/CDKN1A axis. Front Oncol. 11:6796342021. View Article : Google Scholar
|
|
104
|
Song W, Fei F, Qiao F, Weng Z, Yang Y, Cao
B, Yue J, Xu J, Zheng M and Li J: ALKBH5-mediated
N6-methyladenosine modification of TRERNA1 promotes DLBCL
proliferation via p21 downregulation. Cell Death Discov. 8:252022.
View Article : Google Scholar :
|
|
105
|
Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y,
Mei Y, Tan Y, Li X, Zeng Z, et al: LNCAROD is stabilized by m6A
methylation and promotes cancer progression via forming a ternary
complex with HSPA1A and YBX1 in head and neck squamous cell
carcinoma. Mol Oncol. 14:1282–1296. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Lee AW, Ma BB, Ng WT and Chan AT:
Management of nasopharyngeal carcinoma: Current practice and future
perspective. J Clin Oncol. 33:3356–3364. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL,
Lv JW, Huang XD, Liu RQ, Chen F, He XJ, et al: Long noncoding RNA
FAM225A promotes nasopharyngeal carcinoma tumorigenesis and
metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and
Upregulate ITGB3. Cancer Res. 79:4612–4626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Dong Z and Cui H: The emerging roles of
RNA modifications in Glioblastoma. Cancers (Basel). 12:7362020.
View Article : Google Scholar
|
|
109
|
Chang YZ, Chai RC, Pang B, Chang X, An SY,
Zhang KN, Jiang T and Wang YZ: METTL3 enhances the stability of
MALAT1 with the assistance of HuR via m6A modification and
activates NF-kB to promote the malignant progression of
IDH-wildtype glioma. Cancer Lett. 511:36–46. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Roman BR, Morris LG and Davies L: The
thyroid cancer epidemic, 2017 perspective. Curr Opin Endocrinol
Diabetes Obes. 24:332–336. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Dong L, Geng Z, Liu Z, Tao M, Pan M and Lu
X: IGF2BP2 knockdown suppresses thyroid cancer progression by
reducing the expression of long non-coding RNA HAGLR. Pathol Res
Pract. 225:1535502021. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yuan C, Liu C, Zhao S, Zhang X, Jia H,
Chen B, Zhang M, Zheng Y, Zhou J and Bo Y: The role of
N6-Methyladenosine-associated lncRNAs in the immune
microenvironment and prognosis of colorectal cancer. J Oncol.
2022:46893962022. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhang M, Zhang J and Liu Y: Comprehensive
analysis of molecular features, prognostic values, and immune
landscape association of m6A-regulated immune-related lncRNAs in
smoking-associated lung squamous cell carcinoma. Front Genet.
13:8874772022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Du QY, Huo FC, Du WQ, Sun XL, Jiang X,
Zhang LS and Pei DS: METTL3 potentiates progression of cervical
cancer by suppressing ER stress via regulating m6A modification of
TXNDC5 mRNA. Oncogene. 41:4420–4432. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Li HB, Huang G, Tu J, Lv DM, Jin QL, Chen
JK, Zou YT, Lee DF, Shen JN and Xie XB: METTL14-mediated
epitranscriptome modification of MN1 mRNA promote tumorigenicity
and all-trans-retinoic acid resistance in osteosarcoma.
EBioMedicine. 82:1041422022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhao Q, Zhao Y, Hu W, Zhang Y, Wu X, Lu J,
Li M, Li W, Wu W, Wang J, et al: m6A RNA modification
modulates PI3K/Akt/mTOR signal pathway in gastrointestinal cancer.
Theranostics. 10:9528–9543. 2020. View Article : Google Scholar :
|
|
117
|
Lu M, Zhan H, Liu B, Li D, Li W, Chen X
and Zhou X: N6-methyladenosine-related non-coding RNAs are
potential prognostic and immunotherapeutic responsiveness
biomarkers for bladder cancer. EPMA J. 12:589–604. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Liu L, Lei X, Fang Z, Tang Y, Meng J and
Wei Z: LITHOPHONE: Improving lncRNA methylation site prediction
using an ensemble predictor. Front Genet. 11:5452020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng
X, Xiong F, Guo C, Wu X, Li Y, et al: Emerging role of
tumor-related functional peptides encoded by lncRNA and circRNA.
Mol Cancer. 19:222020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Kong S, Tao M, Shen X and Ju S:
Translatable circRNAs and lncRNAs: Driving mechanisms and functions
of their translation products. Cancer Lett. 483:59–65. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Park EG, Pyo SJ, Cui Y, Yoon SH and Nam
JW: Tumor immune microenvironment lncRNAs. Brief Bioinform.
23:bbab5042022. View Article : Google Scholar :
|
|
122
|
Zhang Z, Chen LQ, Zhao YL, Yang CG,
Roundtree IA, Zhang Z, Ren J, Xie W, He C and Luo GZ: Single-base
mapping of m6A by an antibody-independent method. Sci
Adv. 5:eaax02502019. View Article : Google Scholar
|
|
123
|
Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N,
Han D, Dominissini D, Dai Q, Pan T and He C: High-resolution
N(6)-methyladenosine (m(6) A) map using
photo-cross-linking-assisted m(6) A sequencing. Angew Chem Int Ed
Engl. 54:1587–1590. 2015. View Article : Google Scholar
|
|
124
|
Linder B, Grozhik AV, Olarerin-George AO,
Meydan C, Mason CE and Jaffrey SR: Single-nucleotide-resolution
mapping of m6A and m6Am throughout the transcriptome. Nat Methods.
12:767–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Meyer KD: DART-seq: An antibody-free
method for global m6A detection. Nat Methods.
16:1275–1280. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Shu X, Cao J, Cheng M, Xiang S, Gao M, Li
T, Ying X, Wang F, Yue Y, Lu Z, et al: A metabolic labeling method
detects m6A transcriptome-wide at single base
resolution. Nat Chem Biol. 16:887–895. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Wang Y, Xiao Y, Dong S, Yu Q and Jia G:
Antibody-free enzyme-assisted chemical approach for detection of
N6-methyladenosine. Nat Chem Biol. 16:896–903. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhou Y, Zeng P, Li YH, Zhang Z and Cui Q:
SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based
on sequence-derived features. Nucleic Acids Res. 44:e912016.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Jia CZ, Zhang JJ and Gu WZ:
RNA-MethylPred: A high-accuracy predictor to identify
N6-methyladenosine in RNA. Anal Biochem. 510:72–75. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Li GQ, Liu Z, Shen HB and Yu DJ:
TargetM6A: Identifying N6-Methyladenosine Sites from RNA
sequences via Position-specific nucleotide propensities and a
support vector machine. IEEE Trans Nanobioscience. 15:674–682.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S,
Zheng LL, Qu LH and Yang JH: RMBase v2.0: Deciphering the map of
RNA modifications from epitranscriptome sequencing data. Nucleic
Acids Res. 46:D327–D334. 2018. View Article : Google Scholar :
|
|
132
|
Zheng Y, Nie P, Peng D, He Z, Liu M, Xie
Y, Miao Y, Zuo Z and Ren J: m6AVar: A database of functional
variants involved in m6A modification. Nucleic Acids Res.
46:D139–D145. 2018. View Article : Google Scholar :
|
|
133
|
Zhang Y and Hamada M: DeepM6ASeq:
Prediction and characterization of m6A-containing sequences using
deep learning. BMC Bioinformatics. 19:5242018. View Article : Google Scholar
|
|
134
|
Han Y, Feng J, Xia L, Dong X, Zhang X,
Zhang S, Miao Y, Xu Q, Xiao S, Zuo Z, et al: CVm6A: A visualization
and exploration database for m6As in cell lines. Cells.
8:1682019. View Article : Google Scholar
|
|
135
|
Liu K, Cao L, Du P and Chen W:
im6A-TS-CNN: Identifying the N6-Methyladenine site in
multiple tissues by using the convolutional neural network. Mol
Ther Nucleic Acids. 21:1044–1049. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Deng S, Zhang H, Zhu K, Li X, Ye Y, Li R,
Liu X, Lin D, Zuo Z and Zheng J: M6A2Target: A comprehensive
database for targets of m6A writers, erasers and readers. Brief
Bioinform. 22:bbaa0552021. View Article : Google Scholar
|