Introduction
Currently, pancreatic ductal adenocarcinoma (PDAC)
is the seventh commonest cause of deaths in the world (1). Due to delayed diagnosis and a lack of
effective therapeutic regimens, the 5-year survival rate in PDAC is
~6% (2). Surgical resection may be
a potential therapeutic method for patients with early-stage
disease, but it is insufficient in advanced-stage disease (3). The poor prognosis highlights the need
to identify more molecular markers to improve early diagnosis and
reduce mortality. Thus, more in-depth research is necessary to
determine the mechanisms underlying the pathogenesis of PDAC.
Ubiquitylation is an important post-translational
modification that helps maintain protein homeostasis in eukaryotic
cells via the marking of proteins by ubiquitin, which are
subsequently eliminated by the proteasome (4-7). E2
enzymes, which are core constituents of the ubiquitin system, can
influence the signal generation in ubiquitylation and alter the
eventual consequences of substrate proteins (8). Ubiquitin-conjugating enzyme E2K
(UBE2K) is a type of class II E2 group enzyme, also referred to as
E2-25K or HIP-2, which can contribute to the combination of
K48-linked ubiquitin chains (9,10).
UBE2K serves a number of roles in various diseases. For instance,
UBE2K serves important roles in nervous system diseases, including
Huntington's disease (HD), Alzheimer's disease (AD), Parkinson's
disease (PD), and amyotrophic lateral sclerosis (ALS) (11-14).
Findings from previous studies have also demonstrated that UBE2K
serves vital roles in tumors and can be a target in cancer therapy
(15,16). For example, ixazomib exhibits
antitumor potential by targeting UBE2K in myeloma cells (15) and the overexpression of UBE2K
promotes gastric cancer progression in vitro and in
vivo (16). However, its
functions in PDAC remain to be elucidated.
RNA-binding proteins (RBPs) can regulate the fate of
RNA by modulating their stabilization and degradation to influence
gene expression in cancer cells (17). Insulin-like growth factor 2 RNA
binding protein 3 (IGF2BP3) belongs to the insulin like growth
factor 2 RNA binding protein family (18), which broadly participates in the
progression of multiple tumors. In recent studies, IGF2BP3 was
shown to promote malignant tumors progression by encouraging the
stabilization of downstream targets. For example, it enhanced the
stability of RCC2 expression in myeloid leukemia, cooperated with
LINC00958 to maintain the expression of E2F3 in endometrial
carcinoma, interacted with METTL3 to influence the expression of
PD-L1 to promote immune escape in breast carcinoma, and sustained
the expression of SIX4 in ovarian tumor cells (19-22).
However, whether IGF2BP3 regulates UBE2K expression in PDAC has yet
to be reported.
The present study found that UBE2K expression was
higher in PDAC cell lines and tissues compared with that in normal
cell lines and tissues. It further showed that UBE2K overexpression
encouraged cell proliferation and stemness-like phenotype, while
UBE2K knockdown inhibited the proliferation and stemness-like
phenotype. In addition, IGF2BP3 was found to be an RNA-binding
protein that enhanced UBE2K RNA stability, thus promoting UBE2K
expression. Finally, IGF2BP3 and UBE2K were demonstrated to form a
functional axis through functional rescue experiments.
Collectively, the results revealed the oncogenic roles of the
IGF2BP3/UBE2K axis in the malignant progression of PDAC, which
might provide novel insights into targeted therapy for PDAC.
Materials and methods
Databases
The gene expression profiling interactive analysis
(GEPIA) database (http://gepia.cancer-pku.cn/) can analyze the RNA
sequencing data of tumors and normal samples from The Cancer Genome
Atlas (TCGA) and the GTEx projects by clicking to obtain gene
expression profile, correlation analysis and patient survival
probability (23). The present
study detected the expression levels of UBE2K between 179 PAAD
tumor tissues and 171 corresponding normal tissues. In addition,
the overall survival rate (OS) and the correlation between IGF2BP3
and UBE2K in PAAD were also analyzed in GEPIA.
The Kaplan Meier plotter (https://kmplot.com) is a web tool to assess the
survival probability in tumor samples including PDAC from the GEO
and TCGA databases. The present study searched the OS and relapse
free survival rate (RFS) between higher and lower UBE2K RNA
expression levels in PDAC patients.
Cells culture
AsPC-1, BxPC-3, PANC-1, SW1990, and HPNE cells were
maintained in the Key Laboratory of Tumor Molecular Diagnosis and
Individualized Medicine of Zhejiang Province. PANC-1, HPNE, and
SW1990 cells were cultured in DMEM (Biological Industries).
Meanwhile, AsPC-1 and BxPC-3 cells were cultured in 1640 medium
(Biological Industries). The cells were all cultured in media
supplemented with 10% fetal bovine serum (Biological Industries)
supplemented with 1% penicillin/streptomycin in an incubator with
5% CO2 at 37°C condition.
Cells transduction and transfection
Lentivirus for UBE2K overexpression or knockdown was
purchased from Genomeditech Biotechnology (Shanghai) Co., Ltd. 293T
cells were used as the interim cell line to generate the packaged
lentivirus. The concentration of short hairpin (sh)NC (5′-TTC TCC
GAA CGT GTC ACG T-3′), shUBE2K#1 (5′-TGA CTC TCC GCA CGG TAT
TAT-3′), and shUBE2K#2 (5′-GCG AAT CAA GCG GGA GTT CAA-3′) was
1×108 TU/ml, 1.06×108 and 2.11×108
TU/ml, respectively. BxPC-3 and PANC-1 cells were infected by virus
with an MOI of 20 in a biological safety cabinet at room
temperature. Polybrene (6 µg/ml) was used to improve
transfect efficiency. After 72 h, stably transduced cells were
selected by treatment with blasticidin S (30 µg/ml) at 37°C
for 2 weeks. Scramble short interfering (si)RNA and siRNAs
targeting IGF2BP3 were purchased from Shanghai GenePharma Co., Ltd.
The sequences of the siRNAs were: siNC (sense: 5′-UUC UCC GAA CGU
GUC ACG UTT-3′, antisense: 5′-ACG UGA CAC GUU CGG AGA ATT-3′),
siIGF2BP3#1 (sense: 5′-GGC AAA GGA UUC GGA AAC UTT-3′, antisense:
5′-AGU UUC CGA AUC CUU UGC CTT-3′) and siIGF2BP3#2 (sense: 5′-GGC
UCA GGG AAG AAU UUA UTT-3′, antisense: 5′-AUA AAU UCU UCC CUG AGC
CTT-3′). The IGF2BP3 and PCDNA3.1 plasmids were maintained in the
Key Laboratory of Tumor Molecular Diagnosis and Individualized
Medicine of Zhejiang Province. Cells were transfected with 10
µl (20 µM) siRNA or 2 µg plasmid using the
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) based on the manufacturer's instructions in a
biological safety cabin at room temperature for 10-15 min.
Subsequent experiments were performed after the cells had been
incubated at 37°C for 48 h.
Reverse transcription-quantitative (RT-q)
PCR
Entire RNAs were extracted from cells (~90%
confluency)and tissues using an RNA quick purification kit (ES
science; cat. no. RN001) according to the manufacturer's
instructions. Nano-Drop One (Thermo Fisher Scientific, Inc.) was
used to confirm the concentration and quality of RNAs, for which
the ratio of absorption at 260-280 nm was between 1.9-2.2.
Following this, complementary DNA (cDNA) was synthesized using
s1000 (Bio-Rad Laboratories, Inc.) with a prime script RT reagent
kit (Takara Bio, Inc.; cat. no. RR037A-1) according to the
manufacturer's protocol. RT-qPCR was conducted using SYBR Green
(Shanghai Yeasen Biotechnology Co., Ltd.; cat. no. 11184ES08)
according to the manufacturer's protocol by 7500 real-time PCR
system under the following conditions: Holding stage at 95°C for 2
min, cycling stage 40 cycles at 95°C for 10 sec, and 60°C for 30
sec, the melt curve stage at 95°C for 15 sec, 60°C for 30 sec and
95°C for 5 sec. Each experiment was repeated three times. For the
calculation of relative RNA expression, the IGF2BP3 and UBE2K
expression levels were normalized to GAPDH expression using
2−ΔΔCq method (24).
The sequences of primer used were as follows: UBE2K forward 5′-CGC
GGT GCA GCG AAT C-3′ and reverse 5′-TTG CTC GTC TCC TCG CTC TT-3′.
IGF2BP3 forward 5′-CCA TGT GAT TTG CCT CTG CG-3′ and reverse 5′-ACT
GGG TCT GTT TGG TGA TGT-3′ and GAPDH forward 5′-GTC TCC TCT GAC TTC
AAC AGC G-3′ and reverse 5′-ACC ACC CTG TTG CTG TAG CCA A-3′.
Western blotting
Entire protein content of cells and tissues was
extracted using lysis buffer (Beyotime Institute of Biotechnology;
cat. no. P0013) plus phenylmethylsulfonyl fluoride solution
(Beyotime Institute of Biotechnology; cat. no. ST507-10 ml) and
protease inhibitor cocktail (bimake; cat. no. B14002) which were
protease inhibitors. The protein concentration was determined using
the Pierce BCA protein assay kit (Thermo Fisher Scientific, Inc.;
cat. no. 23225). After boiling for 10 min at 95°C, equal quantities
of protein (20-40 µg) were separated by 8-10% SDS-PAGE and
transferred to PVDF membranes (MilliporeSigma; cat. no. IPVH00010).
Then the 0.45 µm membrane was blocked in 5% skimmed milk for
1 h at room temperature. After washing, the membrane was cut into
pieces using a page-ruler pre-stained protein ladder (Thermo Fisher
Scientific, Inc.; cat. no. 26617) and based on the molecular weight
of the proteins. The pieces of the membrane were treated overnight
with primary antibodies, including anti-UBE2K (Proteintech Group,
Inc.; cat. no. 11834-3-AP, 1:2,000. CST Biological Reagents Co.,
Ltd.; cat. no. 8226; 1:1,000), anti-IGF2BP3 (Proteintech Group,
Inc.; cat. no. 14642-1-AP; 1:3,000), anti-NANOG (CST Biological
Reagents Co., Ltd.; cat. no. 4903; 1:2,000), anti-LIN28B (CST
Biological Reagents Co., Ltd.; cat. no. 4196; 1:1,000), GAPDH
(Affinity Biosciences; cat. no. AF7021; 1:6,000), and anti-β-ACTIN
(Affinity Biosciences; cat. no. AF7018; 1:5,000) at 4°C. Then, the
pieces were washed three times with TBST (containing 0.1%
Tween-20), and treated with the goat anti-rabbit IgG secondary
antibody (Thermo Fisher Scientific, Inc.; cat. no. 31460, 1:20,000)
for 1 h at room temperature. This was followed by repeated washing
with TBST three times, and the pieces were incubated in Clarity™
Western ECL Substrate (Bio-Rad Laboratories, Inc.) away from light
several minutes for further visualization. Finally, the protein
content was analyzed by ImageJ software (version 1.52v; National
Institutes of Health).
Cell Counting Kit-8 assay
To measure the cell proliferation ability,
5×103 cells were seeded in a 96-well plate. After 24,
48, and 72 h, 100 µl complete medium and 10 µl of the
CCK-8 assay reagent (Shanghai Yeasen Biotechnology Co., Ltd.; cat.
no. 40203ES80) were added to each well. The cells were incubated
for >1 h, and the absorbance at 450 nm was measured.
Colony formation assay
To detect the colony-forming ability of the cells,
750 cells were seeded in 6-well plates. Then, the plates were
maintained in complete medium for 12 days. When the colonies were
formed, the cells were fixed with polyformaldehyde for 30 min at
room temperature and then stained with 0.5% of crystal violet for 1
h at room temperature. Finally, colonies (>50 cells) visible on
the surface were manually counted.
Sphere formation assay
To measure the cell stemness, 5×104 cells
were seeded in ultralow attachment six-well plates. The cells were
cultured in DME/F-12 (1:1) medium (Biological Industries)
containing epidermal growth factor (10 ng/ml), basic fibroblast
growth factor (10 ng/ml) and N2. Following this, the spheres were
cultured for 14 days and then imaged by inverted fluorescence
microscope (Nikon) for further analysis.
RNA stability assay
BxPC-3 cells were transfected with siRNAs (siNC,
siIGF2BP3#1 and siIGF2BP3#2) in 6-well plates using
Lipofectamine® 3000 transfection reagent (Thermo Fisher
Scientific, Inc.) as above. After 48 h, actinomycin D was used to
treat the cells for 0, 6, 12, and 18 h at 37°C. Total RNA was
isolated and subjected to RT-qPCR to analyze gene expression as
above. GAPDH was used as an endogenous control.
Subcutaneous xenografts in nude mice
A total of 23 4-week old female BALB/C athymic nude
mice (17-21 g) were acquired from the Shanghai SLAC laboratory
Animal Co., Ltd. The mice were maintained in SPF conditions which
included 22°C room temperature, 55% humidity and 12-h light/dark
cycle and had free access to food and water. UBE2K
stable-expression BxPC-3 cells (UBE2K) and the corresponding
control cells (NC) were harvested and resuspended in serum-free
1640 medium (4×106 cells/150 µl). UBE2K
stable-knockdown BXPC3 cells (shUBE2K#1, shUBE2K#2) and their
corresponding control cells (shNC) were also harvested and
resuspended in serum-free 1640 medium (4×106 cells/150
µl). Then, 150 µl of the cell suspension was
subcutaneously injected into the right flank of each nude mouse.
After injection, the tumor was measured every 5 days and the tumor
volume was calculated as follows: tumor volume
(mm3)=(length x width2)/2. At the appropriate
time, the mice were sacrificed by CO2 euthanasia with
60% per minute of container volume replacement. Then the tumors
were harvested for western blotting and RT-qPCR analysis. The above
experiments on nude mice were approved by the animal ethics
committee at Zhejiang Provincial People's Hospital (approval no.
A20220046).
Statistical analysis
Each experiment was repeated at least three times,
and GraphPad Prism was used for statistical analysis. Continuous
variables were presented in term of mean and standard error of mean
(SEM). Differences between two groups were assessed using
two-tailed t tests. Differences between multiple groups were
assessed using one way ANOVA followed by Dunnett's test. P<0.05
was considered to indicate a statistically significant
difference.
Results
UBE2K is expressed at high levels and
indicates poor prognosis in PDAC
UBE2K was upregulated in PDAC compared with that in
corresponding normal tissues, as analyzed from the data obtained
from the GEPIA database (Fig. 1A).
To comprehensively explore the expression of UBE2K in PDAC, the RNA
and protein expression levels of UBE2K in PDAC cell lines (AsPC-1,
BxPC-3, PANC-1, and SW1990) and human pancreatic ductal HPNE cells
were detected. As shown in Fig. 1B and
C, the RNA and protein levels of UBE2K were higher in PDAC cell
lines compared with that in HPNE cells. Subsequently, the clinical
prognosis of UBE2K expression in PDAC was analyzed using GEPIA and
the Kaplan-Meier plotter database. Higher expression of UBE2K was
associated with a shorter overall survival of patients with PDAC
(Fig. 1D and E). Additionally, the
Kaplan-Meier plotter database also showed that higher UBE2K
expression was associated with a poorer RFS in PDAC (Fig. 1F). Overall, UBE2K may be a
potentially unfavorable biomarker for PDAC diagnosis.
 | Figure 1UBE2K is upregulated in tissues and
cell lines of PDAC and predicts a poor prognosis. (A) UBE2K was
highly expressed in PDAC tissues (left, red) compared with normal
tissues (right, black) in GEPIA. (B) Reverse
transcription-quantitative PCR and (C) western blotting results
indicated that UBE2K was highly expressed in PDAC cell lines
(AsPC-1, BxPC-3, PANC-1 and SW1990) compared with HPNE in RNA and
protein levels. The databases of (D) GEPIA and (E) Kaplan-Meier
plotter indicated higher UBE2K expression correlated to lower
overall survival in PDAC patients. (F) Kaplan-Meier plotter
survival analysis indicated that patients with higher expression of
UBE2K exhibited lower relapse free survival rate.
*P<0.05, **P<0.01,
***P<0.001. UBE2K, ubiquitin-conjugating enzyme E2K;
PDAC, pancreatic ductal adenocarcinoma; GEPIA, gene expression
profiling interactive analysis; HPNE, normal human pancreatic
cells; HR, hazard ratio. |
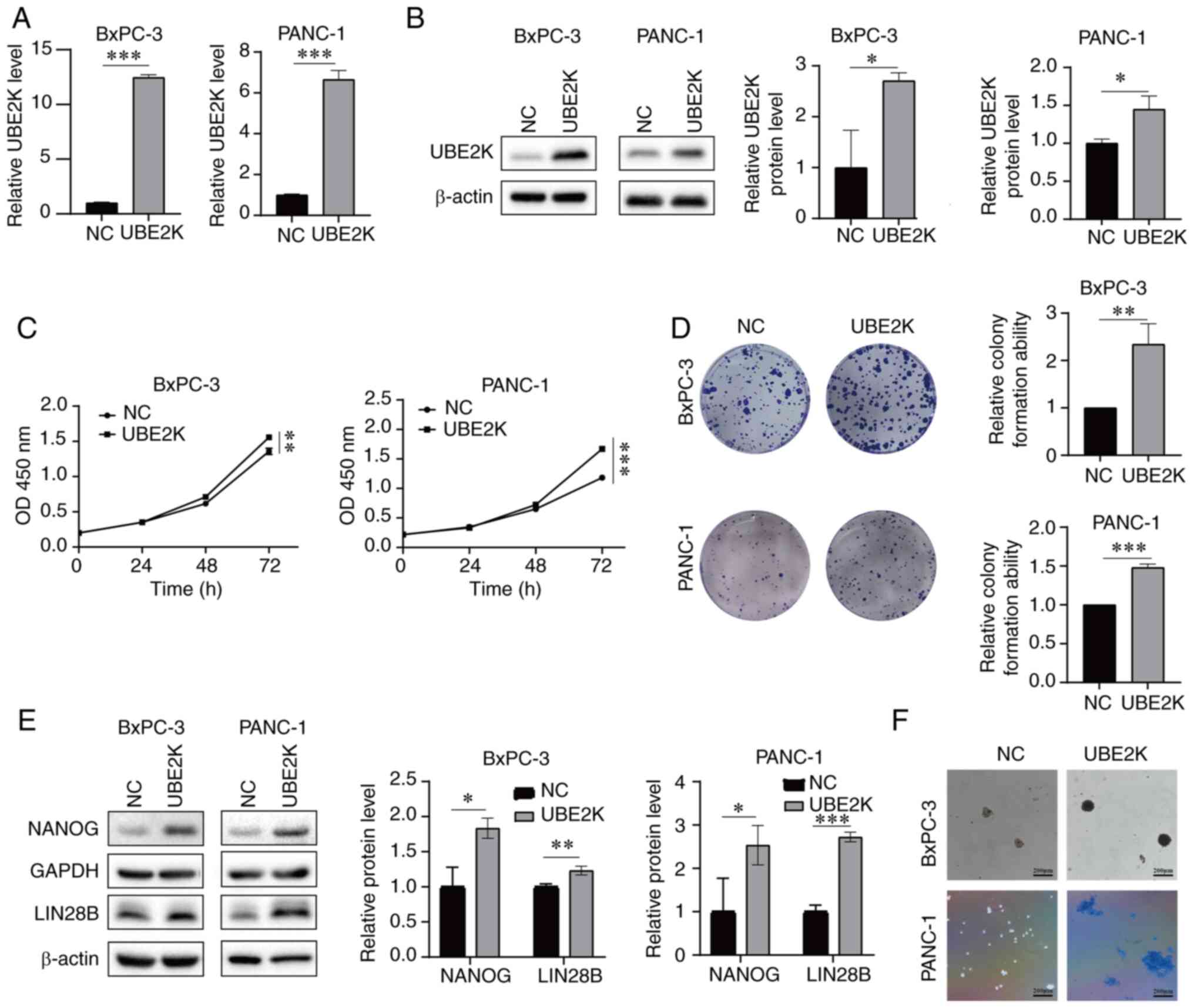
UBE2K overexpression promotes cell
proliferation and PDAC stemness
To explore the function of UBE2K in PDAC, stably
UBE2K-overexpressing BxPC-3 and PANC-1 cells were generated using
lentiviruses. RT-qPCR and western blotting were performed to verify
the UBE2K overexpression rate in both RNA and protein levels
(Fig. 2A and B). The CCK-8 and
colony formation assays showed that UBE2K overexpression promoted
PDAC cell growth in vitro (Fig.
2C and D). A significant upregulation of stemness-related
protein expression was observed in UBE2K-overexpressing cells
(Fig. 2E). In addition, UBE2K
enhanced the sphere formation ability of PDAC cells, since the
sphere was larger after UBE2K overexpression (Fig. 2F). Collectively, these data
indicated that UBE2K overexpression promoted cell proliferation and
stemness of PDAC in vitro.
UBE2K knockdown suppresses cell
proliferation and stemness in PDAC
To further confirm the role of UBE2K in PDAC, UBE2K
knockdown was induced in BxPC-3 and PANC-1cells using lentiviral
vector. The RT-qPCR and western blotting results indicated that
UBE2K was stably knocked down in PDAC cells (Fig. 3A and B). The knockdown of UBE2K
restrained the proliferation of PDAC cells, as revealed in the
CCK-8 and colony formation assays (Fig. 3C and D). In contrast to the results
of UBE2K overexpression, the knockdown of UBE2K suppressed the
protein expression of stemness-related genes, including NANOG and
LIN28B in BxPC-3 and PANC-1 cells (Fig. 3E). Consistently, UBE2K inhibition
also reduced the sphere formation ability (Fig. 3F), as the spheres reduced in size
upon UBE2K knockdown. Collectively, these data suggested that the
knockdown of UBE2K could reduce cell proliferation and stemness in
PDAC.
UBE2K promotes PDAC growth in vivo
To investigate the oncogenic function of UBE2K in
PDAC growth in vivo, a subcutaneous xenograft nude mouse
model was established using BxPC-3 cells. The tumor was larger in
the UBE2K overexpression group than in the control group (Fig. 4A). During the experiment, we
measured the length and width every 5 days. The results revealed
that the volume was larger in the UBE2K overexpression group
(Fig. 4B). The tumor weight in the
UBE2K overexpression group was also greater than that in the
control group (Fig. 4C). Finally,
the RNA and protein levels of UBE2K in the tumor tissues of nude
mice were measured. The RNA and protein expression levels of UBE2K
were higher in the UBE2K overexpression group (Fig. 4D and E).
To further verify the roles of UBE2K in vivo,
shNC and shUBE2K cells were then injected into the nude mice. It
was found that tumors from the shUBE2K groups were smaller than
their control group counterparts (Fig.
4F). As expected, the volume and weight of tumors in the
shUBE2K groups were also less than their control group counterparts
(Fig. 4G and H). Additionally, the
RNA and protein expression levels were also lower in the shUBE2K
groups (Fig. 4I and J). To sum up,
the results suggested that UBE2K is essential for PDAC tumor growth
in vivo.
UBE2K is regulated by IGF2BP3 in
PDAC
The above results indicated the prognostic and
oncogenic roles of UBE2K in PDAC. However, the molecular regulation
was unclear. As previously reported, IGF2BP3 is an RBP, which
broadly participates in cancer progression by regulating its
downstream targets. For example, IGF2BP3 stabilizes the long
noncoding RNA CDKN2B-AS1 to promote proliferation, migration and
invasion in renal clear cell carcinoma (25). IGF2BP3 cooperates with RMB15 to
stabilize the expression of TMBOM6 in laryngeal cancer (26). IGF2BP3 has been shown to serve as a
potential unfavorable factor for PDAC (27). As revealed by the analysis of the
GEPIA database, IGF2BP3 expression was positively associated with
UBE2K expression and statistically significant in PDAC (Fig. 5A). Thus, it was hypothesized that
IGF2BP3 might regulate UBE2K expression in PDAC. To elucidate the
regulation between IGF2BP3 and UBE2K, IGF2BP3 plasmids were
transfected into BxPC-3 cells and the overexpression efficiency of
IGF2BP3 measured by RT-qPCR and western blotting. As shown in
Fig. 5B and C, the overexpression
of IGF2BP3 led to the upregulation of UBE2K at the RNA and protein
levels. By contrast, the siRNA-mediated knockdown of IGF2BP3
decreased the RNA and protein expression of UBE2K (Fig. 5D and E). To determine if IGF2BP3
affects the RNA stability of UBE2K, IGF2BP3 siRNA was transfected
in BxPC-3 cells and the cells treated with actinomycin D. The
results demonstrated that IGF2BP3 knockdown notably decreased the
RNA stability of UBE2K (Fig. 5F).
Therefore, IGF2BP3 stabilized UBE2K to increase its expression.
IGF2BP3/UBE2K forms a functional axis in
regulating the cell growth of PDAC
To explore whether IGF2BP3 contributes to the
functional phenotype of the cell induced by UBE2K, IGF2BP3 was
knockeddown by siRNA followed by overexpression of UBE2K. IGF2BP3
inhibition significantly decreased the cell proliferation induced
by UBE2K overexpression, as revealed in the CCK-8 assay (Fig. 6A-C). Results of the colony
formation assay (Fig. 6D) further
confirmed that the knockdown of IGF2BP3 attenuated UBE2K induced
growth in BxPC-3 and PANC-1 cells. In addition, IGF2BP3 was also
re-expressed into the UBE2K knockdown cells (Fig. 6E and F). CCK-8 and colony formation
assays showed that IGF2BP3 overexpression could successfully
reverse the cell growth suppression caused by UBE2K knockdown
(Fig. 6G and H). In conclusion,
IGF2BP3/UBE2K formed a functional axis in regulating cell growth in
PDAC.
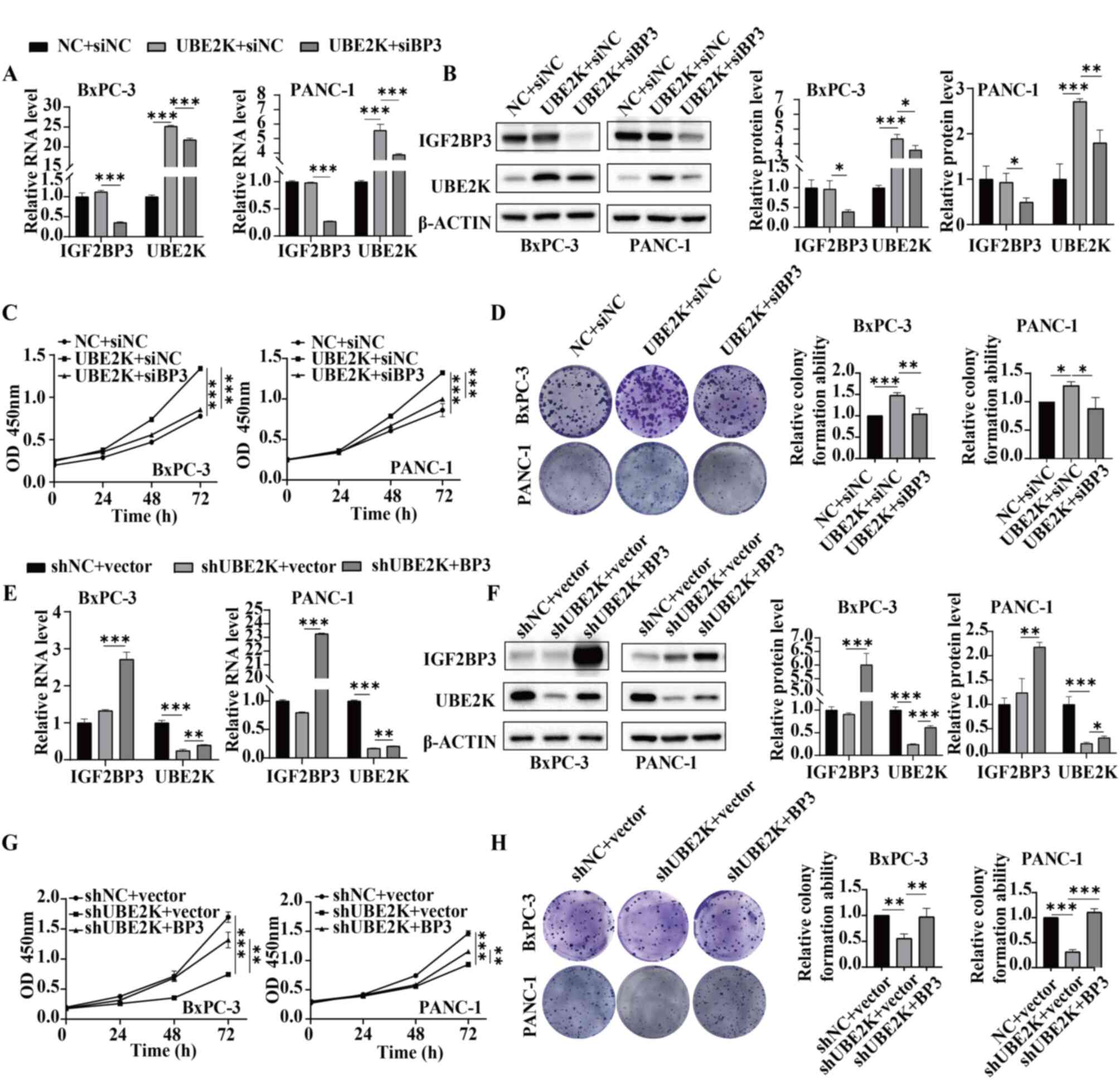 | Figure 6The effect of UBE2K can be revered by
IGF2BP3 in PDAC. (A) Reverse transcription-quantitative PCR and (B)
western blotting results in the groups of NC + siNC, UBE2K + siNC
and UBE2K + siIGF2BP3. The (C) CCK-8 and (D) colony formation
assays results in the groups of NC + siNC, UBE2K + siNC and UBE2K +
siIGF2BP3. (E) Reverse transcription-quantitative PCR and (F)
western blotting results in the groups of shNC + vector, shUBE2K +
vector and shUBE2K + IGF2BP3. The (G) CCK-8 and (H) colony
formation assays results in the groups of shNC + vector, shUBE2K +
vector and shUBE2K + IGF2BP3. *P<0.05,
**P<0.01, ***P<0.001. UBE2K,
ubiquitin-conjugating enzyme E2K; PDAC, pancreatic ductal
adenocarcinoma; NC, negative control; si, short interfering; sh,
short hairpin. |
Discussion
PDAC is an extremely malignant tumor with a poor
5-year overall survival (2). It
has a debilitating effect on the quality of life of afflicted
individuals. Thus far, clear early-stage symptoms of PDAC have yet
to be determined (28). Thus, it
is critical to have thorough awareness of the molecular mechanisms
underlying PDAC pathogenesis and then identify potential
therapeutic targets for PDAC treatment.
Ubiquitin is highly conserved in eukaryotic cells
and is a modified molecule composed of 76 amino acids through a
cascade reaction of ubiquitin-activating enzyme (E1),
ubiquitin-conjugating enzyme (E2) and ubiquitin-protein ligase (E3)
that covalently binds and marks its target substrates (29). Subsequently, the
ubiquitination-modified substrate protein is degraded by the 26S
proteasome (29). UBE2K is a
ubiquitin-coupled E2 enzyme. It serves essential roles in the
progression of human neurodegenerative diseases (11-14).
In PD, UBE2K is expressed at high levels in the patient's brain and
is found to regulate the JNK signaling pathway in Aβ neurotoxicity
(11). Meanwhile, the deficiency
of UBE2K expression contributes to motor function impairment, which
shows that UBE2K can serve as a promising biomarker for PD
diagnosis (13). In types of human
cancer, UBE2K is reported to be upregulated in gastric cancer (GC)
and a high expression of UBE2K is associated with poor prognosis in
patients with GC (16). It is also
reported that UBE2K, acting as an oncogene, is downregulated by
ixazomib and further demonstrated to be involved in
ixazomib-induced cell apoptosis, cell cycle arrest, reactive oxygen
species production and inhibition of myeloma cell proliferation
(15).
The current study explored the function and
molecular mechanism of UBE2K in the malignant progression of PDAC.
First, it revealed that UBE2K was expressed at high levels in PDAC
tissues and cells compared with normal tissues and normal human
pancreatic ductal cells. High UBE2K expression predicted the poor
prognosis of patients with PDAC. Second, the overexpression of
UBE2K promoted the growth and stemness of PDAC cells, whereas the
loss of UBE2K expression suppressed the growth and stemness of PDAC
cells in vitro, which was also confirmed in vivo in
subcutaneous xenograft mouse models. Third, the RNA-binding protein
IGF2BP3 was found to be an upstream regulator of UBE2K that
enhanced its expression by stabilizing UBE2K RNA. The
overexpression or knockdown of IGF2BP3 led to the upregulation or
downregulation of UBE2K at both RNA and protein levels. In
addition, IGF2BP3 overexpression rescued cell growth inhibition
caused by UBE2K knockdown in BxPC-3 and PANC-1 cells in CCK8 and
colony plate experiments, whereas IGF2BP3 knockdown attenuated cell
growth promotion induced by UBE2K overexpression. This indicated
that IGF2BP3 and UBE2K may form the IGF2BP3/UBE2K axis in PDAC
cells. Collectively, the findings illustrated the oncogenic role of
UBE2K in PDAC in vitro and in vivo and clarified the
IGF2BP3-mediated upregulation of UBE2K by enhancing the RNA
stability of UBE2K. Further, the findings of the present study
demonstrated that IGF2BP3 and UBE2K form a functional axis
(IGF2BP3/UBE2K axis) that participates in the malignant progression
of PDAC. This indicates the potential roles of the IGF2BP3/UBE2K
axis in the prognosis of PDAC and as a therapeutic target in this
disease.
However, the present study had some limitations.
First, it did not analyze the association between UBE2K expression
and tumor staging in patients with PDAC. Second, UBE2K is a
ubiquitin ligase and its potential downstream targets regulated by
ubiquitination need to be explored further.
In conclusion, the present study reported the
function of the IGF2BP3/UBE2K axis in regulating PDAC cell growth
and stemness for the first time to the best of the authors'
knowledge. The findings elucidated a novel molecular mechanism
underlying UBE2K function in PDAC progression. This can help
improve the theoretical basis of PDAC carcinogenesis. In addition,
the IGF2BP3/UBE2K axis might be a promising therapeutic target for
PDAC.
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XH and XT designed the study. WF conducted RT-qPCR,
western blotting, CCK8, colony assay and wrote the original draft
of the manuscript. XL, QL and JZ performed the experiments on nude
mice. JG and JZ helped in the sphere formation assays. XH and XT
confirm the authenticity of all the raw data and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal experiments were approved (approval no.
A20220046) by the Animal Ethics Committee of Zhejiang Provincial
People's Hospital.
Patient consent for publication
Not applicable.
Completing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Research Foundation of
Education of Zhejiang Province (grant no. Y202249315), National
Natural Science Foundation of China (grant no. 82103295), Natural
Science Foundation of Zhejiang Province (grant no. LQ22H160062),
and Medical and Health Science Technology Project of Zhejiang
Province (grant nos. 2019RC105 and 2022KY516).
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nevala-Plagemann C, Hidalgo M and
Garrido-Laguna I: From state-of-the-art treatments to novel
therapies for advanced-stage pancreatic cancer. Nat Rev Clin Oncol.
17:108–123. 2020. View Article : Google Scholar
|
|
4
|
Grice GL and Nathan JA: The recognition of
ubiquitinated proteins by the proteasome. Cell Mol Life Sci.
73:3497–3506. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maxwell BA, Gwon Y, Mishra A, Peng J,
Nakamura H, Zhang K, Kim HJ and Taylor JP: Ubiquitination is
essential for recovery of cellular activities after heat shock.
Science. 372:eabc35932021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Salas-Lloret D and Gonzalez-Prieto R:
Insights in post-translational modifications: Ubiquitin and SUMO.
Int J Mol Sci. 23:32812022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Swatek KN and Komander D: Ubiquitin
modifications. Cell Res. 26:399–422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Middleton AJ, Teyra J, Zhu J, Sidhu SS and
Day CL: Identification of ubiquitin variants that inhibit the E2
ubiquitin conjugating enzyme, Ube2k. ACS Chem Biol. 16:1745–1756.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Middleton AJ and Day CL: The molecular
basis of lysine 48 ubiquitin chain synthesis by Ube2K. Sci Rep.
5:167932015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cook BW, Lacoursiere RE and Shaw GS:
Recruitment of ubiquitin within an E2 chain elongation complex.
Biophys J. 118:1679–1689. 2020.
|
|
11
|
Song S, Kim SY, Hong YM, Jo DG, Lee JY,
Shim SM, Chung CW, Seo SJ, Yoo YJ, Koh JY, et al: Essential role of
E2-25K/Hip-2 in mediating amyloid-beta neurotoxicity. Mol Cell.
12:553–563. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Pril R, Fischer DF, Roos RA and van
Leeuwen FW: Ubiquitin-conjugating enzyme E2-25K increases aggregate
formation and cell death in polyglutamine diseases. Mol Cell
Neurosci. 34:10–19. 2007. View Article : Google Scholar
|
|
13
|
Su J, Huang P, Qin M, Lu Q, Sang X, Cai Y,
Wang Y, Liu F, Wu R, Wang X, et al: Reduction of HIP2 expression
causes motor function impairment and increased vulnerability to
dopaminergic degeneration in Parkinson's disease models. Cell Death
Dis. 9:10202018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tak YJ and Kang S: The E2
ubiquitin-conjugating enzyme HIP2 is a crucial regulator of quality
control against mutant SOD1 proteotoxicity. Biochim Biophys Acta
Mol Basis Dis. 1868:1663162022. View Article : Google Scholar
|
|
15
|
Wang Q, Dong Z, Su J, Huang J, Xiao P,
Tian L, Chen Y, Ma L and Chen X: Ixazomib inhibits myeloma cell
proliferation by targeting UBE2K. Biochem Biophys Res Commun.
549:1–7. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu J, Tian B, Yang J, Huo H, Song Z, Yu J
and Gu Y: Reduction of Hip2 suppresses gastric cancer cell
proliferation, migration, invasion and tumorigenesis. Transl Cancer
Res. 9:774–785. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Coppin L, Leclerc J, Vincent A, Porchet N
and Pigny P: Messenger RNA life-cycle in cancer cells: Emerging
role of conventional and non-conventional RNA-Binding proteins? Int
J Mol Sci. 19:6502018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mancarella C and Scotlandi K: IGF2BP3 from
physiology to cancer: Novel discoveries, unsolved issues, and
future perspectives. Front Cell Dev Biol. 7:3632020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang N, Shen Y, Li H, Chen Y, Zhang P,
Lou S and Deng J: The m6A reader IGF2BP3 promotes acute myeloid
leukemia progression by enhancing RCC2 stability. Exp Mol Med.
54:194–205. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Kong F, Ma J, Miao J, Su P, Yang
H, Li Q and Ma X: IGF2BP3 enhances the mRNA stability of E2F3 by
interacting with LINC00958 to promote endometrial carcinoma
progression. Cell Death Discov. 8:2792022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W,
Guo W, Wu X, Pu C, Hu X, et al: METTL3/IGF2BP3 axis inhibits tumor
immune surveillance by upregulating N (6)-methyladenosine
modification of PD-L1 mRNA in breast cancer. Mol Cancer. 21:602022.
View Article : Google Scholar
|
|
22
|
Han J and Hu X: IGF2BP3stabilized SIX4
promotes the proliferation, migration, invasion and tube formation
of ovarian cancer cells. Mol Med Rep. 26:2322022. View Article : Google Scholar
|
|
23
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45(W1):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xie X, Lin J, Fan X, Zhong Y, Chen Y, Liu
K, Ren Y, Chen X, Lai D, Li X, et al: LncRNA CDKN2B-AS1 stabilized
by IGF2BP3 drives the malignancy of renal clear cell carcinoma
through epigenetically activating NUF2 transcription. Cell Death
Dis. 12:2012021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Tian L, Li Y, Wang J, Yan B, Yang
L, Li Q, Zhao R, Liu M, Wang P and Sun Y: RBM15 facilitates
laryngeal squamous cell carcinoma progression by regulating TMBIM6
stability through IGF2BP3 dependent. J Exp Clin Cancer Res.
40:802021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang L, Zhang S, Li H and Xu Y, Wu Q, Shen
J, Li T and Xu Y: Quantification of m6A RNA methylation modulators
pattern was a potential biomarker for prognosis and associated with
tumor immune microenvironment of pancreatic adenocarcinoma. BMC
Cancer. 21:8762021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hershko A: Ubiquitin: Roles in protein
modification and breakdown. Cell. 34:11–12. 1983. View Article : Google Scholar : PubMed/NCBI
|