1. Introduction
Multiple myeloma (MM) is a type of tumor
characterized by the malignant proliferation of plasma cells.
Numerous plasma cells in the bone marrow proliferate and secrete
monoclonal immunoglobulin or its fragment (M protein), which may
lead to clinical symptoms such as anemia, renal insufficiency, bone
destruction and hypercalcemia. According to global statistical
reports, MM is the second most common cancer type of the
hematological system (1). With the
aging of the global population, the incidence of MM has increased
by 126% from 1990 to 2016, causing severe health issues (1). In recent years, progress in MM
treatment, such as proteasome inhibitors, immunomodulators,
immunotherapy and autologous stem cell transplantation, has
markedly improved patients' survival (2). However, MM is a disease that is
difficult to cure. Even if the disease is controlled by treatment,
MM relapse and drug resistance may eventually lead to death
(3).
Following the proposal of the 'Human Genome Project'
and the development of sequencing technology, 90% of the genome was
discovered to be transcribed into RNA, most of which could not be
translated into proteins and only 2% of which were mRNAs that
contribute to protein coding (4).
RNAs that do not encode proteins are called non-coding RNAs
(ncRNAs). ncRNAs were considered 'waste' during transcription, but
numerous studies have confirmed that they have important roles in
regulating gene expression and the progression of neoplastic
diseases (5). Long ncRNAs
(lncRNAs), with >200 nt in length, belong to the family of
ncRNAs whose function has not been fully defined.
LncRNAs are poorly conserved among species and have
high tissue, cell and spatiotemporal specificities (6), making them potential tumor
biomarkers. Evidence suggests that lncRNAs may either promote or
suppress the progression of human neoplastic diseases (7). Although lncRNAs rarely encode
proteins, they may act on nearby molecules or targets. LncRNAs in
the cytoplasm bind to ribosomes, degrade and regulate mRNA, mediate
regulation of RNA terminal specific structural sequences and act as
bait for microRNA (miRNA/miR) and prevent miRNA from degrading
targeted mRNA (8-10). LncRNAs regulate mRNA stability and
may also act as scaffoldings to regulate protein interactions
(8-10). LncRNAs in the nucleus may bind to
chromatin regulatory factors, transcription factors and chromatin
to affect transcriptional activation, transcriptional inhibition
and post-transcriptional regulation through cis- or trans-actions
and regulate gene expression at the pre-transcriptional,
transcriptional and post-transcriptional levels (10-12).
The present review summarizes the latest findings of
lncRNAs in the pathogenesis of MM and the complex regulatory
network of lncRNAs and discusses the roles of lncRNAs in diagnostic
and treatment strategies for MM, laying a foundation to further the
understanding of the pathogenesis of MM, the development of highly
specific diagnostic and prognostic tools and effective treatment
strategies.
2. LncRNAs may act as oncogenes to promote
MM tumor progression
Most lncRNAs are upregulated in MM, promoting
proliferation, DNA protection, adhesion, migration and invasion of
MM tumor cells through various mechanisms by inhibiting apoptosis
and remodeling the tumor microenvironment (TME) to facilitate the
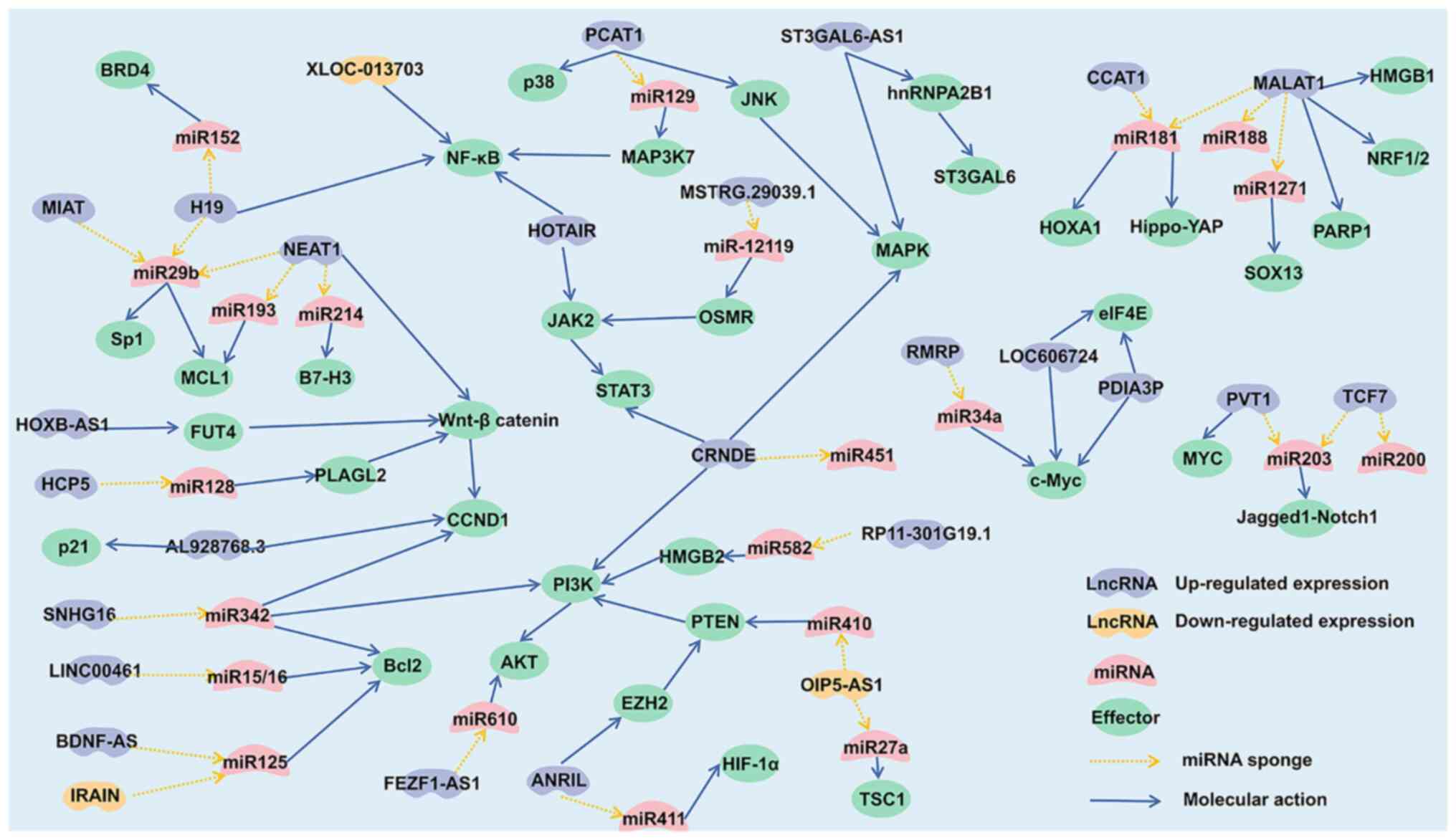
growth of tumor cells (Table I).
LncRNAs with cancer-promoting effects in MM were screened and a
gene regulatory network was constructed (Fig. 1), which helps understand the
pathogenesis of MM.
 | Table IMolecular mechanisms of multiple
myeloma-associated lncRNAs. |
Table I
Molecular mechanisms of multiple
myeloma-associated lncRNAs.
| LncRNA | Author, year | Direction of
differential expression | ceRNA target | Downstream
regulatory targets | Function | (Refs.) |
|---|
| MALAT1 | Sun, 2019 | Up-regulated | miR-181a-5p | Hippo/YAP | Proliferation,
adherence | (15) |
| Liu, 2021 | Up-regulated | miR-188-5p | n.a. | Proliferation, DNA
replication | (16) |
| Hu, 2018 | Up-regulated | n.a. | PARP1, LIG3 | Apoptosis, DNA
repair, drug resistance | (52) |
| Gao, 2017 | Up-regulated | n.a. | HMGB1/Beclin-1,
HMGB1/LC3B | Apoptosis | (56) |
| Liu, 2020 | Up-regulated | miR-1271-5p | SOX13 | Invasion,
glycolysis | (60) |
| Amodio, 2018 | Up-regulated | n.a. | NRF1/2 | Apoptosis | (126) |
| H19 | Zheng, 2020 | Up-regulated | miR-152-3p | BRD4 | Proliferation, cell
cycle | (18) |
| Sun, 2017 | Up-regulated | n.a. | NF-κB, IL-8 | Proliferation,
colony formation | (38) |
| Pan, 2019 | Up-regulated | miR-29b-3p | MCL1 | Drug resistance,
apoptosis | (117) |
| PCAT1 | Shen, 2020 | Up-regulated | miR-129 | MAP3K7/NF-κB | Proliferation | (20) |
| Shen, 2019 | Up-regulated | n.a. | p38, JNK/MAPK | Proliferation, drug
resistance | (39) |
| PVT1 | Yang, 2018 | Up-regulated | miR-203a | n.a. | Proliferation | (22) |
| Handa, 2020 | Up-regulated | n.a. | MYC | Proliferation | (40) |
| ANRIL | Wang, 2020 | Up-regulated | miR-411-3p | HIF-1α | Proliferation, stem
cell-like properties of tumors | (23) |
| Yang, 2021 | Up-regulated | n.a. | EZH2/PTEN/AKT | Drug
resistance | (100) |
| CRNDE | Meng, 2017 | Up-regulated | miR-451 | n.a. | Proliferation | (24) |
| David, 2021 | Up-regulated | n.a. | IL-6/STAT, RAS,
MAPK, PI3K/AKT | Drug
resistance | (121) |
| HCP5 | Liu, 2021 | Up-regulated | miR-128-3p |
PLAGL2/Wnt/β-catenin/CCND1 | Proliferation | (25) |
| TCF7 | Liu, 2021 | Up-regulated | miR-203 | Jagged1/Notch1 | Proliferation | (26) |
| Ding, 2021 | Up-regulated | miR-200c | n.a. | Proliferation | (27) |
| RMRP | Xiao, 2019 | Up-regulated | miR-34a-5p | c-Myc | Proliferation | (28) |
| MSTRG.29039.1 | Liu, 2021 | Up-regulated | miR-12119 | OSMR/JAK2/
STAT3 | Proliferation | (29) |
| RP11-301G19.1 | Wang, 2022 | Up-regulated | miR-582-5p | HMGB2/PI3K/
AKT | Proliferation, cell
cycle | (30) |
| LINC01234 | Chen, 2019 | Up-regulated | miR-124-3p | GRB2 | Proliferation, cell
cycle | (31) |
| LINC00461 | Deng, 2019 | Up-regulated | miR-15/16 | Bcl2 | Proliferation | (32) |
| BDNF-AS | Chu, 2022 | Up-regulated | miR-125-5p | Bcl2 | Proliferation | (33) |
| UCA1 | Yang, 2019 | Up-regulated | miR-1271-5p | HGF | Proliferation | (34) |
| FEZF1-AS1 | Li, 2018 | Up-regulated | miR-610 | AKT3 | Proliferation, cell
cycle | (35) |
| CCAT1 | Chen, 2018 | Up-regulated | miR-181a-5p | HOXA1 | Proliferation, cell
cycle | (36) |
| MIAT | Fu, 2019 | Up-regulated | miR-29b | MCL1, Sp1 | Proliferation, drug
resistance | (37) |
| ST3GAL6-AS1 | Ronchetti,
2020 | Up-regulated | n.a. | MAPK,
ubiquitination protein | Proliferation | (41) |
| Shen, 2021 | Up-regulated | n.a. | hnRNPA2B1/
ST3GAL6 | Adherence,
migration, invasion | (63) |
| NEAT1 | Geng, 2018 | Up-regulated | n.a. | Wnt/β-catenin | Proliferation,
migration, invasion | (42) |
| Taiana, 2020 | Up-regulated | n.a. | RAD51B, CHK1, CHK2,
RPA32, BRCA1 | Apoptosis, DNA
repair | (55) |
| Gao, 2020 | Up-regulated | miR-214 | B7-H3 | Remodeling the
TME | (66) |
| Che, 2021 | Up-regulated | miR-29b-3p | Sp1 | Drug
resistance | (118) |
| Wu, 2018 | Up-regulated | miR-193a | MCL1 | Drug
resistance | (120) |
| HOTAIR | Zhu, 2019 | Up-regulated | n.a. | NF-κB | Proliferation | (43) |
| Guan, 2019 | Up-regulated | n.a. | JAK2/STAT3 | Drug
resistance | (107) |
| LUCAT1 | Liu, 2020 | Up-regulated | n.a. | TGF-β | Proliferation, cell
cycle | (44) |
| AL928768.3 | Shen, 2022 | Up-regulated | n.a. | CDK2, CCND1,
p21 | Proliferation, cell
cycle | (45) |
| HOXB-AS1 | Chen, 2020 | Up-regulated | n.a. |
FUT4-Wnt-β-catenin | Proliferation | (46) |
| LBX2-AS1 | Jia, 2021 | Up-regulated | n.a. | LBX2 | Proliferation | (47) |
| SNHG16 | Yang, 2020 | Up-regulated | miR-342-3p | Caspase3, caspase9,
Foxa3a, Bax, Bcl2, CCND1, PI3K-AKT | Apoptosis, cell
cycle | (57) |
| SOX2OT | Yu, 2020 | Up-regulated | miR-144-3p | c-MET | Tumor
progression | (61) |
| LINC01606 | He, 2021 | Up-regulated | miR-579-3p | n.a. | Migration,
invasion | (62) |
| LOC606724 | Wang, 2022 | Up-regulated | n.a. | eIF4E, c-Myc | Remodeling the
TME | (67) |
| PDIA3P | Yang, 2018 | Up-regulated | n.a. | c-Myc, G6PD | Drug
resistance | (116) |
| LINC01003 | Wu, 2021 | Down-regulated | miR-33a-5p | PIM1 | Proliferation,
adherence | (68) |
| OIP5-AS1 | Yang, 2017 | Down-regulated | miR-410 | PTEN/PI3K/
AKT/KLF10 | Cell cycle | (69) |
| Wang, 2020 | Down-regulated | miR-27a-3p | TSC1 | Apoptosis,
migration, invasion | (70) |
| DANCR | Wu, 2021 | Down-regulated | miR-135b-5p | KLF9 | Migration,
invasion | (71) |
| IRAIN | Jiang, 2019 | Down-regulated | miR-125b | n.a. | Proliferation | (72) |
| XLOC-013703 | Pu, 2019 | Down-regulated | n.a. | IL-6/NF-κB | Apoptosis, cell
cycle | (73) |
| BM742401 | Li, 2020 | Down-regulated | n.a. | n.a. | Homing,
migration | (74) |
| PRAL | Xiao, 2018 | Down-regulated | miR-210 | BMP2 | Drug
resistance | (79) |
LncRNAs promote the proliferation of MM
tumor cells
MM is characterized by the malignant clonal
proliferation of plasma cells (13) and most oncogenic lncRNAs promote MM
cell proliferation.
LncRNAs bind to miRNAs and act as competing
endogenous RNAs (ceRNAs) to regulate miRNA expression, an important
molecular mechanism of the lncRNA regulatory network.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is
the most widely studied lncRNA, which is upregulated in breast
cancer, cervical cancer, colorectal cancer, lung cancer and other
cancer types, promoting tumorigenesis (14). Silencing MALAT1 significantly
inhibited MM cell proliferation (15,16).
Sun et al (15)
demonstrated that the MALAT1/miR-181a-5p/Hippo-Yes-associated
protein (Hippo-YAP) pathway and silencing MALAT1 increased the
expression of the targeted miR-181a-5p and activated the Hippo-YAP
signaling pathway, inhibiting cell proliferation. Liu et al
(16) reported another
MALAT1/miR-188-5p pathway, in which MALAT1 negatively regulates
miR-188-5p expression, promoting DNA replication and the transition
from the G1 to S phase of the cell cycle. H19 imprinted maternally
expressed transcript (H19) is also one of the 'hot genes' regulated
by lncRNAs, which is abnormally highly expressed in oral squamous
cell carcinoma, hepatocellular carcinoma, breast cancer, bladder
cancer and other malignant cancers (17). Zheng et al (18) demonstrated that H19 silencing in
tumor cells led to bromodomain containing 4 (BRD4)-mediated
upregulation of proliferation-related signals, resulting in the
inhibition of cell proliferation and cell cycle arrest at G1 phase
and confirming the H19/miR-152-3p/BRD4 pathway. Prostate
cancer-associated transcript 1 (PCAT1) promotes cell proliferation
in various tumor types, such as bladder cancer, esophageal squamous
cell carcinoma and lung cancer (19). Overexpression of PCAT1 by plasmid
vector in MM decreased miR-129 levels and upregulated
mitogen-activated protein kinase kinase kinase 7 (MAP3K7),
activating the nuclear factor κB (NF-κB) pathway and leading to
PCAT1/miR-129/MAP3K7/NF-κB signaling (20). Similarly, plasmacytoma variant
translocation 1(PVT1) is a widely studied lncRNA (21). PVT1 is highly expressed in myeloma
cells; the level of miR-203a is reduced through the targeted action
of ceRNA and the PVT1/miR-203 pathway promotes cell proliferation
(22). Antisense noncoding RNA in
the INK4 locus (ANRIL) downregulates miR-411-3p and upregulates
hypoxia-inducible factor 1α (HIF-1α), forming the
ANRIL/miR-411-3p/HIF-1α pathway to promote the malignant
proliferation of tumor cells and tumor stem cell-like
characteristics (23). Colorectal
neoplasia differentially expressed (CRNDE) negatively targets
miR-451, forming a CRNDE/miR-451 pathway that promotes cell
proliferation (24). Human
leukocyte antigen complex P5 (HCP5) acts on miR-128-3p through a
'molecular sponge' to increase pleomorphic adenoma gene like-2
expression and activate the Wnt/β-catenin/cyclin D1 (CCND1)
signaling pathway, forming the HCP5/miR-128-3p/Wnt/β-catenin/CCND1
pathway (25). Elevated
transcription factor 7 (TCF7) levels may promote MM cell
proliferation. Liu et al (26) confirmed the
TCF7/miR-203/Jagged1/Notch1 pathway and Ding et al (27) confirmed another regulatory pathway,
TCF7/miR-200c. The RNA component of mitochondrial RNA processing
endoribonuclease (RMRP) may regulate cell proliferation through the
RMRP/miR-34a-5p/c-Myc pathway, and c-Myc locates in the RMRP
promoter region to promote RMRP transcription, forming a circular
pathway (28). In addition to
these 'hot spot' lncRNA studies, numerous newly discovered lncRNAs
have a cancer-promoting effect in MM.MSTRG.29039.1 reduces the
inhibitory effect of miR-12119 on the oncostatin M receptor (OSMR),
and OSMR upregulation activates the Janus kinase 2 (JAK2)/signal
transducer and activator of transcription 3 (STAT3) signaling
pathway, forming the MSTRG.29039.1/miR-12119/OSMR/JAK2/STAT3
pathway (29). RP11-301G19.1
upregulates high-mobility group protein B2 (HMGB2), the target gene
of miR-582-5p, promotes the phosphorylation of phosphoinositide
3-kinase (PI3K) and protein kinase B (AKT) and activates the
RP11-301G19.1/miR-582-5p/PI3K/Akt pathway (30). LINC01234 promotes MM cell
proliferation through the LINC01234/miR-124-3p/growth factor
receptor-bound protein 2 pathway (31). LINC00461 promotes MM cell
proliferation through the LINC00461/miR-15/B-cell lymphoma 2
(Bcl-2) and LINC00461/miR-16/Bcl-2 pathways (32). In addition, there are several
lncRNAs in MM that target miRNAs to regulate downstream proteins
and promote cell proliferation through the 'molecular sponge'
function. For instance, brain-derived neurotrophic factor-antisense
(BDNF-AS)/miR-125-5p/Bcl-2 (33),
urothelial cancer associated 1 (UCA1)/miR-1271-5p/hepatocyte growth
factor (34), FEZ family zinc
finger 1 antisense RNA 1/miR-610/AKT serine/threonine kinase 3
(35), colon cancer-associated
transcript 1 (CCAT1)/miR-181a-5p/homeobox A1 (36), myocardial infarction-associated
transcript (MIAT)/miR-29b/myeloid cell leukemia-1 (MCL1) and
MIAT/miR-29b/Sp1 transcription factor (Sp1) (37).
In addition to targeting miRNAs, lncRNAs may
directly mediate protein expression or activate signaling pathways,
thus promoting myeloma cell proliferation. H19 directly activates
the NF-κB signaling pathway and upregulates the secretion of the
downstream cytokine IL-8, thus promoting cell proliferation and
colony formation (38). PCAT1
directly activates the p38 and c-Jun N-terminal kinase
(JNK)/mitogen-activated protein kinase (MAPK) signaling pathways,
promoting the proliferation and survival of MM cells (39). PVT1 regulates MYC expression at the
transcriptional level, and both PVT1 and MYC genes are regulated by
BRD4 (40). Ronchetti et al
(41) reported that ST3
β-galactoside α-2,3-sialyltransferase 6-antisense RNA1
(ST3GAL6-AS1) silencing decreased MAPK phosphorylation and
ubiquitination, thereby inhibiting cell proliferation. Geng et
al (42) demonstrated that
nuclear enriched abundant transcript 1 (NEAT1) overexpression
increased the expression of proteins related to the Wnt/β-catenin
signaling pathway, suggesting that NEAT1 is able to mediate the
Wnt/β-catenin signaling pathway to regulate cell proliferation.
Similarly, HOX transcript antisense RNA (HOTAIR) was indicated to
activate the NF-κB signaling pathway in myeloma cells (43), and lung cancer-associated
transcript 1 (LUCAT1) activated the transforming growth factor-β
signaling pathway (44).
AL928768.3, which directly acts on cyclin-dependent kinase 2 and
CCND1, reduces cyclin suppressor gene p21 to avoid cell cycle
arrest in G0/G1 phase, which is conducive to cell proliferation
(45). LncRNAs may also enhance
mRNA stability and promote the proliferation of myeloma cells by
improving the expression of target genes. Chen et al
(46) reported that HOXB cluster
antisense RNA 1 was able to enhance the interaction between
ELAV-like RNA binding protein 1 and fucosyltransferase 4 (FUT4)
proteins, promote the stability of FUT4 mRNA and thus activate the
Wnt-β-catenin signaling pathway. Ladybird homeobox 2 (LBX2)-AS1
improved the stability of LBX2 mRNA and increased its expression of
LBX2, thus promoting myeloma progression (47).
LncRNAs inhibit apoptosis of MM
cells
During normal cell proliferation, the G1/S
checkpoint of the cell cycle actively recognizes the integrity of
DNA replication. If DNA damage occurs, the cell cycle stops at the
G1 phase and the cell becomes unable to enter the S phase to start
the DNA repair process (48). If
the damage is so severe that it outpaces the cell's ability to
repair DNA, apoptosis is triggered in the cell (49). Due to the lack of 'functional' p53,
the key regulator of the G1/S checkpoint, the cell cycle of tumor
cells is accelerated to rapidly enter the S phase, but DNA damage,
such as DNA double-strand break (DSB), single strand break (SSB)
and interchain crosslinking may have toxic effects on cells
(50). Recent studies indicated
that tumor cells enhance their DNA repair ability to avoid
apoptosis (51). LncRNAs in
myeloma cells may act as oncogenes to mediate the DNA repair
process, thus having a role in DNA protection and anti-apoptosis.
As a protein scaffold, MALAT1 directly binds poly (ADP-ribose)
polymerase 1 (PARP1) to form functional complexes, and it
indirectly binds DNA ligase 3 (LIG3) to enhance the alternative
non-homologous end joining DNA repair pathway (52,53).
PARP1 is a protein closely related to DSB and SSB in the process of
DNA repair, which may catalyze PAR and induce apoptosis. MALAT1
binding to PARP1 may reduce PAR signaling and inhibit the release
of PARP1, thus reducing cell apoptosis (54). NEAT1 mediates a variety of DNA
repair mechanisms, including the homologous recombination signaling
pathway, mismatch repair and nucleotide resection repair. NEAT1
silencing downregulated DNA repair-related genes, such as RAD51
recombinase paralog B, checkpoint kinase 1 (CHK1), CHK2, 32-kDa
subunit of human RPA and breast cancer gene 1, and it significantly
inhibited the DNA repair ability of myeloma cells (55).
LncRNAs in MM cells may also directly regulate miRNA
or protein expression and inhibit tumor cell apoptosis. MALAT1
decreases HMGB1 ubiquitination, inhibits the degradation of HMGB1
after its translation and promotes the expression of Beclin-1 and
microtubule associated protein 1 light chain 3 β proteins, thus
inhibiting apoptosis (56).
Through the effect of ceRNA, small nucleolar RNA host gene 16
(SNHG16) regulates the expression of miR-342-3p, downregulates the
levels of caspase 3, caspase 9, forkhead transcription factor O
subfamily member 3a and Bax, and upregulates the levels of Bcl-2,
CCND1, PI3K and AKT, promoting the transition of the cell cycle
from the G1 phase to the S phase (57).
LncRNAs enhance the adhesion, invasion
and energy metabolism of MM cells
The adhesion and invasion ability of tumor cells
endows the process of tumor metastasis and promotes the development
of tumors (58). Rapid and
abnormal proliferation of tumor cells requires a large amount of
energy metabolism. Normal cells are mainly powered by the oxidative
phosphorylation of ATP; however, there is widespread hypoxia in
tumor tissues, which cannot effectively carry out the oxidative
phosphorylation process. Therefore, the energy supply of tumor
cells may occur in an anaerobic environment, and the energy
metabolism proceeds through glycolysis via the
glucose-pyruvate-lactic acid pathway (59). Studies have indicated that
MM-associated lncRNAs may enhance the adhesion, invasion and
glycolytic abilities of tumor cells. MALAT1 forms the
MALAT1/miR-1271-5p/SRY-box transcription factor 13 (SOX13) pathway,
promoting MM cell invasion and glycolysis (60). SOX2 overlapping transcript (SOX2OT)
forms the SOX2OT/miR-144-3p/c-MET pathway and promotes MM
progression (61). LINC01606 forms
part of the LINC01606/miR-579-3p pathway, promoting the migration
and invasion of myeloma cells (62). ST3GAL6-AS1 recruits heterogeneous
nuclear ribonucleoprotein A2/B1 protein, inhibits the degradation
of ST3GAL6 mRNA, upregulates ST3GAL6 protein, and promotes cell
adhesion, migration and invasion (63).
LncRNAs reshape the TME in MM cells
The TME is an environment for the survival of tumor
cells, containing fibroblasts, immune cells, endothelial cells,
adipocytes, neurons and other non-neoplastic cells, as well as the
components of the extracellular matrix, such as chemokines,
cytokines and exosomes (64). The
TME is an important regulatory factor in tumor angiogenesis,
continuous proliferation, migration, invasion and immune escape.
Various cytokines and exosomes reshape the TME and maintain an
environment conducive to tumor growth. Cells in the TME are also
stimulated by cytokines and undergo phenotypic changes, further
promoting tumor development (65).
MM-associated lncRNAs participate in the remodeling of the TME. For
instance, NEAT1 regulates B7-H3 by mediating the expression of
miR-214 through a 'molecular sponge' effect. B7-H3 activates the
JAK2-STAT3 signaling pathway to regulate macrophage polarization in
the TME. NEAT1 promotes the polarization of M2-type
tumor-associated macrophages through the NEAT1/miR-214/B7-H3
pathway (66). LncRNAs in the TME
may also affect MM cells. Wang et al (67) reported that adipocytes in the TME
secrete exosomes enriched with LOC606724 and SNHG1, and apoptosis
of MM cells is significantly inhibited after phagocytosis of
exosomes. LOC606724 may act as a 'molecular scaffold' to connect
eukaryotic translation initiation factor 4E (eIF4E) and c-Myc.
eIF4E is a key factor in protein translation and LOC606724 may
promote the synthesis of c-Myc mediated by eIF4E.
3. LncRNAs may act as tumor suppressor genes
to inhibit MM tumor progression
Although studies have indicated that most lncRNAs
promote cancer occurrence and development, certain lncRNAs function
as tumor suppressor genes and their expression is downregulated in
MM (Table I; Fig. 1). MM-related tumor suppressor
lncRNAs inhibit tumor cell proliferation and migration through
ceRNA and promote apoptosis. LINC01003 inhibits tumor cell adhesion
and proliferation through the LINC01003/miR-33a-5p/Pim-1
proto-oncogene, serine/threonine kinase pathway (68). Studies have indicated that
Opa-interacting protein 5-antisense RNA 1 (OIP5-AS1) may have a
role in cancer inhibition through multiple regulatory pathways, and
the OIP5-AS1/miR-410/phosphatase and tensin homolog (PTEN)/PI3K/AKT
pathway regulates the expression of downstream KLF transcription
factor 10 (KLF10) to arrest cell cycle progression (69). The OIP5-AS1/miR-27a-3p/tuberous
sclerosis 1 pathway inhibits tumor cell migration and invasion and
promotes apoptosis (70).
Differentiation antagonizing non-protein coding RNA (DANCR) forms
the DANCR/miR-135b-5p/KLF9 pathway, which reduces tumor cell
viability, migration and invasion (71). Insulin-like growth factor receptor
antisense imprinted non-protein coding RNA (IRAIN) forms the
IRAIN/miR-125b pathway to inhibit tumor cell proliferation and
promote apoptosis (72). LncRNAs
may also directly regulate signaling pathways in tumor cells.
XLOC-013703 reduces the secretion of interleukin 6 (IL-6) and
inhibits the activation of the NF-κB signaling pathway, thus
causing cell cycle arrest and accelerating apoptosis (73). The low expression of MM-associated
lncRNAs in tumor tissues may be related to hypermethylation of the
promoter region. Li et al (74) demonstrated that BM742401 inhibits
the homing and migration of MM cells but does not affect cell
proliferation or apoptosis. The failure of BM742401's anti-cancer
function is due to the hypermethylation of its promoter region. A
demethylation agent promoted BM742401 expression and restored its
anti-tumor effect.
4. LncRNAs are closely related to MM tumor
progression and patient prognosis
Both oncogenic and tumor suppressor lncRNAs have a
key role in the incidence and development of MM, suggesting that
lncRNA expression is closely related to MM progression and the
prognosis of patients (Table II).
The expression of cancer-promoting lncRNAs was significantly higher
in the intermediate and late stages of MM than in the early stage;
lncRNA expression was positively associated with the level of tumor
pathogenicity factors and negatively associated with the survival
time of patients and complete remission (CR). However, tumor
suppressor lncRNAs have the opposite effects.
 | Table IIAssociation between multiple
myeloma-associated lncRNAs and clinicopathological factors. |
Table II
Association between multiple
myeloma-associated lncRNAs and clinicopathological factors.
| LncRNA | Author, year | Expression | Pathological
indicators | Clinical
manifestations and cytogenetics | Prognosis | (Refs.) |
|---|
| ST3GAL6-AS1 | Shen, 2021 Shen,
2018 | Up-regulated | DS, ISS, R-ISS,
infiltration of plasma cells | n.a. | n.a. | (63,76) |
| H19 | Zheng, 2020 Sun,
2017 Pan, 2018 | Up-regulated | DS, ISS | Bone disease | OS | (18,38,77) |
| TUG1 | Yin, 2019 | Up-regulated | DS, ISS, R-ISS,
β2-MG, albumin, globulin | n.a. | n.a. | (78) |
| BDNF-AS | Chu, 2022 | Up-regulated | DS, ISS | n.a. | OS | (33) |
| MIAT | Fu, 2019 | Up-regulated | DS, ISS, IgH,
IgL | Overall cytogenetic
risk | OS | (37) |
| ANRIL | Yin, 2021 Yang,
2021 | Up-regulated | ISS,
β2-MG | n.a. | OS, PFS, CR | (85,100) |
| MSTRG.29039.1 | Liu, 2021 | Up-regulated | ISS,
β2-MG, LDH, infiltration of plasma cells | n.a. | n.a. | (29) |
| NEAT1 | Gao, 2020 Yu,
2020 | Up-regulated | ISS,
β2-MG, LDH | n.a. | OS, PFS, CR,
ORR | (66,86) |
| HCP5 | Liu, 2021 | Up-regulated | ISS | n.a. | PFS | (25) |
| UCA1 | Sedlarikova,
2017 | Up-regulated | ISS, albumin,
IgM | t(4;14),
Del(13q14), 1q21 amplification | OS | (87) |
| PCAT1 | Zhao, 2021 | Up-regulated | ISS, R-ISS,
β2-MG, LDH | Bone disease,
Del(17p) | OS, PFS | (88) |
| TCF7 | Liu, 2021 Ding,
2021 | Up-regulated | ISS,
β2-MG | t(14;16) | OS, EFS, CR | (26,27) |
| NR-046683 | Dong, 2019 | Up-regulated | ISS,
β2-MG | n.a. | PFS | (89) |
| AL928768.3 | Shen, 2022 | Up-regulated | ISS | n.a. | n.a. | (45) |
| LUCAT1 | Liu,2020 | Up-regulated | ISS | n.a. | OS | (44) |
| ANGPTL1-3 | Zhou, 2022 | Up-regulated | ISS, R-ISS | Del(17p),
t(4;14) | PFS, CR | (90) |
| CCAT1 | Chen, 2018 | Up-regulated | ISS | n.a. | OS | (36) |
| CCAT2 | Xu, 2020 | Up-regulated | ISS,
β2-MG | Kidney disease | n.a. | (91) |
| PRINS | Sedlarikova,
2018 | Up-regulated | Infiltration of
plasma cells | t(4;14) | n.a. | (95) |
| RMRP | Xiao, 2019 | Up-regulated | n.a. | n.a. | OS, DFS | (28) |
| HCP5 | Liu, 2021 | Up-regulated | n.a. | n.a. | OS | (25) |
| CRNDE | Meng, 2017 | Up-regulated | n.a. | n.a. | OS | (24) |
| LINC01606 | He, 2021 | Up-regulated | n.a. | n.a. | OS | (62) |
| LINC00461 | Deng, 2019 | Up-regulated | n.a. | n.a. | OS | (32) |
| LOC606724 | Wang, 2022 | Up-regulated | n.a. | n.a. | OS, CR | (67) |
| SNHG1 | Wang, 2022 | Up-regulated | n.a. | n.a. | OS, CR | (67) |
| OIP5-AS1 | Wang, 2020 | Down-regulated | ISS, IMWG risk
stratification | n.a. | OS | (70) |
| XLOC-013703 | Pu, 2019 | Down-regulated | DS, R-ISS,
β2-MG | n.a. | n.a. | (73) |
| PRAL | Xiao, 2018 | Down-regulated | DS, ISS | n.a. | OS, DFS | (79) |
| BM742401 | Li, 2020 | Down-regulated | n.a. | n.a. | OS | (74) |
LncRNAs are associated with pathological
indicators of MM
Owing to the variety of clinical manifestations of
MM and its numerous variants, there are currently multiple
diagnostic criteria and staging systems for MM. Durie-Salmon
staging (DS staging) was the first MM staging system and is most
widely used (75). ST3GAL6-AS1
(76), H19 (77), taurine-upregulated gene1 (TUG1)
(78), p53 regulation associated
lncRNA (PRAL) (79), BDNF-AS
(33), XLOC-013703 (73) and MIAT (37) were associated with the DS stage of
MM. Subsequent studies have indicated that β2 microglobulin (β2-MG)
may affect MM malignancy and patient prognosis and become a
reliable predictor of the survival time of patients with MM
(80,81). Albumin may mediate IL-6 expression
and affect MM cell proliferation and tumor malignancy, and serum
albumin is an important prognostic factor for MM (82,83).
In 2005, the International Myeloma Foundation proposed a new
International Staging System (ISS) based on β2-MG and albumin
levels (84). Previous studies
have confirmed that most MM-related lncRNAs are associated with ISS
staging. Examples for this are ANRIL (85), ST3GAL6-AS1 (63), MSTRG.29039.1 (29), H19 (77), NEAT1 (86), HCP5 (25), UCA1 (87), PCAT1 (88), PRAL (79), TCF7 (26), NR-046683 (89), AL928768.3 (45), BDNF-AS (33), LUCAT1 (44), OIP5-AS1 (70), angiopoietin-like (ANGPTL)1-3
(90), MIAT (37), CCAT1 (36), CCAT2 (91) and TUG1 (78). Other studies have found that ANRIL
(85), MSTRG.29039.1 (29), NEAT1 (86), TUG1 (78), PCAT1 (88), TCF7 (26), NR-046683 (89), CCAT2 (91) and XLOC-013703 (73) were correlated with serum β2-MG
levels in patients with MM. Furthermore, UCA1 (87) and TUG1 (78) were associated with serum albumin
levels in patients with MM. Lactate dehydrogenase (LDH) is not a
characteristic indicator of MM, but elevated LDH indicates a
significantly poor prognosis (92,93).
MSTRG.29039.1 (29), NEAT1
(86) and PCAT1 (88) are associated with serum LDH levels.
In 2015, the International Myeloma Working Group (IMWG) published
the Revised International Staging System (R-ISS) based on LDH
levels (94). Studies have
confirmed that ST3GAL6-AS1 (76),
PCAT1 (88), ANGPTL1-3 (90), XLOC-013703 (73) and TUG1 (78) are related to the R-ISS stage. In
addition, OIP5-AS1 is related to the risk stratification proposed
by IMWG (70). ST3GAL6-AS1
(76), MSTRG.29039.1 (29) and PRINS (95) were related to the infiltration
level of plasma cells. TUG1 expression is associated with serum
globulin levels (78), UCA1 with
serum IgM (87) and MIAT with
serum IgH and IgL (37).
LncRNAs associated with the clinical
manifestations of MM
Deregulated proliferation and extensive infiltration
of malignant plasma cells in the bone marrow may cause bone issues,
such as bone pain and pathological fractures. About two out of
three patients with MM seek treatment for a bone disease as their
first symptom (96). H19 (18) and PCAT1 (88) were associated with MM-related bone
diseases. The accumulation of monoclonal immunoglobulin secreted by
malignant plasma cells may seriously interfere with renal tubular
function, resulting in renal damage manifestations such as renal
dysfunction, proteinuria and hematuria, as well as increased serum
creatinine and urea nitrogen (97). CCAT2 is associated with
MM-associated kidney disease (91).
LncRNAs are associated with cytogenetic
abnormalities of MM
The IMWG proposed that cytogenetic abnormalities
worsen the prognosis of patients with MM and suggests that
Del(17p), t(4;14), t(14;16), as well as other factors, should be
included as reference factors in the diagnosis of MM and
cytogenetic abnormalities maybe detected using fluorescence in
situ hybridization technology for risk stratification (98). It was reported that Del(17p) is
related to PCAT1 (88) and
ANGPTL1-3 (90); t(4;14) is
related to UCA1 (87), ANGPTL1-3
(90) and psoriasis
susceptibility-related RNA gene induced by stress (PRINS) (95); UCA1 is also associated with
Del(13q14) and 1q21 amplification (87); t(14;16) is related to TCF7
(27); and MIAT is associated with
overall cytogenetic risk (37).
LncRNAs are associated with the prognosis
of patients with MM
The overall survival (OS) rate of patients with MM
has improved since 1970, but the prognosis is still not ideal due
to the high recurrence and drug resistance of MM (99). Therefore, a new method of risk
stratification is required to accurately assess the prognosis.
MM-associated lncRNAs are associated with tumor progression and may
be potential indicators of disease and prognosis. Elevated
expression of 'cancer-promoting lncRNAs' is associated with poor
prognosis and short survival. Studies suggested that ANRIL
(100), LOC606724 (67), SNHG1 (67), H19 (38), NEAT1 (66), HCP5 (25), UCA1 (87), PCAT1 (88), PRAL (79), TCF7 (27), BM742401 (74), BDNF-AS (33), CRNDE (24), LINC01606 (62), LUCAT1 (44), LINC00461 (32), OIP5-AS1 (70), CCAT1 (36), MIAT (37) and RMRP (28) were associated with OS in patients
with MM. In addition, certain lncRNAs are associated with other
survival indicators. For instance, PRAL (79) and RMRP (28) were correlated with disease-free
survival, ANRIL (85), NEAT1
(66), HCP5 (25), PCAT1 (88), NR-046683 (89) and ANGPTL1-3 (90) with progression-free survival and
TCF7 with event-free survival (27). For hematological malignancies,
including MM, treatment response is directly correlated with the
survival time of patients. During treatment, to prolong survival,
CR is considered the basic condition for effective treatment
(101,102). Detection of the expression levels
of MM-related lncRNAs to assess CR in patients may help to estimate
the prognosis. High expression of cancer-promoting lncRNAs, such as
ANRIL (85), LOC606724 (67), SNHG1 (67), NEAT1 (86), TCF7 (26) and ANGPTL1-3 (90), may predict low CR rates. NEAT1 was
negatively correlated with the overall response rate (86).
5. LncRNAs are potential markers for
diagnosing liquid biopsies in patients with MM
At present, liquid biopsy is a popular research
field for cancer diagnosis. It may diagnose the disease without
invasive surgery or examination and effectively reduce patients'
pain and economic burden (103).
Bone marrow aspiration is a traditional examination method for MM
diagnosis. However, for patients with MM who require an early
differential diagnosis and regular examination during treatment,
bone marrow aspiration is an invasive examination that may cause
great pain and have low repeatability. Furthermore, MM is a
multi-focal disease with significant spatial heterogeneity and
extramedullary disease, and the results of bone marrow aspiration
may be biased (104,105). MM-associated lncRNAs are
abnormally expressed in the peripheral blood of patients, and
compared with conventional clinical indicators, their sensitivity
and specificity are similar or even better, exhibiting specific
biomarker characteristics, and they may thus be potential
diagnostic indicators for MM (Table
III).
 | Table IIIDiagnostic value of MM-associated
lncRNAs in liquid biopsy. |
Table III
Diagnostic value of MM-associated
lncRNAs in liquid biopsy.
| LncRNA | Author, year | Number of
cases | Expression | Sensitivity, % | Specificity, % | AUC | (Refs.) |
|---|
| TUG1 | Yin, 2019 | 110 MM patients/98
healthy controls | Up-regulated | 65.5 | 94.9 | 0.792 | (78) |
| PCAT1 | Shen, 2017 | 60 MM patients/48
healthy controls | Up-regulated | 71.7 | 93.8 | 0.892 | (106) |
| H19 | Pan, 2018 | 80 MM patients/67
healthy controls | Up-regulated | 77.5 | 88.1 | 0.888 | (77) |
| HOTAIR | Guan, 2019 | 118 MM patients/78
healthy controls | Up-regulated | 70.1 | 79.9 | 0.798 | (107) |
| LINC01606 | He, 2021 | 72 MM patients/68
healthy controls | Up-regulated | 85.3 | 72.4 | 0.862 | (62) |
| PRINS | Sedlarikova,
2018 | 50 MM patients/30
healthy controls | Up-regulated | 80.8 | 76.9 | 0.753 | (95) |
| LBX2-AS1 | Jia, 2021 | 60 MM patients/60
healthy controls | Up-regulated | n.a. | n.a. | 0.753 | (47) |
| XLOC-013703 | Pu, 2019 | 107 MM patients/60
healthy controls | Down-regulated | 89.7 | 90.9 | 0.940 | (73) |
β2-MG and globulin are commonly used to diagnose MM.
A study reported that the serum TUG1 levels in patients with MM
were significantly higher than those in healthy individuals, with
better sensitivity and specificity (65.5 and 94.9%, respectively),
and even better than β2-MG (65.5 and 79.6%, respectively) and
globulin (54.5 and 69.4%, respectively) (78). To study the stability of lncRNAs in
serum, after the first detection of TUG1 levels, the authors placed
the serum samples at room temperature for 24 h or repeatedly froze
and thawed them 10 times, and TUG1 was measured. The results
indicated no significant changes in the TUG1 levels from the two
measurements, unaffected by harsh conditions. It has been suggested
that lncRNAs in the serum have good stability. Shen et al
(106) reported that the
sensitivity of serum PCAT1 (71.7%) was higher than that of the
common MM indices, β2-MG (48.3%), LDH (15.0%), κ light chain
(25.0%) and λ light chain (28.3%), with similar specificity. The
sensitivity and specificity of β2-MG combined with PCAT1 were 85
and 88%, respectively. Studies also suggested that the sensitivity
and specificity of serum H19 (77), HOTAIR (107), LINC01606 (62) and XLOC-013703 (73) were 77.5 and 88.1%, 70.1 and 79.9%,
85.3 and 72.4%, and 89.7 and 90.9%, respectively. LBX2-AS1 was
reported to be an effective diagnostic marker for MM (47). In addition to free lncRNAs in
serum, lncRNAs in serum exosomes are potential diagnostic markers.
Exosomes are biological vesicles that encapsulate tumor derivatives
and have roles in information exchange and substance transfer. The
peripheral molecular membrane endows exosomes with high stability
and is not easily damaged by interference from the external
environment (108). LncRNAs
wrapped in exosomes maybe detected and applied. Sedlarikova et
al (95) reported that PRINS
in peripheral blood exosomes of patients with MM was significantly
increased, and the sensitivity and specificity for the diagnosis of
MM were 80.8 and 76.9%, respectively.
6. LncRNAs regulate drug resistance in MM
cells
The continuous development of new anti-MM drugs in
the past 20 years has significantly improved the prognosis of
patients and the average survival time has been extended from 3-4
years to 7-8 years (109,110). There is currently no cure for MM
and initial anti-MM treatment is active and effective. However,
relapse inevitably occurs over time and after each relapse, MM
becomes more aggressive and resistant to the initial treatment
regimen, leading to recurrent/refractory MM (111). Understanding the underlying
mechanisms of MM resistance is essential for studying the
pathogenesis of relapsed/refractory MM and developing more
effective treatment strategies.
MM secretes proteins in large quantities, which
relies on proteasomal degradation of misfolded and aggregated
proteins. When the function of the proteasome is inhibited, the
excessive accumulation of proteins in MM cells may trigger
apoptosis, so proteasome inhibitors are used as the first-line
standard therapy for MM (112,113). Bortezomib, a new-generation
proteasome inhibitor, is the first drug approved by the US Food and
Drug Administration (FDA) to treat relapsed/refractory MM, marking
a breakthrough in anti-MM therapy (114). Bortezomib inhibits the activation
of anti-apoptotic proteins downstream of the NF-κB signaling
pathway and prevents the degradation of pro-apoptotic proteins,
thus accelerating apoptosis in MM cells (115). Studies have indicated that
MM-related lncRNAs regulate the resistance of MM cells to
bortezomib, resulting in drug resistance. ANRIL interacts with the
enhancer of zeste 2 polycomb repressive complex 2 subunit in the MM
cell nucleus to regulate the post-translational modification of the
downstream target PTEN, resulting in epigenetic silencing of the
PTEN promoter region binding to H3K27me3, thus increasing the
phosphorylation of AKT and the resistance of MM cells to
bortezomib, and reducing bortezomib-induced apoptosis (100). PCAT1 directly targets the
downstream p38 and JNK-MAPK signaling pathways, reducing the
sensitivity of MM cells to bortezomib (39). By interacting with the oncogene
c-Myc, protein disulfide isomerase family A member 3 pseudogene 1
regulates the transactivation activity of c-Myc and binds to the
promoter of glucose 6-phosphate dehydrogenase (G6PD) to increase
G6PD expression, thereby increasing pentose phosphate pathway (PPP)
flux. PPP produces NADPH in MM cells to enhance bortezomib
resistance (116). MM-associated
lncRNAs also regulate the expression of miRNAs through the
classical 'molecular sponge' action, thus making cells resistant to
bortezomib. H19 targets miR-29b-3p to promote the expression of
MCL1, which inhibits apoptosis, leading to drug resistance
(117). NEAT1 forms the
NEAT1/miR-29b-3p/Sp1 pathway to enhance drug resistance of MM
cells, and Sp1, as a transcription factor, targets the promoter
region binding to NEAT1 to induce the transcription of NEAT1,
eventually forming a feedback pathway (118). MIAT forms the MIAT/miR-29b
pathway to enhance bortezomib resistance in MM cells (37). As a tumor suppressor lncRNA, PRAL
was downregulated in MM cells and the PRAL/miR-210/bone
morphogenetic protein 2 (BMP2) pathway was used to mediate the
upregulation of BMP2 by targeting miR-210 to enhance the
therapeutic effect of bortezomib on MM cells (79).
In addition to increasing bortezomib resistance,
MM-associated lncRNAs may mediate resistance to other drugs.
Dexamethasone is the most widely used glucocorticoid in MM therapy
and may degrade poly (ADP) nucleotides, reduce the mitochondrial
transmembrane potential and induce MM cell apoptosis (119). NEAT1 promotes MCL1 expression
through the NEAT1/miR-193a/MCL1 pathway and the resistance of MM
cells to dexamethasone (120).
CRNDE activates the IL-6 signaling pathway, enhances the activity
of downstream STAT, RAS, MAPK and PI3K/AKT pathways, and prevents
dexamethasone-induced apoptosis in MM cells, resulting in drug
resistance and disease recurrence (121). HOTAIR activates the JAK2/STAT3
signaling pathway and enhances the resistance to dexamethasone in
MM cells (107). Hu et al
(52) reported that MALAT1 was
increased in bortezomib-, mefalam- and adriamycin-resistant cell
lines, and silencing MALAT1 rendered drug-resistant cell lines
sensitive to the corresponding drugs, suggesting that MALAT1
regulates the resistance of MM cells to bortezomib, mefalam and
adriamycin.
7. LncRNAs may be a new treatment target for
MM
Currently, the focus of drug research for MM
treatment involves small chemicals and biomacromolecules (122). However, the non-targeting of
small chemical molecules and the difficulty of biomacromolecules
penetrating cell membranes limit their potential applications
(123). Conventional therapy for
tumors frequently has a temporary therapeutic effect, followed by a
reduced response because it targets disease-related proteins rather
than transcribed genes (124). In
contrast, nucleic acid therapy may achieve sustained therapeutic
effects and even cure disease by introducing, inhibiting, replacing
and editing the relevant DNA or RNA. Therefore, nucleic acid
therapy may be used as an alternative or complementary therapy to
chemotherapy (125). Most nucleic
acid treatments in clinical trials are performed in four ways:
Antisense oligonucleotides (ASO), short interfering RNA, lipid
nanoparticles and adeno-associated virus carriers. ASO is the
application of a short oligonucleotide-binding target RNA, causing
RNase H-dependent RNA degradation. Several ASO drugs have been
approved by the FDA for the treatment of spinal muscular atrophy,
cytomegalovirus retinitis and muscular dystrophy (124).
Previous studies have indicated that MM-related
lncRNAs are crucial for tumorigenesis and development. Nucleic acid
therapy may degrade 'cancer-promoting lncRNAs' or enhance the
effect of 'cancer-suppressing lncRNAs', providing a new direction
for anti-MM therapy. Amodio et al (126) demonstrated that MALAT1 was an
intracellular lncRNA with significantly high expression in MM
cells, which promoted proteasomal degradation of damaged and
misfolded proteins and inhibited apoptosis through upregulation of
the proteasome transcription activator NF-E2 related factor-1/2
(NRF1/2), and NRF1 binds to the promoter of MALAT1 to form a
counter-activated feedback pathway. After LNA gapmeR-ASO was
applied to target MALAT1 degradation, H3K27Me3 shifted from the
promoter region of Kelch-like ECH-associated protein 1 (KEAP1) to
reduce KEAP1 methylation. Increased KEAP1 expression reduced NRF1/2
and proteasome levels, promoting apoptosis. In the above study, LNA
gapmeR-ASO not only tolerated nucleases and had a good target
affinity, but also showed no toxicity which proved that LNA
gapmeR-ASO was an ideal nucleic acid therapy route. LNA gapmeR-ASO
targeting MALAT1 enhanced the sensitivity of MM cells to
bortezomib, suggesting that they may be used alone to induce
apoptosis or in conjunction with bortezomib in MM treatment. Hu
et al (52) reported that
MALAT1 promotes DNA repair and anti-apoptosis through the
MALAT1/PARP1/LIG3 pathway, and silencing MALAT1 enhanced the toxic
effect of bortezomib on MM cells. To further investigate ASO
targeting MALAT1 in MM treatment, they combined ASO and MALAT1 with
the nanomaterial single-wall carbon nanotube (SWCNT) to enhance
targeting affinity and drug stability. After injection of
SWCNT-ASO-MALAT1 into the mouse model of MM, the drug concentrated
near the tumor, reducing the tumor burden and significantly
prolonging the survival of the mice, suggesting that SWCNT-ASO
targeting MALAT1 may effectively inhibit tumor growth in
vivo without significant toxicity.
8. Bioinformatics assisting lncRNA
research
With the innovation of gene chips and
high-throughput sequencing technology, lncRNA research has been
continuously improved. The application of sequencing databases and
network tools combined with bioinformatics methods is the current
trend in medical research (127).
The conventional bioinformatics approach for lncRNA research
involves first obtaining the whole-genome transcripts. Second,
filter conditions are set to screen for transcripts that do not
encode proteins. The expression levels of protein-coding genes and
lncRNAs are obtained from RNA sequencing, and lncRNA quantity is
analyzed by calculation. Finally, the functions of lncRNAs are
predicted based on the co-expression networks between lncRNAs and
protein-coding genes, the interactions between lncRNAs and RNAs,
and the interactions between lncRNAs and proteins (128-130). With the development of lncRNA
research, a large amount of experimental data has continued to
emerge. Bioinformatics and mathematical algorithms are applied to
effectively store experimental information in the databases, which
are continuously maintained and updated, which is helpful for
researchers to directly use the databases for experimental designs
and avoid starting from the transcriptome sequencing analysis for
each study, thereby effectively saving time and cost.
This section briefly describes the lncRNA-related
databases that are widely used at present. The LNCipedia database
integrates the information of several human lncRNA databases,
including LncRNAdb, Broad Institute, Ensembl and Gencode, and
provides the sequence, annotation, structure and miRNA combination
information of lncRNAs. It currently contains 127,802 transcripts
and 56,946 genes (131). The
LNCBook database launched a new version (LNCBook 2.0) in June 2022,
providing information on lncRNA expression, sequence alignment,
classification, coding ability prediction, methylation, variation,
lncRNA-miRNA interactions and lncRNA-protein interactions (132). It is one of the most abundant
databases that provides human lncRNA information. The LncSEA
database provides detailed information on >50,000 human lncRNAs,
including expression, methylation, disease relationships, tumor
markers, subcellular localization and transcription factors
(133). The LncExpDB database is
not limited to diseases and it covers multiple biological
components, such as normal tissues, normal cell lines, tumor cell
lines, organ development, cell differentiation and exosomes;
101,293 lncRNAs have been included, including the annotation and
predictive function (134). The
deepBase database was updated with the deepBase 3.0 version in
January 2021, providing information on lncRNA expression,
evolution, function prediction, prognosis and other information on
tissues, cancers and exosomes (135). The TANRIC database integrates
lncRNA information in tissues and cell lines from The Cancer Genome
Atlas and Cancer Cell Line Encyclopedia databases, providing lncRNA
annotation, expression, clinical indicators and prognosis
information (136). The LnCAR
database is based on 52,300 samples from 10 types of cancer in the
Gene Expression Omnibus database, providing differential expression
of lncRNAs, clinical indicators and prognosis information (137). The LncRNADisease 2.0 database
focuses on association analysis between lncRNAs and diseases,
providing 205,959 association scores (138). The LncMAP database provides
regulatory networks among lncRNAs, transcription factors and genes
in >20 types of tumors (139).
The lncRNA SNP database focuses on single nucleotide polymorphism
information in human and mouse lncRNAs (140). The Lnc2Meth database focuses on
the association analysis of lncRNAs and DNA methylation in human
diseases (141).
Most studies use bioinformatics to initially screen
target lncRNAs and predict their functions for studying MM-related
lncRNAs, which is then verified by in vitro and in
vivo experiments. Due to the lack of validation, only a small
number of studies have been solely based on bioinformatics data
analysis. Todoerti et al (142) screened the data of patients with
MM with molecular aberrations and clinical information from public
databases and indicated that MIAT was positively correlated with
cytogenetic indicators t(4;14), del(1p), del(13q) and
hyperdiploidy, and high MIAT expression suggested poor OS. In MM
pathogenesis, genes encoding ribosome, immune response, mitotic
spindle, apoptosis and p53 pathway are upregulated in cases with
high MIAT expression. By contrast, DNA repair-related genes and MYC
target genes are downregulated. Regarding drug resistance, MIAT
expression increased in MM cells with drug resistance, or MM
relapsed after bortezomib treatment. Todoerti et al
(142) conducted a comprehensive
study of MIAT but did not perform any experimental verification.
Zhou et al (143)
downloaded a large amount of gene expression data and clinical
information of patients with MM from the GEO database and
identified 59 lncRNAs associated with OS; four of them were
independent risk factors for predicting OS, RP4-803J11.2 and
RP1-43E13.2 were upregulated, and ZFY-AS1 and RP11-553L6.5
downregulated. Functional enrichment analysis suggested that
RP4-803J11.2, RPP1-43E13.2, ZFY-AS1 and RP11-553L6.5 were involved
in the cell cycle, chromatin modification, DNA replication,
microtubule process, DNA repair and RNA processing in MM.
9. Summary and outlook
In recent years, researchers have identified
numerous lncRNAs abnormally expressed in tumors, making lncRNAs a
research hotspot. The present review discussed recent studies on
MM-associated lncRNAs, emphasizing their roles in tumor development
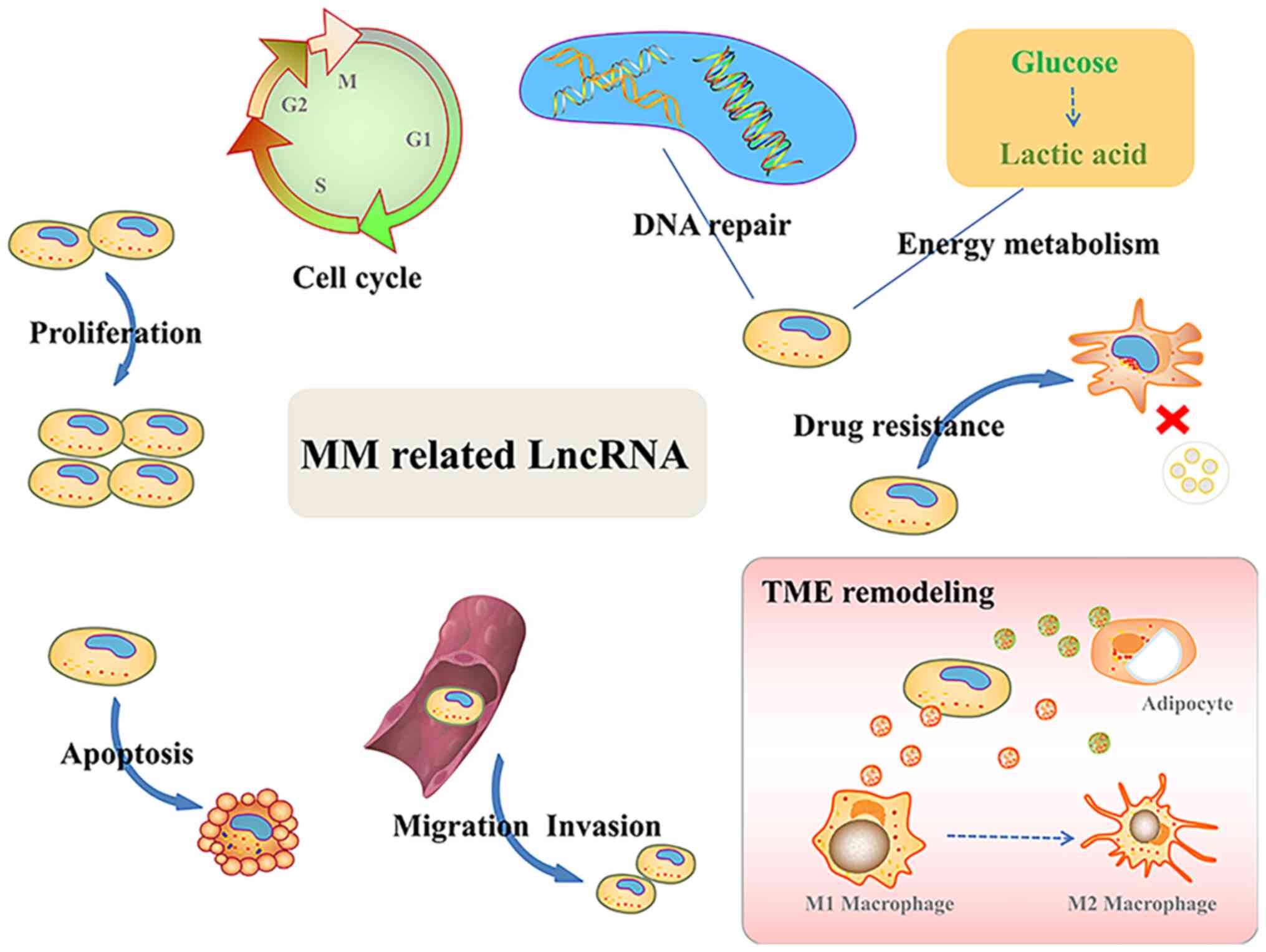
(Fig. 2). MM-associated lncRNAs
change the biological features of tumor cells, such as
proliferation, apoptosis, adhesion, invasion, energy metabolism,
therapeutic resistance and TME reshaping. LncRNAs are closely
related to pathological indicators and prognosis of MM and are
potential biomarkers and reference molecules for disease risk
stratification. Regarding the molecular mechanisms, lncRNAs exert
their effects through ceRNA interactions, binding proteins and
transcription factors, acting as 'molecular scaffolds', mRNA
stabilizers, mediating cell signaling pathways, epigenetic gene
regulation and other pathways. By determining the specific
regulatory mechanisms of lncRNAs, targeted therapies for MM using
nucleic acids may avoid frequent drug resistance and disease
relapse.
However, in the face of tens of thousands of
lncRNAs and the complex and huge molecular regulatory networks
behind them, the current understanding of lncRNAs remains
incomplete. There is insufficient evidence for lncRNA as a mature
tumor diagnostic marker, which requires to be further explored in
large-sample studies and with multi-disease stratification. A
single lncRNA cannot drive the biological functions of tumor cells
and the same signaling pathway does not function alone. It is
necessary to further explore the synergistic effects of multiple
lncRNAs, signaling pathways and acting proteins, and to enrich and
expand the molecular regulatory networks of lncRNAs, which will
deepen our understanding of the pathogenesis of MM. The present
study provides a solid foundation and new insight for developing
novel biomarkers and targeted lncRNA therapeutics.
Availability of data and materials
Data sharing is not applicable.
Authors' contributions
CY wrote the manuscript. YL, JS and SW drew the
figures. YH performed the literature review. KC and MS designed the
study and approved the final version of the manuscript for
publication. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81873455 and 82070222) and the
Training Program for Young and Middle-aged Health Science and
Technology Innovation Leaders in Henan Province (grant no.
YXKC2022015).
References
|
1
|
Cowan AJ, Allen C, Barac A, Basaleem H,
Bensenor I, Curado MP, Foreman K, Gupta R, Harvey J, Hosgood HD, et
al: Global burden of multiple myeloma: A systematic analysis for
the global burden of disease study 2016. JAMA Oncol. 4:1221–1227.
2018.
|
|
2
|
Kumar SK, Dimopoulos MA, Kastritis E,
Terpos E, Nahi H, Goldschmidt H, Hillengass J, Leleu X, Beksac M,
Alsina M, et al: Natural history of relapsed myeloma, refractory to
immunomodulatory drugs and proteasome inhibitors: A multicenter
IMWG study. Leukemia. 31:2443–2448. 2017.
|
|
3
|
van de Donk N, Pawlyn C and Yong KL:
Multiple myeloma. Lancet. 397:410–427. 2021.
|
|
4
|
Birney E, Stamatoyannopoulos JA, Dutta A,
Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis
ET, Thurman RE, et al: Identification and analysis of functional
elements in 1% of the human genome by the ENCODE pilot project.
Nature. 447:799–816. 2007.
|
|
5
|
Yan H and Bu P: Non-coding RNA in cancer.
Essays. Biochem. 65:625–639. 2021.
|
|
6
|
Winkle M, Kluiver JL, Diepstra A and van
den Berg A: Emerging roles for long noncoding RNAs in B-cell
development and malignancy. Crit Rev Oncol Hematol. 120:77–85.
2017.
|
|
7
|
Li Y, Li G, Guo X, Yao H, Wang G and Li C:
Non-coding RNA in bladder cancer. Cancer Lett. 485:38–44. 2020.
|
|
8
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021.
|
|
9
|
Zhu J, Fu H, Wu Y and Zheng X: Function of
lncRNAs and approaches to lncRNA-protein interactions. Sci China
Life Sci. 56:876–885. 2013.
|
|
10
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018.
|
|
11
|
Sun Q, Hao Q and Prasanth KV: Nuclear long
noncoding RNAs: Key regulators of gene expression. Trends Genet.
34:142–157. 2018.
|
|
12
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017.
|
|
13
|
Kazandjian D: Multiple myeloma
epidemiology and survival: A unique malignancy. Semin Oncol.
43:676–681. 2016.
|
|
14
|
Goyal B, Yadav SRM, Awasthee N, Gupta S,
Kunnumakkara AB and Gupta SC: Diagnostic, prognostic, and
therapeutic significance of long non-coding RNA MALAT1 in cancer.
Biochim Biophys Acta Rev Cancer. 1875:1885022021.
|
|
15
|
Sun Y, Jiang T, Jia Y, Zou J, Wang X and
Gu W: LncRNA MALAT1/miR-181a-5p affects the proliferation and
adhesion of myeloma cells via regulation of Hippo-YAP signaling
pathway. Cell Cycle. 18:2509–2523. 2019.
|
|
16
|
Liu H, Chi Z, Jin H and Yang W: MicroRNA
miR-188-5p as a mediator of long non-coding RNA MALAT1 regulates
cell proliferation and apoptosis in multiple myeloma.
Bioengineered. 12:1611–1626. 2021.
|
|
17
|
Ghafouri-Fard S, Esmaeili M and Taheri M:
H19 lncRNA: Roles in tumorigenesis. Biomed Pharmacother.
123:1097742020.
|
|
18
|
Zheng JF, Guo NH, Zi FM and Cheng J: Long
noncoding RNA H19 promotes tumorigenesis of multiple myeloma by
activating BRD4 signaling by targeting MicroRNA 152-3p. Mol Cell
Biol. 40:e00382–19. 2020.
|
|
19
|
Xiong T, Li J, Chen F and Zhang F: PCAT-1:
A novel oncogenic long non-coding RNA in human cancers. Int J Biol
Sci. 15:847–856. 2019.
|
|
20
|
Shen X, Kong S, Yang Q, Yin Q, Cong H,
Wang X and Ju S: PCAT-1 promotes cell growth by sponging miR-129
via MAP3K7/NF-κB pathway in multiple myeloma. J Cell Mol Med.
24:3492–3503. 2020.
|
|
21
|
Onagoruwa OT, Pal G, Ochu C and Ogunwobi
OO: Oncogenic role of PVT1 and therapeutic implications. Front
Oncol. 10:172020.
|
|
22
|
Yang M, Zhang L, Wang X, Zhou Y and Wu S:
Down-regulation of miR-203a by lncRNA PVT1 in multiple myeloma
promotes cell proliferation. Arch Med Sci. 14:1333–1339. 2018.
|
|
23
|
Wang M, Zhao HY, Zhang JL, Wan DM, Li YM
and Jiang ZX: Dysregulation of LncRNA ANRIL mediated by miR-411-3p
inhibits the malignant proliferation and tumor stem cell like
property of multiple myeloma via hypoxia-inducible factor 1α. Exp
Cell Res. 396:1122802020.
|
|
24
|
Meng YB, He X, Huang YF, Wu QN, Zhou YC
and Hao DJ: Long noncoding RNA CRNDE promotes multiple myeloma cell
growth by suppressing miR-451. Oncol Res. 25:1207–1214. 2017.
|
|
25
|
Liu Q, Ran R, Song M, Li X, Wu Z, Dai G
and Xia R: LncRNA HCP5 acts as a miR-128-3p sponge to promote the
progression of multiple myeloma through activating
Wnt/β-catenin/cyclin D1 signaling via PLAGL2. Cell Biol Toxicol.
38:979–993. 2022.
|
|
26
|
Liu H, Shen Y, Xu Y, Wang L, Zhang C,
Jiang Y, Hong L, Huang H and Liu H: lncRNA transcription factor 7
is related to deteriorating clinical features and poor prognosis in
multiple myeloma, and its knockdown suppresses disease progression
by regulating the miR-203-mediated Jagged1-Notch1 signaling
pathway. Oncol Lett. 21:4122021.
|
|
27
|
Ding T, Deng R and Huang T: Long
non-coding RNA T cell factor 7 is associated with increased disease
risk and poor prognosis, and promotes cell proliferation,
attenuates cell apoptosis and miR-200c expression in multiple
myeloma. Oncol Lett. 21:1292021.
|
|
28
|
Xiao X, Gu Y, Wang G and Chen S: c-Myc,
RMRP, and miR-34a-5p form a positive-feedback loop to regulate cell
proliferation and apoptosis in multiple myeloma. Int J Biol
Macromol. 122:526–537. 2019.
|
|
29
|
Liu Z, Han M, Meng N, Luo J and Fu R:
lncRNA MSTRG.29039.1 promotes proliferation by sponging
hsa-miR-12119 via JAK2/STAT3 pathway in multiple myeloma. Oxid Med
Cell Longev. 2021:99694492021.
|
|
30
|
Wang F, Luo Y, Zhang L, Younis M and Yuan
L: The LncRNA RP11-301G19.1/miR-582-5p/HMGB2 axis modulates the
proliferation and apoptosis of multiple myeloma cancer cells via
the PI3K/AKT signalling pathway. Cancer Gene Ther. 29:292–303.
2022.
|
|
31
|
Chen X, Liu Y, Yang Z, Zhang J, Chen S and
Cheng J: LINC01234 promotes multiple myeloma progression by
regulating miR-124-3p/GRB2 axis. Am J Transl Res. 11:6600–6618.
2019.
|
|
32
|
Deng M, Yuan H, Liu S, Hu Z and Xiao H:
Exosome-transmitted LINC00461 promotes multiple myeloma cell
proliferation and suppresses apoptosis by modulating microRNA/BCL-2
expression. Cytotherapy. 21:96–106. 2019.
|
|
33
|
Chu M, Fan Y, Wu L, Ma X, Sao J, Yao Y,
Zhuang W and Zhang C: Knockdown of lncRNA BDNF-AS inhibited the
progression of multiple myeloma by targeting the
miR-125a/b-5p-BCL2axis. Immun Ageing. 19:32022.
|
|
34
|
Yang Y and Chen L: Downregulation of
lncRNA UCA1 facilitates apoptosis and reduces proliferation in
multiple myeloma via regulation of the miR-1271-5p/HGF axis. J Chin
Med Assoc. 82:699–709. 2019.
|
|
35
|
Li QY, Chen L, Hu N and Zhao H: Long
non-coding RNA FEZF1-AS1 promotes cell growth in multiple myeloma
via miR-610/Akt3 axis. Biomed Pharmacother. 103:1727–1732.
2018.
|
|
36
|
Chen L, Hu N, Wang C, Zhao H and Gu Y:
Long non-coding RNA CCAT1 promotes multiple myeloma progression by
acting as a molecular sponge of miR-181a-5p to modulate HOXA1
expression. Cell Cycle. 17:319–329. 2018.
|
|
37
|
Fu Y, Liu X, Zhang F, Jiang S, Liu J and
Luo Y: Bortezomib-inducible long non-coding RNA myocardial
infarction associated transcript is an oncogene in multiple myeloma
that suppresses miR-29b. Cell Death Dis. 10:3192019.
|
|
38
|
Sun Y, Pan J, Zhang N, Wei W, Yu S and Ai
L: Knockdown of long non-coding RNA H19 inhibits multiple myeloma
cell growth via NF-κB pathway. Sci Rep. 7:180792017.
|
|
39
|
Shen X, Shen P, Yang Q, Yin Q, Wang F,
Cong H, Wang X and Ju S: Knockdown of long non-coding RNA PCAT-1
inhibits myeloma cell growth and drug resistance via p38 and JNK
MAPK pathways. J Cancer. 10:6502–6510. 2019.
|
|
40
|
Handa H, Honma K, Oda T, Kobayashi N,
Kuroda Y, Kimura-Masuda K, Watanabe S, Ishihara R, Murakami Y,
Masuda Y, et al: Long Noncoding RNA PVT1 is regulated by
bromodomain protein BRD4 in multiple myeloma and is associated with
disease progression. Int J Mol Sci. 21:71212020.
|
|
41
|
Ronchetti D, Todoerti K, Vinci C, Favasuli
V, Agnelli L, Manzoni M, Pelizzoni F, Chiaramonte R, Platonova N,
Giuliani N, et al: Expression pattern and biological significance
of the lncRNA ST3GAL6-AS1 in multiple myeloma. Cancers (Basel).
12:7822020.
|
|
42
|
Geng W, Guo X, Zhang L, Ma Y, Wang L, Liu
Z, Ji H and Xiong Y: Resveratrol inhibits proliferation, migration
and invasion of multiple myeloma cells via NEAT1-mediated
Wnt/β-catenin signaling pathway. Biomed Pharmacother. 107:484–494.
2018.
|
|
43
|
Zhu BZ and Lin L: Effects of lncRNA HOTAIR
on proliferation and apoptosis of myeloma cells through NF-κB
pathway. Eur Rev Med Pharmacol Sci. 23:10042–10048. 2019.
|
|
44
|
Liu Z, Gao H, Peng Q and Yang Y: Long
noncoding RNA LUCAT1 promotes multiple myeloma cell growth by
regulating the TGF-β signaling pathway. Technol Cancer Res Treat.
19:15330338209457702020.
|
|
45
|
Shen Q, Jiang Q, Cong Z, Zhou Y, Huang X,
Zhu L, Xu X and Qian J: Knockdown of lncRNA AL928768.3 inhibits
multiple myeloma cell proliferation by inducing cell cycle arrest
in G0/G1 phase. Ann Transl Med. 10:1722022.
|
|
46
|
Chen R, Zhang X and Wang C: LncRNA
HOXB-AS1 promotes cell growth in multiple myeloma via FUT4 mRNA
stability by ELAVL1. J Cell Biochem. 121:4043–4051. 2020.
|
|
47
|
Jia H, Liu Y, Lv S, Qiao R, Zhang X, Niu
F, Shang W, Liu S, Dong J and Zhang Z: LBX2-AS1 as a novel
diagnostic biomarker and therapeutic target facilitates multiple
myeloma progression by enhancing mRNA stability of LBX2. Front Mol
Biosci. 8:7065702021.
|
|
48
|
Halazonetis TD, Gorgoulis VG and Bartek J:
An oncogene-induced DNA damage model for cancer development.
Science. 319:1352–1355. 2008.
|
|
49
|
Kar S: Unraveling cell-cycle dynamics in
cancer. Cell Systems. 2:8–10. 2016.
|
|
50
|
Basu AK: DNA damage, mutagenesis and
cancer. Int J Mol Sci. 19:9702018.
|
|
51
|
Larsen BD, Benada J, Yung PYK, Bell RAV,
Pappas G, Urban V, Ahlskog JK, Kuo TT, Janscak P, Megeney LA, et
al: Cancer cells use self-inflicted DNA breaks to evade growth
limits imposed by genotoxic stress. Science. 376:476–483. 2022.
|
|
52
|
Hu Y, Lin J, Fang H, Fang J, Li C, Chen W,
Liu S, Ondrejka S, Gong Z, Reu F, et al: Targeting the
MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in
multiple myeloma. Leukemia. 32:2250–2262. 2018.
|
|
53
|
Sharma S, Javadekar SM, Pandey M,
Srivastava M, Kumari R and Raghavan SC: Homology and enzymatic
requirements of microhomology-dependent alternative end joining.
Cell Death Dis. 6:e16972015.
|
|
54
|
Huambachano O, Herrera F, Rancourt A and
Satoh MS: Double-stranded DNA binding domain of poly(ADP-ribose)
polymerase-1 and molecular insight into the regulation of its
activity. J Biol Chem. 286:7149–7160. 2011.
|
|
55
|
Taiana E, Favasuli V, Ronchetti D,
Todoerti K, Pelizzoni F, Manzoni M, Barbieri M, Fabris S,
Silvestris I, Gallo Cantafio ME, et al: Long non-coding RNA NEAT1
targeting impairs the DNA repair machinery and triggers anti-tumor
activity in multiple myeloma. Leukemia. 34:234–244. 2020.
|
|
56
|
Gao D, Lv AE, Li HP, Han DH and Zhang YP:
LncRNA MALAT-1 elevates HMGB1 to promote autophagy resulting in
inhibition of tumor cell apoptosis in multiple myeloma. J Cell
Biochem. 118:3341–3348. 2017.
|
|
57
|
Yang X, Huang H, Wang X, Liu H, Liu H and
Lin Z: Knockdown of lncRNA SNHG16 suppresses multiple myeloma cell
proliferation by sponging miR-342-3p. Cancer Cell Int.
20:382020.
|
|
58
|
Hamidi H and Ivaska J: Every step of the
way: Integrins in cancer progression and metastasis. Nat Rev
Cancer. 18:533–548. 2018.
|
|
59
|
Ganapathy-Kanniappan S and Geschwind JF:
Tumor glycolysis as a target for cancer therapy: Progress and
prospects. Mol Cancer. 12:1522013.
|
|
60
|
Liu N, Feng S, Li H, Chen X, Bai S and Liu
Y: Long non-coding RNA MALAT1 facilitates the tumorigenesis,
invasion and glycolysis of multiple myeloma via miR-1271-5p/SOX13
axis. J Cancer Res Clin Oncol. 146:367–379. 2020.
|
|
61
|
Tianhua Y, Dianqiu L, Xuanhe Z, Zhe Z and
Dongmei G: Long non-coding RNA Sox2 overlapping transcript (SOX2OT)
promotes multiple myeloma progression via microRNA-143-3p/c-MET
axis. J Cell Mol Med. 24:5185–5194. 2020.
|
|
62
|
He X, Fan X, Zhang B, Wu L and Wu X:
Expression of LINC01606 in multiple myeloma and its effect on cell
invasion and migration. Am J Transl Res. 13:8777–8786. 2021.
|
|
63
|
Shen Y, Feng Y, Li F, Jia Y, Peng Y, Zhao
W, Hu J and He A: lncRNA ST3GAL6-AS1 promotes invasion by
inhibiting hnRNPA2B1-mediated ST3GAL6 expression in multiple
myeloma. Int J Oncol. 58:52021.
|
|
64
|
Xiao Y and Yu D: Tumor microenvironment as
a therapeutic target in cancer. Pharmacol Ther. 221:1077532021.
|
|
65
|
Hinshaw DC and Shevde LA: The tumor
microenvironment innately modulates cancer progression. Cancer Res.
79:4557–4566. 2019.
|
|
66
|
Gao Y, Fang P, Li WJ, Zhang J, Wang GP,
Jiang DF and Chen FP: LncRNA NEAT1 sponges miR-214 to regulate M2
macrophage polarization by regulation of B7-H3 in multiple myeloma.
Mol Immunol. 117:20–28. 2020.
|
|
67
|
Wang Z, He J, Bach DH, Huang YH, Li Z, Liu
H, Lin P and Yang J: Induction of m6A methylation in
adipocyte exosomal LncRNAs mediates myeloma drug resistance. J Exp
Clin Cancer Res. 41:42022.
|
|
68
|
Wu L, Xia L, Chen X, Ruan M, Li L and Xia
R: Long non-coding RNA LINC01003 suppresses the development of
multiple myeloma by targeting miR-33a-5p/PIM1 axis. Leuk Res.
106:1065652021.
|
|
69
|
Yang N, Chen J, Zhang H, Wang X, Yao H,
Peng Y and Zhang W: LncRNA OIP5-AS1 loss-induced microRNA-410
accumulation regulates cell proliferation and apoptosis by
targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple
myeloma. Cell Death Dis. 8:e29752017.
|
|
70
|
Wang Y, Wang H, Ruan J, Zheng W, Yang Z
and Pan W: Long non-coding RNA OIP5-AS1 suppresses multiple myeloma
progression by sponging miR-27a-3p to activate TSC1 expression.
Cancer Cell Int. 20:1552020.
|
|
71
|
Wu L, Xia L, Jiang H, Hu Y, Li L, Xu L and
Xia R: Long non-coding RNA DANCR represses the viability, migration
and invasion of multiple myeloma cells by sponging miR-135b-5p to
target KLF9. Mol Med Rep. 24:6492021.
|
|
72
|
Jiang Y, Chen J and Chen G: Long noncoding
RNA IRAIN acts as tumor suppressor via miR-125b in multiple
myeloma. Oncol Lett. 18:6787–6794. 2019.
|
|
73
|
Pu J, Huang H, Su J, Yuan J, Cong H, Wang
X and Ju S: Decreased expression of long noncoding RNA XLOC_013703
promotes cell growth via NF-κB pathway in multiple myeloma. IUBMB
Life. 71:1240–1251. 2019.
|
|
74
|
Li Z, Kumar S, Jin DY, Calin GA, Chng WJ,
Siu KL, Poon MW and Chim CS: Epigenetic silencing of long
non-coding RNA BM742401 in multiple myeloma: Impact on prognosis
and myeloma dissemination. Cancer Cell Int. 20:4032020.
|
|
75
|
Fechtner K, Hillengass J, Delorme S, Heiss
C, Neben K, Goldschmidt H, Kauczor HU and Weber MA: Staging
monoclonal plasma cell disease: Comparison of the Durie-Salmon and
the Durie-Salmon PLUS staging systems. Radiology. 257:195–204.
2010.
|
|
76
|
Shen Y, Feng Y, Chen H, Huang L, Wang F,
Bai J, Yang Y, Wang J, Zhao W, Jia Y, et al: Focusing on long
non-coding RNA dysregulation in newly diagnosed multiple myeloma.
Life Sci. 196:133–142. 2018.
|
|
77
|
Pan Y, Chen H, Shen X, Wang X, Ju S, Lu M
and Cong H: Serum level of long noncoding RNA H19 as a diagnostic
biomarker of multiple myeloma. Clin Chim Acta. 480:199–205.
2018.
|
|
78
|
Yin Q, Shen X, Cui X and Ju S: Elevated
serum lncRNA TUG1 levels are a potential diagnostic biomarker of
multiple myeloma. Exp Hematol. 79:47–55.e42. 2019.
|
|
79
|
Xiao G, Li Y, Wang Y, Zhao B, Zou Z, Hou
S, Jia X, Liu X, Yao Y, Wan J, et al: LncRNA PRAL is closely
related to clinical prognosis of multiple myeloma and the
bortezomib sensitivity. Exp Cell Res. 370:254–263. 2018.
|
|
80
|
Cowan AJ, Green DJ, Kwok M, Lee S, Coffey
DG, Holmberg LA, Tuazon S, Gopal AK and Libby EN: Diagnosis and
management of multiple myeloma: A review. JAMA. 327:464–477.
2022.
|
|
81
|
Rossi D, Fangazio M, De Paoli L, Puma A,
Riccomagno P, Pinto V, Zigrossi P, Ramponi A, Monga G and Gaidano
G: Beta-2-microglobulin is an independent predictor of progression
in asymptomatic multiple myeloma. Cancer. 116:2188–2200. 2010.
|
|
82
|
Kim JE, Yoo C, Lee DH, Kim SW, Lee JS and
Suh C: Serum albumin level is a significant prognostic factor
reflecting disease severity in symptomatic multiple myeloma. Ann
Hematol. 89:391–397. 2010.
|
|
83
|
Kyle RA, Gertz MA, Witzig TE, Lust JA,
Lacy MQ, Dispenzieri A, Fonseca R, Rajkumar SV, Offord JR, Larson
DR, et al: Review of 1027 patients with newly diagnosed multiple
myeloma. Mayo Clin Proc. 78:21–33. 2003.
|
|
84
|
Greipp PR, San Miguel J, Durie BG, Crowley
JJ, Barlogie B, Bladé J, Boccadoro M, Child JA, Avet-Loiseau H,
Kyle RA, et al: International staging system for multiple myeloma.
J Clin Oncol. 23:3412–34202. 2005.
|
|
85
|
Yin Y, Yang W, Zhang L, Liu K and Luo Z:
Long non-coding RNA ANRIL and its target microRNAs (microRNA-34a,
microRNA-125a and microRNA-186) relate to risk stratification and
prognosis in multiple myeloma. Hematology. 26:160–169. 2021.
|
|
86
|
Yu H, Peng S, Chen X, Han S and Luo J:
Long non-coding RNA NEAT1 serves as a novel biomarker for treatment
response and survival profiles via microRNA-125a in multiple
myeloma. J Clin Lab Anal. 34:e233992020.
|
|
87
|
Sedlarikova L, Gromesova B, Kubaczkova V,
Radova L, Filipova J, Jarkovsky J, Brozova L, Velichova R, Almasi
M, Penka M, et al: Deregulated expression of long non-coding RNA
UCA1 in multiple myeloma. Eur J Haematol. 99:223–233. 2017.
|
|
88
|
Zhao P and Zhao X: Baseline lncRNA PCAT1
high expression and its longitude increment during induction
therapy predict worse prognosis in multiple myeloma patients. J
Clin Lab Anal. 35:e239242021.
|
|
89
|
Dong H, Jiang S, Fu Y, Luo Y, Gui R and
Liu J: Upregulation of lncRNA NR_046683 serves as a prognostic
biomarker and potential drug target for multiple myeloma. Front
Pharmacol. 10:452019.
|
|
90
|
Zhou F and Guo L: Lncrna ANGPTL1-3 and its
target microRNA-30a exhibit potency as biomarkers for bortezomib
response and prognosis in multiple myeloma patients. Hematology.
27:596–602. 2022.
|
|
91
|
Xu H, Yin Q, Shen X and Ju S: Long
non-coding RNA CCAT2 as a potential serum biomarker for diagnosis
and prognosis of multiple myeloma. Ann Hematol. 99:2159–2171.
2020.
|
|
92
|
Dimopoulos MA, Barlogie B, Smith TL and
Alexanian R: High serum lactate dehydrogenase level as a marker for
drug resistance and short survival in multiple myeloma. Ann Intern
Med. 115:931–935. 1991.
|
|
93
|
Shouval R, Teper O, Fein JA, Danylesko I,
Shem Tov N, Yerushalmi R, Avigdor A, Vasilev E, Magen H, Nagler A,
et al: LDH and renal function are prognostic factors for long-term
outcomes of multiple myeloma patients undergoing allogeneic
hematopoietic stem cell transplantation. Bone Marrow Transplant.
55:1736–1743. 2020.
|
|
94
|
Palumbo A, Avet-Loiseau H, Oliva S,
Lokhorst HM, Goldschmidt H, Rosinol L, Richardson P, Caltagirone S,
Lahuerta JJ, Facon T, et al: Revised international staging system
for multiple myeloma: A report from international myeloma working
group. J Clin Oncol. 33:2863–2869. 2015.
|
|
95
|
Sedlarikova L, Bollova B, Radova L,
Brozova L, Jarkovsky J, Almasi M, Penka M, Kuglík P, Sandecká V,
Stork M, et al: Circulating exosomal long noncoding RNA PRINS-First
findings in monoclonal gammopathies. Hematol Oncol. 36:786–791.
2018.
|
|
96
|
Terpos E, Zamagni E, Lentzsch S, Drake MT,
García-Sanz R, Abildgaard N, Ntanasis-Stathopoulos I, Schjesvold F,
de la Rubia J, Kyriakou C, et al: Treatment of multiple
myeloma-related bone disease: Recommendations from the Bone Working
Group of the International Myeloma Working Group. Lancet Oncol.
22:e119–e130. 2021.
|
|
97
|
Dimopoulos MA, Sonneveld P, Leung N,
Merlini G, Ludwig H, Kastritis E, Goldschmidt H, Joshua D, Orlowski
RZ, Powles R, et al: International Myeloma working group
recommendations for the diagnosis and management of myeloma-related
renal impairment. J Clin Oncol. 34:1544–1557. 2016.
|
|
98
|
Sonneveld P, Avet-Loiseau H, Lonial S,
Usmani S, Siegel D, Anderson KC, Chng WJ, Moreau P, Attal M, Kyle
RA, et al: Treatment of multiple myeloma with high-risk
cytogenetics: A consensus of the International Myeloma Working
Group. Blood. 127:2955–2962. 2016.
|
|
99
|
Röllig C, Knop S and Bornhäuser M:
Multiple myeloma. Lancet. 385:2197–2208. 2015.
|
|
100
|
Yang LH, Du P, Liu W, An LK, Li J, Zhu WY,
Yuan S, Wang L and Zang L: LncRNA ANRIL promotes multiple myeloma
progression and bortezomib resistance by EZH2-mediated
epigenetically silencing of PTEN. Neoplasma. 68:788–797. 2021.
|
|
101
|
Paiva B, van Dongen JJ and Orfao A: New
criteria for response assessment: Role of minimal residual disease
in multiple myeloma. Blood. 125:3059–3068. 2015.
|
|
102
|
Gay F, Larocca A, Wijermans P, Cavallo F,
Rossi D, Schaafsma R, Genuardi M, Romano A, Liberati AM,
Siniscalchi A, et al: Complete response correlates with long-term
progression-free and overall survival in elderly myeloma treated
with novel agents: Analysis of 1175 patients. Blood. 117:3025–3031.
2011.
|
|
103
|
Ignatiadis M, Sledge GW and Jeffrey SS:
Liquid biopsy enters the clinic-implementation issues and future
challenges. Nat Rev Clin Oncol. 18:297–312. 2021.
|
|
104
|
Allegra A, Cancemi G, Mirabile G, Tonacci
A, Musolino C and Gangemi S: Circulating tumour cells, cell free
DNA and tumour-educated platelets as reliable prognostic and
management biomarkers for the liquid biopsy in multiple myeloma.
Cancers (Basel). 14:41362022.
|
|
105
|
Wallington-Beddoe CT and Mynott RL:
Prognostic and predictive biomarker developments in multiple
myeloma. J Hematol Oncol. 14:1512021.
|
|
106
|
Shen X, Zhang Y, Wu X, Guo Y, Shi W, Qi J,
Cong H, Wang X, Wu X and Ju S: Upregulated lncRNA-PCAT1 is closely
related to clinical diagnosis of multiple myeloma as a predictive
biomarker in serum. Cancer Biomark. 18:257–263. 2017.
|
|
107
|
Guan R, Wang W, Fu B, Pang Y, Lou Y and Li
H: Increased lncRNA HOTAIR expression promotes the chemoresistance
of multiple myeloma to dexamethasone by regulating cell viability
and apoptosis by mediating the JAK2/STAT3 signaling pathway. Mol
Med Rep. 20:3917–3923. 2019.
|
|
108
|
Yu W, Hurley J, Roberts D, Chakrabortty
SK, Enderle D, Noerholm M, Breakefield XO and Skog JK:
Exosome-based liquid biopsies in cancer: Opportunities and
challenges. Ann Oncol. 32:466–477. 2021.
|
|
109
|
Kumar SK, Rajkumar V, Kyle RA, van Duin M,
Sonneveld P, Mateos MV, Gay F and Anderson KC: Multiple myeloma.
Nat Rev Dis Primers. 3:170462017.
|
|
110
|
Kumar SK, Dispenzieri A, Lacy MQ, Gertz
MA, Buadi FK, Pandey S, Kapoor P, Dingli D, Hayman SR, Leung N, et
al: Continued improvement in survival in multiple myeloma: Changes
in early mortality and outcomes in older patients. Leukemia.
28:1122–1128. 2014.
|
|
111
|
Dimopoulos MA, Richardson PG, Moreau P and
Anderson KC: Current treatment landscape for relapsed and/or
refractory multiple myeloma. Nat Rev Clin Oncol. 12:42–54.
2015.
|
|
112
|
Obeng EA, Carlson LM, Gutman DM,
Harrington WJ Jr, Lee KP and Boise LH: Proteasome inhibitors induce
a terminal unfolded protein response in multiple myeloma cells.
Blood. 107:4907–4916. 2006.
|
|
113
|
Davis LN and Sherbenou DW: Emerging
therapeutic strategies to overcome drug resistance in multiple
myeloma. Cancers (Basel). 13:16862021.
|
|
114
|
Chen D, Frezza M, Schmitt S, Kanwar J and
Dou QP: Bortezomib as the first proteasome inhibitor anticancer
drug: Current status and future perspectives. Curr Cancer Drug
Targets. 11:239–253. 2011.
|
|
115
|
Pinto V, Bergantim R, Caires HR, Seca H,
Guimarães JE and Vasconcelos MH: Multiple myeloma: Available
therapies and causes of drug resistance. Cancers (Basel).
12:4072020.
|
|
116
|
Yang X, Ye H, He M, Zhou X, Sun N, Guo W,
Lin X, Huang H, Lin Y, Yao R, et al: LncRNA PDIA3P interacts with
c-Myc to regulate cell proliferation via induction of pentose
phosphate pathway in multiple myeloma. Biochem Biophys Res Commun.
498:207–213. 2018.
|
|
117
|
Pan Y, Zhang Y, Liu W, Huang Y, Shen X,
Jing R, Pu J, Wang X, Ju S, Cong H, et al: LncRNA H19
overexpression induces bortezomib resistance in multiple myeloma by
targeting MCL-1 via miR-29b-3p. Cell Death Dis. 10:1062019.
|
|
118
|
Che F, Ye X, Wang Y, Ma S and Wang X: Lnc
NEAT1/miR-29b-3p/Sp1 form a positive feedback loop and modulate
bortezomib resistance in human multiple myeloma cells. Eur J
Pharmacol. 891:1737522021.
|
|
119
|
Chauhan D and Anderson KC: Mechanisms of
cell death and survival in multiple myeloma (MM): Therapeutic
implications. Apoptosis. 8:337–343. 2003.
|
|
120
|
Wu Y and Wang H: LncRNA NEAT1 promotes
dexamethasone resistance in multiple myeloma by targeting
miR-193a/MCL1 pathway. J Biochem Mol Toxicol. 32:2018 View Article : Google Scholar
|
|
121
|
David A, Zocchi S, Talbot A, Choisy C,
Ohnona A, Lion J, Cuccuini W, Soulier J, Arnulf B, Bories JC, et
al: The long non-coding RNA CRNDE regulates growth of multiple
myeloma cells via an effect on IL6 signalling. Leukemia.
35:1710–1721. 2021.
|
|
122
|
Voorhees PM, Jakubowiak AJ, Kumar SK,
Kanapuru B, Baines AC, Bhatnagar V, Ershler R, Theoret MR, Gormley
NJ and Pazdur R: Perspectives on drug development in multiple
myeloma-looking forward to 2025. Clin Cancer Res. 28:23–26.
2022.
|
|
123
|
Gupta A, Andresen JL, Manan RS and Langer
R: Nucleic acid delivery for therapeutic applications. Adv Drug
Deliv Rev. 178:1138342021.
|
|
124
|
Kulkarni JA, Witzigmann D, Thomson SB,
Chen S, Leavitt BR, Cullis PR and van der Meel R: The current
landscape of nucleic acid therapeutics. Nat Nanotechnol.
16:630–643. 2021.
|
|
125
|
K C RB, Thapa B, Valencia-Serna J,
Aliabadi HM and Uludağ H: Nucleic acid combinations: A new frontier
for cancer treatment. J Control Release. 256:153–169. 2017.
|
|
126
|
Amodio N, Stamato MA, Juli G, Morelli E,
Fulciniti M, Manzoni M, Taiana E, Agnelli L, Cantafio MEG, Romeo E,
et al: Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene
expression of proteasome subunits and triggers anti-multiple
myeloma activity. Leukemia. 32:1948–1957. 2018.
|
|
127
|
Anashkina AA, Leberfarb EY and Orlov YL:
Recent trends in cancer genomics and bioinformatics tools
development. Int J Mol Sci. 22:121462021.
|
|
128
|
Zheng H, Talukder A, Li X and Hu H: A
systematic evaluation of the computational tools for lncRNA
identification. Brief Bioinform. 22:bbab2852021.
|
|
129
|
Duan Y, Zhang W, Cheng Y, Shi M and Xia
XQ: A systematic evaluation of bioinformatics tools for
identification of long noncoding RNAs. RNA. 27:80–98. 2021.
|
|
130
|
Herman AB, Tsitsipatis D and Gorospe M:
Integrated lncRNA function upon genomic and epigenomic regulation.
Mol Cell. 82:2252–2266. 2022.
|
|
131
|
Volders PJ, Anckaert J, Verheggen K,
Nuytens J, Martens L, Mestdagh P and Vandesompele J: LNCipedia 5:
Towards a reference set of human long non-coding RNAs. Nucleic
Acids Res. 47:D135–D139. 2019.
|
|
132
|
Ma L, Cao J, Liu L, Du Q, Li Z, Zou D,
Bajic VB and Zhang Z: LncBook: A curated knowledgebase of human
long non-coding RNAs. Nucleic Acids Res. 47:D128–D134. 2019.
|
|
133
|
Chen J, Zhang J, Gao Y, Li Y, Feng C, Song
C, Ning Z, Zhou X, Zhao J, Feng M, et al: LncSEA: A platform for
long non-coding RNA related sets and enrichment analysis. Nucleic
Acids Res. 49:D969–D980. 2021.
|
|
134
|
Li Z, Liu L, Jiang S, Li Q, Feng C, Du Q,
Zou D, Xiao J, Zhang Z and Ma L: LncExpDB: An expression database
of human long non-coding RNAs. Nucleic Acids Res. 49:D962–D968.
2021.
|
|
135
|
Xie F, Liu S, Wang J, Xuan J, Zhang X, Qu
L, Zheng L and Yang J: deepBase v3.0: Expression atlas and
interactive analysis of ncRNAs from thousands of deep-sequencing
data. Nucleic Acids Res. 49:D877–D883. 2021.
|
|
136
|
Li J, Han L, Roebuck P, Diao L, Liu L,
Yuan Y, Weinstein JN and Liang H: TANRIC: An interactive open
platform to explore the function of lncRNAs in cancer. Cancer Res.
75:3728–3737. 2015.
|
|
137
|
Zheng Y, Xu Q, Liu M, Hu H, Xie Y, Zuo Z
and Ren J: lnCAR: A Comprehensive resource for lncRNAs from cancer
arrays. Cancer Res. 79:2076–2083. 2019.
|
|
138
|
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q and
Dong D: LncRNADisease 2.0: An updated database of long non-coding
RNA-associated diseases. Nucleic Acids Res. 47:D1034–D1037.
2019.
|
|
139
|
Li Y, Li L, Wang Z, Pan T, Sahni N, Jin X,
Wang G, Li J, Zheng X, Zhang Y, et al: LncMAP: Pan-cancer atlas of
long noncoding RNA-mediated transcriptional network perturbations.
Nucleic Acids Res. 46:1113–1123. 2018.
|
|
140
|
Gong J, Liu W, Zhang J, Miao X and Guo AY:
lncRNASNP: A database of SNPs in lncRNAs and their potential
functions in human and mouse. Nucleic Acids Res. 43:D181–D186.
2015.
|
|
141
|
Zhi H, Li X, Wang P, Gao Y, Gao B, Zhou D,
Zhang Y, Guo M, Yue M, Shen W, et al: Lnc2Meth: A manually curated
database of regulatory relationships between long non-coding RNAs
and DNA methylation associated with human disease. Nucleic Acids
Res. 46:D133–D138. 2018.
|
|
142
|
Todoerti K, Ronchetti D, Puccio N,
Silvestris I, Favasuli V, Amodio N, Gentile M, Morabito F, Neri A
and Taiana E: Dissecting the biological relevance and clinical
impact of lncRNA MIAT in multiple myeloma. Cancers (Basel).
13:55182021.
|
|
143
|
Zhou M, Zhao H, Wang Z, Cheng L, Yang L,
Shi H, Yang H and Sun J: Identification and validation of potential
prognostic lncRNA biomarkers for predicting survival in patients
with multiple myeloma. J Exp Clin Cancer Res. 34:1022015.
|