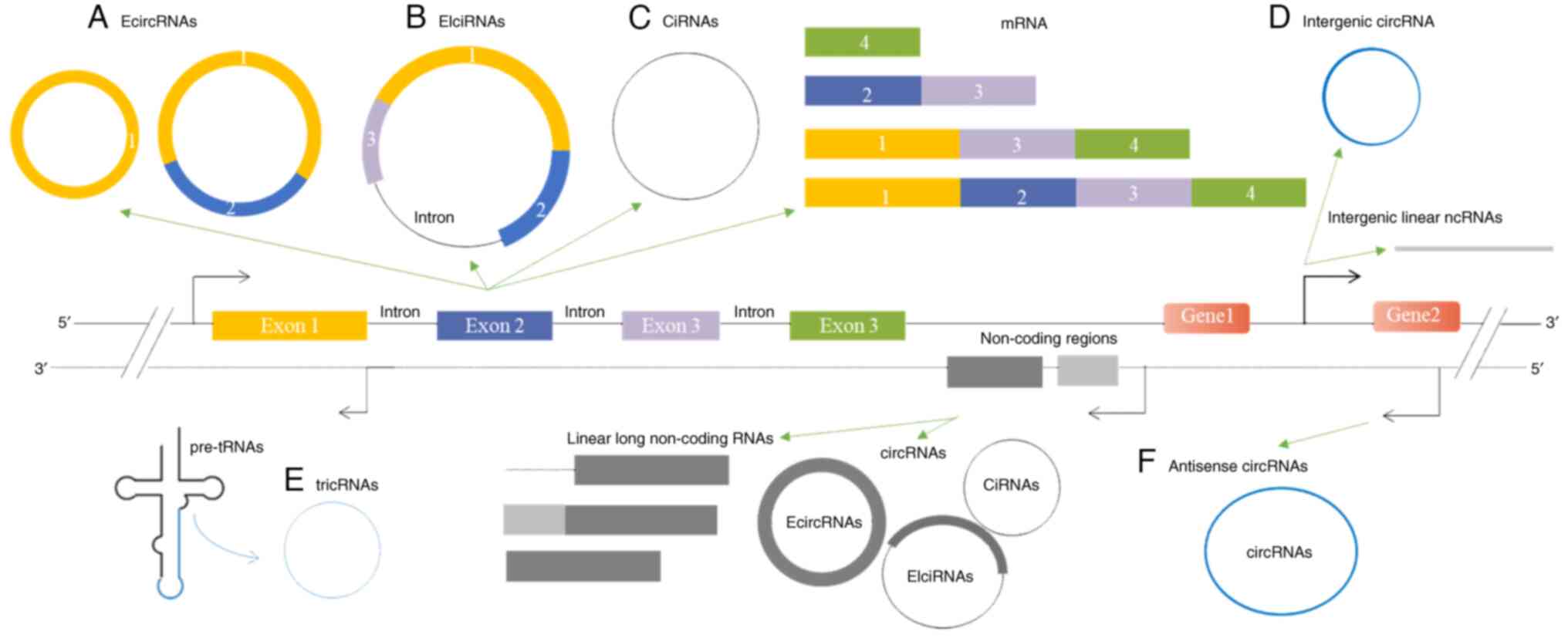
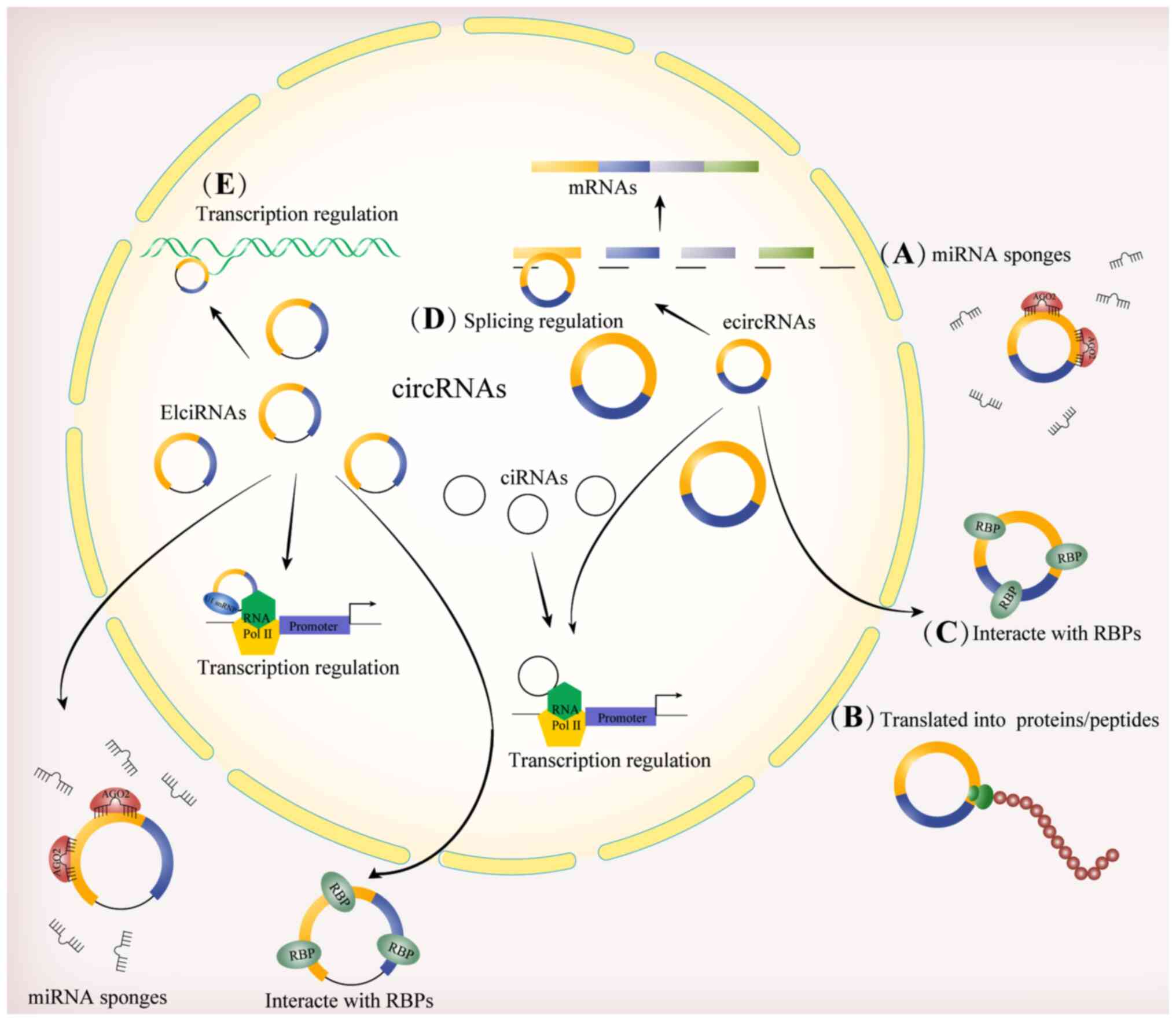
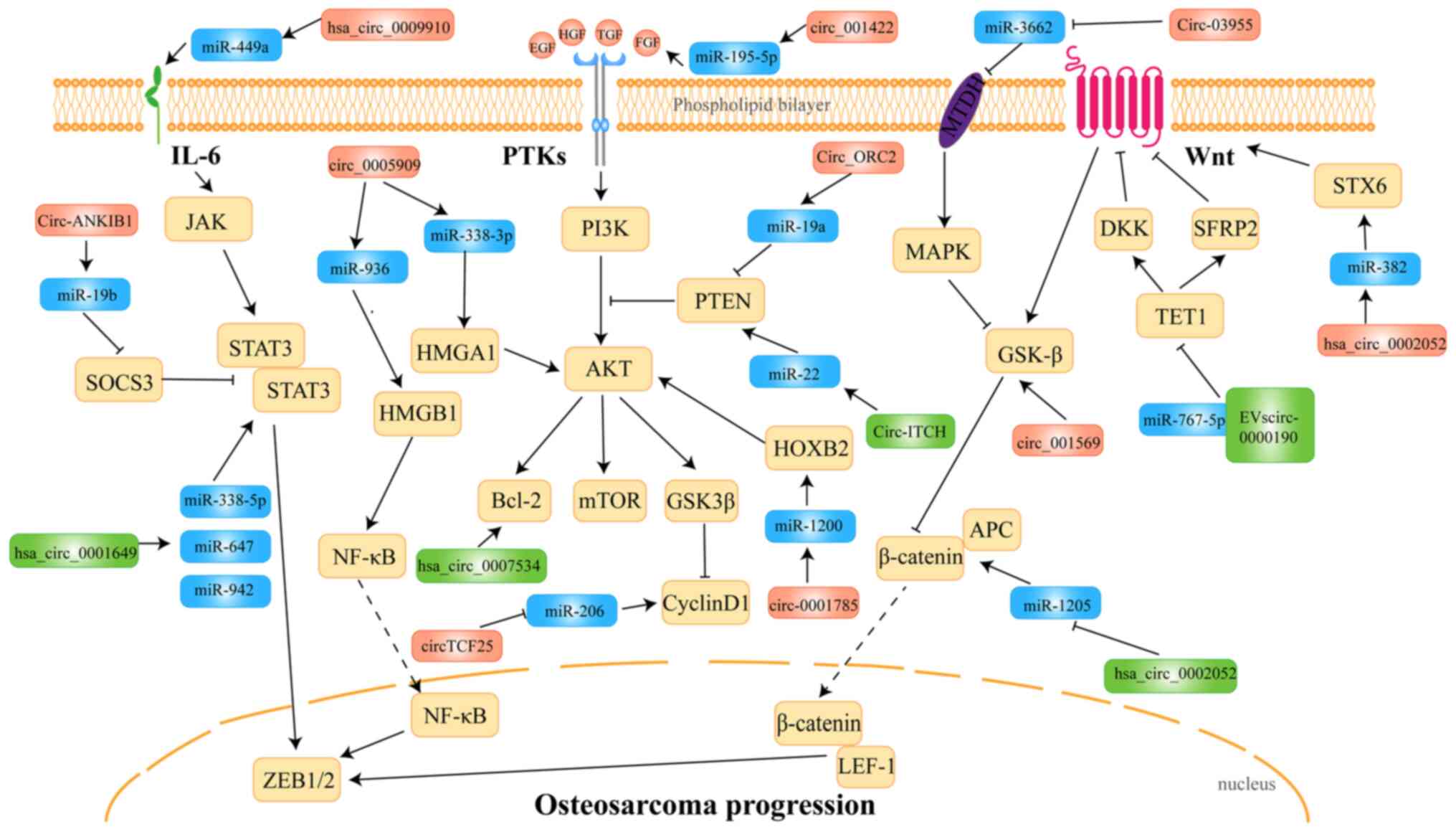
|
1
|
Link MP, Goorin AM, Miser AW, Green AA,
Pratt CB, Belasco JB, Pritchard J, Malpas JS, Baker AR, Kirkpatrick
JA, et al: The effect of adjuvant chemotherapy on relapse-free
survival in patients with osteosarcoma of the extremity. N Engl J
Med. 314:1600–1606. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Damron TA, Ward WG and Stewart A:
Osteosarcoma, chondrosarcoma, and Ewing's sarcoma: National Cancer
Data Base report. Clin Orthop Relat Res. 459:40–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meyers PA, Healey JH, Chou AJ, Wexler LH,
Merola PR, Morris CD, Laquaglia MP, Kellick MG, Abramson SJ and
Gorlick R: Addition of pamidronate to chemotherapy for the
treatment of osteosarcoma. Cancer. 117:1736–1744. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gaspar N, Occean B, Pacquement H, Bompas
E, Bouvier C, Brisse HJ, Castex MP, Cheurfa N, Corradini N, Delaye
J, et al: Results of methotrexate-etoposide-ifosfamide based
regimen (M-EI) in osteosarcoma patients included in the French
OS2006/sarcome-09 study. Eur J Cancer. 88:57–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sadykova LR, Ntekim AI, Muyangwa-Semenova
M, Rutland CS, Jeyapalan JN, Blatt N and Rizvanov AA: Epidemiology
and risk factors of osteosarcoma. Cancer Invest. 38:259–269. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kushlinskii NE, Fridman MV and Braga EA:
Molecular mechanisms and microRNAs in osteosarcoma pathogenesis.
Biochemistry (Mosc). 81:315–328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saraf AJ, Fenger JM and Roberts RD:
Osteosarcoma: Accelerating progress makes for a hopeful future.
Front Oncol. 8:42018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chong ZX, Yeap SK and Ho WY: Unraveling
the roles of miRNAs in regulating epithelial-to-mesenchymal
transition (EMT) in osteosarcoma. Pharmacol Res. 172:1058182021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang J, Yu X, Yan Y, Wang C and Wang W:
PI3K/Akt signaling in osteosarcoma. Clin Chim Acta. 444:182–192.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
ENCODE Project Consortium, . Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen L and Yang L: Regulation of circRNA
biogenesis. Rna Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang G, Liang M, Liu H, Huang J, Li P,
Wang C, Zhang Y, Lin Y and Jiang X: CircRNA hsa_circRNA_104348
promotes hepatocellular carcinoma progression through modulating
miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell
Death Dis. 11:10652020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng X, Huang M, Xing L, Yang R, Wang X,
Jiang R, Zhang L and Chen J: The circRNA circSEPT9 mediated by E2F1
and EIF4A3 facilitates the carcinogenesis and development of
triple-negative breast cancer. Mol Cancer. 19:732020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang N, Lu K, Qu H, Wang H, Chen Y, Shan
T, Ge X, Wei Y, Zhou P and Xia J: CircRBM33 regulates IL-6 to
promote gastric cancer progression through targeting miR-149.
Biomed Pharmacother. 125:1098762020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou P, Xie W, Huang HL, Huang RQ, Tian C,
Zhu HB, Dai YH and Li ZY: circRNA_100859 functions as an oncogene
in colon cancer by sponging the miR-217-HIF-1α pathway. Aging
(Albany NY). 12:13338–13353. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu H, Liu Y, Cheng P, Wang C, Liu Y, Zhou
W, Xu Y and Ji G: CircRNA_0000392 promotes colorectal cancer
progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin
Cancer Res. 39:2832020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arnaiz E, Sole C, Manterola L,
Iparraguirre L, Otaegui D and Lawrie CH: CircRNAs and cancer:
Biomarkers and master regulators. Semin Cancer Biol. 58:90–99.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X, Yang L and Chen L: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li F, Tang H, Zhao S, Gao X, Yang L and Xu
J: Circ-E-Cad encodes a protein that promotes the proliferation and
migration of gastric cancer via the TGF-β/Smad/C-E-Cad/PI3K/AKT
pathway. Mol Carcinog. 62:360–368. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang
H, Song X, Han D, Wang X, Liu Y, et al: circ-EIF6 encodes
EIF6-224aa to promote TNBC progression via stabilizing MYH9 and
activating the Wnt/beta-catenin pathway. Mol Ther. 30:415–430.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng J, Li G, Wang W, Stovall DB, Sui G
and Li D: Circular RNAs with protein-coding ability in oncogenesis.
Biochim Biophys Acta Rev Cancer. 1878:1889092023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sinha T, Panigrahi C, Das D and Chandra
PA: Circular RNA translation, a path to hidden proteome. Wiley
Interdiscip Rev RNA. 13:e16852022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Li X, Xu D, Chen X, Li S, Zhang L,
Chan MTV and Wu WKK: An update on the roles of circular RNAs in
osteosarcoma. Cell Prolif. 54:e129362021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kolakofsky D: Isolation and
characterization of Sendai virus DI-RNAs. Cell. 8:547–555. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Solé C and Lawrie CH: Circular RNAs and
cancer: Opportunities and challenges. Adv Clin Chem. 99:87–146.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Noto JJ, Schmidt CA and Matera AG:
Engineering and expressing circular RNAs via tRNA splicing. Rna
Biol. 14:978–984. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Talhouarne GJS and Gall JG: Lariat
intronic RNAs in the cytoplasm of vertebrate cells. Proc Natl Acad
Sci USA. 115:E7970–E7977. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lu Z, Filonov GS, Noto JJ, Schmidt CA,
Hatkevich TL, Wen Y, Jaffrey SR and Matera AG: Metazoan tRNA
introns generate stable circular RNAs in vivo. RNA. 21:1554–1565.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen X, Han P, Zhou T, Guo X, Song X and
Li Y: circRNADb: A comprehensive database for human circular RNAs
with protein-coding annotations. Sci Rep. 6:349852016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tang Z, Li X, Zhao J, Qian F, Feng C, Li
Y, Zhang J, Jiang Y, Yang Y, Wang Q and Li C: TRCirc: A resource
for transcriptional regulation information of circRNAs. Brief
Bioinform. 20:2327–2333. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao X, Cai Y and Xu J: Circular RNAs:
Biogenesis, mechanism, and function in human cancers. Int J Mol
Sci. 20:39262019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li B and Li X: Overexpression of
hsa_circ_0007534 predicts unfavorable prognosis for osteosarcoma
and regulates cell growth and apoptosis by affecting AKT/GSK-3β
signaling pathway. Biomed Pharmacother. 107:860–866. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu B, Yang T, Wang Z, Zhang Y, Liu S and
Shen M: CircRNA CDR1as/miR-7 signals promote tumor growth of
osteosarcoma with a potential therapeutic and diagnostic value.
Cancer Manag Res. 10:4871–4880. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang H, Yan J, Lang X and Zhuang Y:
Expression of circ_001569 is upregulated in osteosarcoma and
promotes cell proliferation and cisplatin resistance by activating
the Wnt/β-catenin signaling pathway. Oncol Lett. 16:5856–5862.
2018.PubMed/NCBI
|
|
49
|
Deng N, Li L, Gao J, Zhou J, Wang Y, Wang
C and Liu Y: Hsa_circ_0009910 promotes carcinogenesis by promoting
the expression of miR-449a target IL6R in osteosarcoma. Biochem
Biophys Res Commun. 495:189–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wu Z, Shi W and Jiang C: Overexpressing
circular RNA hsa_circ_0002052 impairs osteosarcoma progression via
inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis.
Biochem Biophys Res Commun. 502:465–471. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li X, Sun XH, Xu HY, Pan HS, Liu Y and He
L: Circ_ORC2 enhances the regulatory effect of miR-19a on its
target gene PTEN to affect osteosarcoma cell growth. Biochem
Biophys Res Commun. 514:1172–1178. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li S, Pei Y, Wang W, Liu F, Zheng K and
Zhang X: Circular RNA 0001785 regulates the pathogenesis of
osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2.
Cell Cycle. 18:1281–1291. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ren C, Liu J, Zheng B, Yan P, Sun Y and
Yue B: The circular RNA circ-ITCH acts as a tumour suppressor in
osteosarcoma via regulating miR-22. Artif Cells Nanomed Biotechnol.
47:3359–3367. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang Z, Deng M, Chen L, Wang W, Liu G, Liu
D, Han Z and Zhou Y: Circular RNA Circ-03955 promotes
epithelial-mesenchymal transition in osteosarcoma by regulating
miR-3662/metadherin pathway. Front Oncol. 10:5454602020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ding S, Zhang G, Gao Y, Chen S and Cao C:
Circular RNA hsa_circ_0005909 modulates osteosarcoma progression
via the miR-936/HMGB1 axis. Cancer Cell Int. 20:3052020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang PR, Ren J, Wan JS, Sun R and Li Y:
Circular RNA hsa_circ_0002052 promotes osteosarcoma via modulating
miR-382/STX6 axis. Hum Cell. 33:810–818. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Du YX, Guo LX, Pan HS, Liang YM and Li X:
Circ_ANKIB1 stabilizes the regulation of miR-19b on SOCS3/STAT3
pathway to promote osteosarcoma cell growth and invasion. Hum Cell.
33:252–260. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gao Y, Ma H, Gao Y, Tao K, Fu L, Ren R, Hu
X, Kou M, Chen B, Shi J and Wen Y: CircRNA Circ_0001721 promotes
the progression of osteosarcoma through miR-372-3p/MAPK7 axis.
Cancer Manag Res. 12:8287–8302. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu YP, Wan J, Long F, Tian J and Zhang C:
circPVT1 facilitates invasion and metastasis by regulating
miR-205-5p/c-FLIP axis in osteosarcoma. Cancer Manag Res.
12:1229–1240. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen J, Liu G, Wu Y, Ma J, Wu H, Xie Z,
Chen S, Yang Y, Wang S, Shen P, et al: CircMYO10 promotes
osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to
enhance the transcriptional activity of β-catenin/LEF1 complex via
effects on chromatin remodeling. Mol Cancer. 18:1502019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li S, Pei Y, Wang W, Liu F, Zheng K and
Zhang X: Extracellular nanovesicles-transmitted circular RNA
has_circ_0000190 suppresses osteosarcoma progression. J Cell Mol
Med. 24:2202–2214. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sun D and Zhu D: Circular RNA
hsa_circ_0001649 suppresses the growth of osteosarcoma cells via
sponging multiple miRNAs. Cell Cycle. 19:2631–2643. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yang B, Li L, Tong G, Zeng Z, Tan J, Su Z,
Liu Z, Lin J, Gao W, Chen J, et al: Circular RNA circ_001422
promotes the progression and metastasis of osteosarcoma via the
miR-195-5p/FGF2/PI3K/Akt axis. J Exp Clin Cancer Res. 40:2352021.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang C, Na N, Liu L and Qiu Y: CircRNA
hsa_circ_0005909 promotes cell proliferation of osteosarcoma cells
by targeting miR-338-3p/HMGA1 axis. Cancer Manag Res. 13:795–803.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bai Y, Li Y, Bai J and Zhang Y:
Hsa_circ_0004674 promotes osteosarcoma doxorubicin resistance by
regulating the miR-342-3p/FBN1 axis. J Orthop Surg Res. 16:5102021.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li S, Liu F, Zheng K, Wang W, Qiu E, Pei
Y, Wang S, Zhang J and Zhang X: CircDOCK1 promotes the
tumorigenesis and cisplatin resistance of osteogenic sarcoma via
the miR-339-3p/IGF1R axis. Mol Cancer. 20:1612021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shi Z, Wang K, Xing Y and Yang X:
CircNRIP1 encapsulated by bone marrow mesenchymal stem cell-derived
extracellular vesicles aggravates osteosarcoma by modulating the
miR-532-3p/AKT3/PI3K/AKT axis. Front Oncol. 11:6581392021.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Feng ZH, Zheng L, Yao T, Tao SY, Wei XA,
Zheng ZY, Zheng BJ, Zhang XY, Huang B, Liu JH, et al:
EIF4A3-induced circular RNA PRKAR1B promotes osteosarcoma
progression by miR-361-3p-mediated induction of FZD4 expression.
Cell Death Dis. 12:10252021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li JJ, Xiong MY, Sun TY, Ji CB, Guo RT, Li
YW and Guo HY: CircFAM120B knockdown inhibits osteosarcoma
tumorigenesis via the miR-1205/PTBP1 axis. Aging (Albany NY).
13:23831–23841. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gao Y, Liu C, Zhao X, Liu C, Bi W and Jia
J: hsa_circ_0000006 induces tumorigenesis through miR-361-3p
targeting immunoglobulin-like domains protein 1 (LRIG1) in
osteosarcoma. Ann Transl Med. 9:12422021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang M, Yu GY, Liu G and Liu WD: Circular
RNA circ_0002137 regulated the progression of osteosarcoma through
regulating miR-433-3p/IGF1R axis. J Cell Mol Med. 26:1806–1816.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang W, Wang J, Li Y and Zhao Y:
Circ_0051079 silencing inhibits the malignant phenotypes of
osteosarcoma cells by the TRIM66/Wnt/β-catenin pathway in a
miR-625-5p-dependent manner. J Bone Oncol. 35:1004362022.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Liu Y, Yuan J, Zhang Q, Ren Z, Li G and
Tian R: Circ_0016347 modulates proliferation, migration, invasion,
cell cycle, and apoptosis of osteosarcoma cells via the
miR-661/IL6R axis. Autoimmunity. 55:264–274. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu Y, Qiu G, Luo Y, Li S, Xu Y, Zhang Y,
Hu J, Li P, Pan H and Wang Y: Circular RNA ROCK1, a novel circRNA,
suppresses osteosarcoma proliferation and migration via altering
the miR-532-5p/PTEN axis. Exp Mol Med. 54:1024–1037. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Peng L, Liu Q, Wu T, Li P, Cai Y, Wei X,
Zeng Y, Ye J, Chen P, Huang J and Lin H: Hsa_circ_0087302, a
circular RNA, affects the progression of osteosarcoma via the
Wnt/β-catenin signaling pathway. Int J Med Sci. 19:1377–1387. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang D, Wang Y, Wang H, Yang Y, Li L, Liu
Y and Yin X: Hsa_circ_0000591 drives osteosarcoma glycolysis and
progression by sequestering miR-194-5p and elevating HK2
expression. Clin Exp Pharmacol Physiol. 50:463–475. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Z, Zheng L, Yang L, Chen D, Ren G, Yan
X and Pu J: Hsa_circ_0020378 targets miR-556-5p/MAPK1 to regulate
osteosarcoma cell proliferation and migration. Gene.
856:1471352023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gong Z, Shen P, Wang H, Zhu J, Liang K,
Wang K, Mi Y, Shen S, Fang X and Liu G: A novel circular RNA
circRBMS3 regulates proliferation and metastasis of osteosarcoma by
targeting miR-424-eIF4B/YRDC axis. Aging (Albany NY). 15:1564–1590.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Trang NTN, Lai CY, Tsai HC, Huang YL, Liu
SC, Tsai CH, Fong YC, Tzeng HE and Tang CH: Apelin promotes
osteosarcoma metastasis by upregulating PLOD2 expression via the
Hippo signaling pathway and hsa_circ_0000004/miR-1303 axis. Int J
Biol Sci. 19:412–425. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Luo Y, Yang B, Yuan X and Zheng J:
Silencing circUSP48 suppresses osteosarcoma progression by
regulating the miR-335/smad nuclear interacting protein 1 pathway.
J Clin Lab Anal. 37:e248282023. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Xu L, Duan J, Li M, Zhou C and Wang Q:
Circ_0000253 promotes the progression of osteosarcoma via the
miR-1236-3p/SP1 axis. J Pharm Pharmacol. 75:227–235. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Liu T, Song Z and Gai Y: Circular RNA
circ_0001649 acts as a prognostic biomarker and inhibits NSCLC
progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys
Res Commun. 503:1503–1509. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Jiang Y, Hou J, Zhang X, Xu G, Wang Y,
Shen L, Wu Y, Li Y and Yao L: Circ-XPO1 upregulates XPO1 expression
by sponging multiple miRNAs to facilitate osteosarcoma cell
progression. Exp Mol Pathol. 117:1045532020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Imataka H, Olsen HS and Sonenberg N: A new
translational regulator with homology to eukaryotic translation
initiation factor 4G. EMBO J. 16:817–825. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Morino S, Imataka H, Svitkin YV, Pestova
TV and Sonenberg N: Eukaryotic translation initiation factor 4E
(eIF4E) binding site and the middle one-third of eIF4GI constitute
the core domain for cap-dependent translation, and the C-terminal
one-third functions as a modulatory region. Mol Cell Biol.
20:468–477. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liberman N, Gandin V, Svitkin YV, David M,
Virgili G, Jaramillo M, Holcik M, Nagar B, Kimchi A and Sonenberg
N: DAP5 associates with eIF2β and eIF4AI to promote internal
ribosome entry site driven translation. Nucleic Acids Res.
43:3764–3775. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Lamphear BJ, Kirchweger R, Skern T and
Rhoads RE: Mapping of functional domains in eukaryotic protein
synthesis initiation factor 4G (eIF4G) with picornaviral proteases.
Implications for cap-dependent and cap-independent translational
initiation. J Biol Chem. 270:21975–21983. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hellen CU and Sarnow P: Internal ribosome
entry sites in eukaryotic mRNA molecules. Gene Dev. 15:1593–1612.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
He L, Man C, Xiang S, Yao L, Wang X and
Fan Y: Circular RNAs' cap-independent translation protein and its
roles in carcinomas. Mol Cancer. 20:1192021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Liu Y, Li Z, Zhang M, Zhou H, Wu X, Zhong
J, Xiao F, Huang N, Yang X, Zeng R, et al: Rolling-translated EGFR
variants sustain EGFR signaling and promote glioblastoma
tumorigenicity. Neuro Oncol. 23:743–756. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: circFBXW7 inhibits malignant progression by
sponging miR-197-3p and encoding a 185-aa protein in
triple-negative breast cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang
J, Xia L, Yin Q, Zou B, Zheng J, et al: A novel protein encoded by
circFNDC3B inhibits tumor progression and EMT through regulating
Snail in colon cancer. Mol Cancer. 19:712020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Weigelt CM, Sehgal R, Tain LS, Cheng J,
Eßer J, Pahl A, Dieterich C, Grönke S and Partridge L: An
insulin-sensitive circular RNA that regulates lifespan in
Drosophila. Mol Cell. 79:268–279.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Duan JL, Chen W, Xie JJ, Zhang ML, Nie RC,
Liang H, Mei J, Han K, Xiang ZC, Wang FW, et al: A novel peptide
encoded by N6-methyladenosine modified circMAP3K4 prevents
apoptosis in hepatocellular carcinoma. Mol Cancer. 21:932022.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Song R, Ma S, Xu J, Ren X, Guo P, Liu H,
Li P, Yin F, Liu M, Wang Q, et al: A novel polypeptide encoded by
the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR.
Mol Cancer. 22:162023. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo
X, Hossain MT, Zhu X, Du K, Hu F, et al: A novel protein
AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin
signaling pathway to promote gastric cancer progression. Mol
Cancer. 20:1582021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S,
Xuan Z, Xie L, Qiu S, He Z, et al: A novel protein encoded by
circMAPK1 inhibits progression of gastric cancer by suppressing
activation of MAPK signaling. Mol Cancer. 20:662021. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang C, Zhou X, Geng X, Zhang Y, Wang J,
Wang Y, Jing J, Zhou X and Pan W: Circular RNA hsa_circ_0006401
promotes proliferation and metastasis in colorectal carcinoma. Cell
Death Dis. 12:4432021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Liang ZX, Liu HS, Xiong L, Yang X, Wang
FW, Zeng ZW, He XW, Wu XR and Lan P: A novel NF-κB regulator
encoded by circPLCE1 inhibits colorectal carcinoma progression by
promoting RPS3 ubiquitin-dependent degradation. Mol Cancer.
20:1032021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X,
Zhong J, Zhao Z, Zhao K, Liu D, et al: Circular RNA-encoded
oncogenic E-cadherin variant promotes glioblastoma tumorigenicity
through activation of EGFR-STAT3 signalling. Nat Cell Biol.
23:278–291. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H,
Huang N, Yang X, Xiao F, Liu D, et al: A novel tumor suppressor
protein encoded by circular AKT3 RNA inhibits glioblastoma
tumorigenicity by competing with active phosphoinositide-dependent
kinase-1. Mol Cancer. 18:1312019. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang X, Jian W, Luo Q and Fang L:
CircSEMA4B inhibits the progression of breast cancer by encoding a
novel protein SEMA4B-211aa and regulating AKT phosphorylation. Cell
Death Dis. 13:7942022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412.
2017.PubMed/NCBI
|
|
110
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Conn VM, Hugouvieux V, Nayak A, Conos SA,
Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta
C and Conn SJ: A circRNA from SEPALLATA3 regulates splicing of its
cognate mRNA through R-loop formation. Nat Plants. 3:170532017.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Liu CX and Chen LL: Circular RNAs:
Characterization, cellular roles, and applications. Cell.
185:2016–2034. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Hirano T, Ishihara K and Hibi M: Roles of
STAT3 in mediating the cell growth, differentiation and survival
signals relayed through the IL-6 family of cytokine receptors.
Oncogene. 19:2548–2556. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Liu Y, Liao S, Bennett S, Tang H, Song D,
Wood D, Zhan X and Xu J: STAT3 and its targeting inhibitors in
osteosarcoma. Cell Prolif. 54:e129742021. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
DiDonato JA, Mercurio F and Karin M: NF-κB
and the link between inflammation and cancer. Immunol Rev.
246:379–400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Dolcet X, Llobet D, Pallares J and
Matias-Guiu X: NF-kB in development and progression of human
cancer. Virchows Arch. 446:475–482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Sims GP, Rowe DC, Rietdijk ST, Herbst R
and Coyle AJ: HMGB1 and RAGE in inflammation and cancer. Annu Rev
Immunol. 28:367–388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Porta C, Paglino C and Mosca A: Targeting
PI3K/Akt/mTOR signaling in cancer. Front Oncol. 4:642014.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Gill J and Gorlick R: Advancing therapy
for osteosarcoma. Nat Rev Clin Oncol. 18:609–624. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Xi Y and Chen Y: PTEN plays dual roles as
a tumor suppressor in osteosarcoma cells. J Cell Biochem.
118:2684–2692. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Liau SS, Jazag A and Whang EE: HMGA1 is a
determinant of cellular invasiveness and in vivo metastatic
potential in pancreatic adenocarcinoma. Cancer Res. 66:11613–11622.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Cai Y, Cai T and Chen Y: Wnt pathway in
osteosarcoma, from oncogenic to therapeutic. J Cell Biochem.
115:625–631. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
McQueen P, Ghaffar S, Guo Y, Rubin EM, Zi
X and Hoang BH: The Wnt signaling pathway: Implications for therapy
in osteosarcoma. Expert Rev Anticanc. 11:1223–1232. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Guo Y, Zi X, Koontz Z, Kim A, Xie J,
Gorlick R, Holcombe RF and Hoang BH: Blocking Wnt/LRP5 signaling by
a soluble receptor modulates the epithelial to mesenchymal
transition and suppresses met and metalloproteinases in
osteosarcoma Saos-2 cells. J Orthop Res. 25:964–971. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Duan H, Yan Z, Chen W, Wu Y, Han J, Guo H
and Qiao J: TET1 inhibits EMT of ovarian cancer cells through
activating Wnt/β-catenin signaling inhibitors DKK1 and SFRP2.
Gynecol Oncol. 147:408–417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhou X, Natino D, Qin Z, Wang D, Tian Z,
Cai X, Wang B and He X: Identification and functional
characterization of circRNA-0008717 as an oncogene in osteosarcoma
through sponging miR-203. Oncotarget. 9:22288–22300. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
Overexpressed circPVT1, a potential new circular RNA biomarker,
contributes to doxorubicin and cisplatin resistance of osteosarcoma
cells by regulating ABCB1. Int J Biol Sci. 14:321–330. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lei S and Xiang L: Up-regulation of
circRNA hsa_circ_0003074 expression is a reliable diagnostic and
prognostic biomarker in patients with osteosarcoma. Cancer Manag
Res. 12:9315–9325. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Lin Y, Jewell BE, Gingold J, Lu L, Zhao R,
Wang LL and Lee DF: Osteosarcoma: Molecular pathogenesis and iPSC
modeling. Trends Mol Med. 23:737–755. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Corre I, Verrecchia F, Crenn V, Redini F
and Trichet V: The osteosarcoma microenvironment: A complex but
targetable ecosystem. Cells. 9:9762020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.PubMed/NCBI
|
|
133
|
Xi Y, Fowdur M, Liu Y, Wu H, He M and Zhao
J: Differential expression and bioinformatics analysis of circRNA
in osteosarcoma. Biosci Rep. 39:BSR201815142019. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Yanbin Z and Jing Z: CircSAMD4A
accelerates cell proliferation of osteosarcoma by sponging miR-1244
and regulating MDM2 mRNA expression. Biochem Biophys Res Commun.
516:102–111. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Xie C, Chen B, Wu B, Guo J, Shi Y and Cao
Y: CircSAMD4A regulates cell progression and epithelial-mesenchymal
transition by sponging miR-342-3p via the regulation of FZD7
expression in osteosarcoma. Int J Mol Med. 46:107–118.
2020.PubMed/NCBI
|