Introduction
Malignant neoplasms collectively are the second
leading cause of mortality globally, imposing a significant
economic burden on public healthcare systems around the world
(1). By 2022, China had ~4.82
million new cancer cases, with 3.21 million cases of
cancer-associated mortality (2).
Compared with USA and UK, China has a lower cancer incidence rate
but a higher cancer mortality rate (3). Although there had been a gradual
decline in the incidence of gastric cancer (GC), hepatocellular
carcinoma (HCC) and esophageal cancers, the morbidity and mortality
rates of colorectal cancer (CRC), prostate and breast cancers have
seen a progressive rise (4).
Although notable advances in surgical, radiotherapeutic
chemotherapeutic, targeted therapeutic, immunotherapeutic and
multimodality therapeutic methods have all led to significant
progress in enhancing treatment efficacy (5), the overall therapeutic effect remains
poor in the long-term. Therefore, further studies are required to
explore the mechanisms that regulate tumor development and
responses to therapy.
Non-coding RNAs form a family that includes
microRNAs (miRNA or miR), long non-coding RNAs (lncRNAs) and
circular RNAs, which are classified according to their nucleotide
length (6,7). In particular, lncRNAs are typically
>200 nucleotides in length and can form conserved secondary
structures, although the majority of which do not encode proteins
(8). lncRNAs are structurally
similar to mRNAs, since they also contain promoters and can undergo
splicing, capping, polyadenylation and editing (9). In addition, their expression is
regulated in a time- and space-specific manner (9). lncRNAs have been reported to regulate
gene expression upstream of various biological processes, including
cell proliferation and metabolism (10–13).
lncRNAs can be classified into the following six categories based
on their location on the genome: i) Intergenic lncRNAs, which are
transcripts located between two protein-coding genes; ii) intronic
lncRNAs, which are located within an intron of a coding gene; iii)
bidirectional lncRNAs, which are located within 1 kb of promoters
in the opposite direction from the protein-coding transcript; iv)
enhancer lncRNAs, which are located in the enhancer regions; v)
sense lncRNAs, located in a small segment of a coding gene, which
is the overlapping region of one or several introns or exons; and
vi) antisense-lncRNAs (AS-lncRNAs), which are transcribed from the
antisense strand of protein-coding genes and overlap one or several
introns and exons of the sense sequence (14). AS-lncRNAs are transcribed from the
opposite strand of a protein or non-protein-coding gene (15). AS-lncRNAs have also been previously
observed to serve an important role in the development and
progression of malignancies (16–18).
In the present review, the mechanism underlying the regulation of
target gene expression by AS-lncRNAs was described, before its
downstream consequences in tumor occurrence and development were
summarized. Finally, the possible association between AS-lncRNA
expression and therapy resistance in cancers was reported.
Mechanism underlying antisense-long-stranded
non-coding RNA (AS-lncRNA)-mediated regulation of gene
expression
AS-lncRNAs typically do not translate into proteins
that can exert biological effects, but it can control gene
expression through other means (8). AS-lncRNAs can mainly regulate the
expression of target genes through eight methods (Fig. 1), which are summarized in this
section.
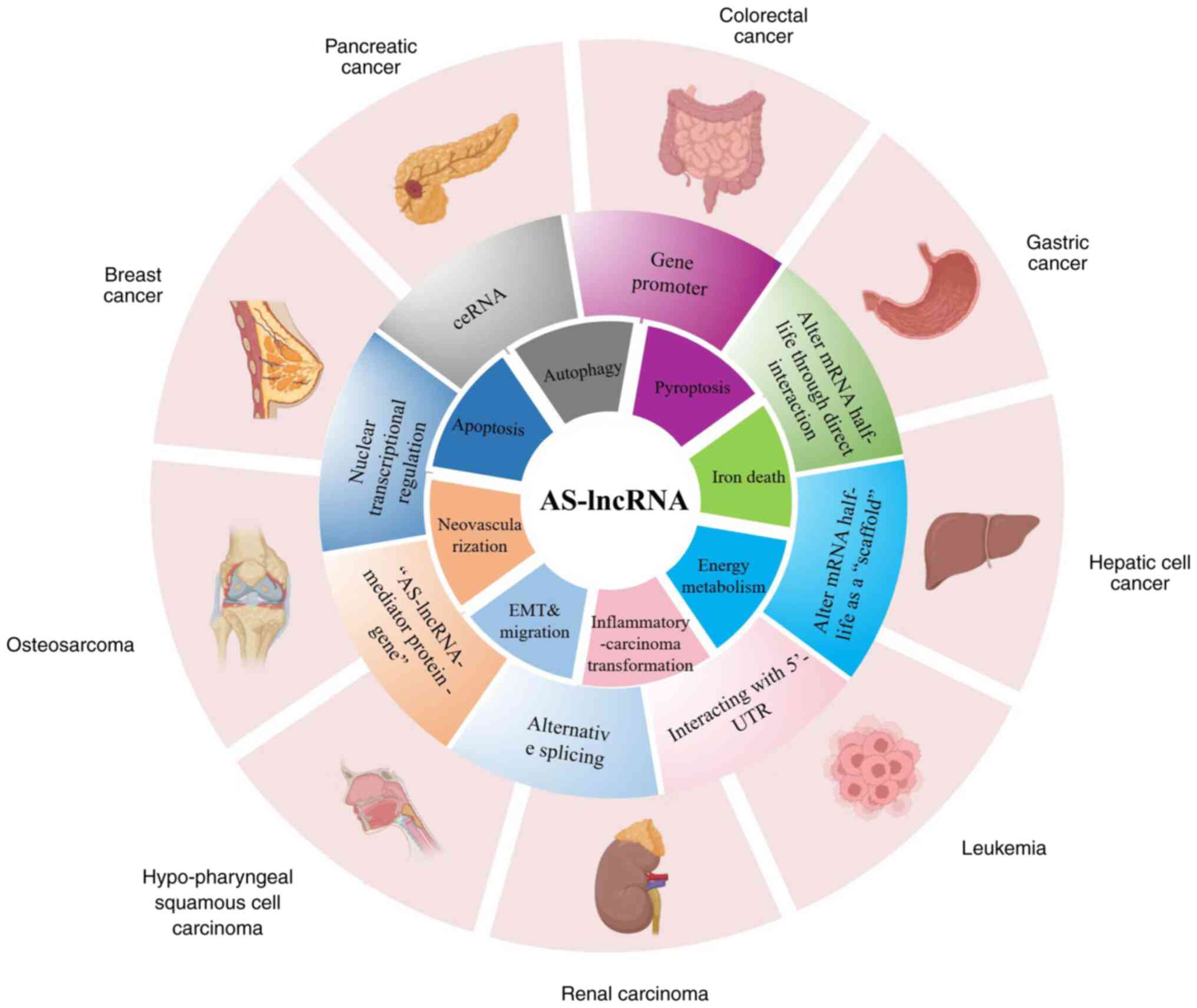 | Figure 1.AS-lncRNAs can regulate tumor gene
expression through endogenous competition mechanism, promoter
interaction, direct interaction with mRNA, acting as ‘scaffold’ to
change the half-life of mRNA, interaction with 5-UTR, regulation of
meaningful mRNA, positive feedback loop of ‘AS-lncRNA-mediator
protein-meaning gene’ model, transcriptional regulation in nucleus
and so on. AS-lncRNAs can regulate tumor by regulating tumor cell
death and proliferation, energy metabolism, inflammatory cancer
transformation, invasion and migration, and regulation of tumor
neovascularization. AS-lncRNA, antisense-long-stranded non-coding
RNA; UTR, untranslated regions; ceRNA, competing endogenous RNA;
EMT, epithelial-mesenchymal transition. |
Through competing endogenous RNAs
(ceRNAs)
A number of studies have previously reported that
there is a regulatory relationship between AS-lncRNAs and miRNAs.
Specifically, AS-lncRNAs can negatively regulate the expression of
miRNAs upstream of the occurrence and development of tumors through
the ceRNA mechanism (19), namely
by serving as an endogenous miRNA sponge (20). Si et al (21) found that zinc finger protein
(ZNF)561-AS1 expression was increased in tissues from patients with
CRC and mediated carcinogenic effects in CRC, which predicted poor
prognosis in patients. Serine and arginine rich splicing factor 6
(SRSF6) is a proto-oncogene that was also found to be overexpressed
in CRC. In addition, ZNF561-AS1 was found to be an upstream
post-transcriptional regulator of SRSF6. Using Starbase and
Targetscan bioinformatics analyses, ZNF561-AS1 and SRSF6 were found
to have potential binding sites for miR-26a-5p and miR-128-3p. The
results of agonaute 2 (ago2)-RNA immunoprecipitation (RIP) assay
and luciferase reporter assays revealed that ZNF561-AS1 was a ceRNA
of miR-26a-5p and miR-128-3p. Further luciferase reporter assays
later found that ZNF561-AS1 knockdown significantly inhibited SRSF6
wild-type plasmid luciferase activity. However, transfection with
miR-26a-5p and/or miR-128-3p inhibitors was able to either
partially or completely rescue such luciferase activity in CRC
cells with ZNF561-AS1 expression knocked down. This led to the
conclusion that ZNF561-AS1 can promote the expression of SRSF6 by
functioning as a sponge for miR-26a-5p and miR-128-3p, thereby
promoting the proliferation of CRC cells. In a different study,
Shuai et al (22) found
that lncRNA motor neuron and pancreas homeobox 1 (MNX1)-AS1
expression was increased in GC tissues, suggesting poor patient
prognosis. Subsequent ago2-RIP assay results suggested that
MNX1-AS1 may serve as a ceRNA. RNA-sequencing and reverse
transcription-quantitative PCR assays found that Bcl-2 expression
was significantly decreased in GC cells with MNX1-AS1 expression
knocked down. MiRanda prediction then revealed that Bcl-2 was
likely to be candidate target to bind to miR-6785-5p. Luciferase
reporter gene assay then found miR-6785-5p to bind to the Bcl-2
3′untranslated region (3′UTR), suggesting that miR-6785-5p can
target Bcl-2 mRNA. Results from both bioinformatics analyses and
luciferase reporter assays suggested that lncRNA MNX1-AS1 can also
serve as a ceRNA of miR-6785-5p to upregulate the expression of
Bcl-2 in GC cells by sponging miR-6785-5p, thereby promoting GC
progression.
Interacting with promoters
A number of pathways that can promote the
co-expression of AS-lncRNAs and nearby protein-coding genes have
been reported (23,24). On the transcriptional level, one
possible mechanism is that AS-lncRNAs share a promoter with
neighboring protein-coding genes (25), where they can regulate the H3K4me3
enrichment state in the promoter regions of genes of interest to in
turn modulate their expression (26). Bartl et al (27) previously reported that the lncRNA
hedgehog-interacting protein 1 (HHIP)-AS1 and HHIP share a
promoter, whereby promoter activity was negatively regulated by DNA
methylation. By contrast, Shuai et al (22) found that lncRNA MNX1-AS1 was
overexpressed in GC tissues, which was not conducive to GC
prognosis. Gene Ontology analysis revealed that the potential
targets of MNX1-AS1 are possibly involved in various biological
processes, such as signal transduction, cell proliferation and cell
death. Chromatin immunoprecipitation (ChIP) experiments
subsequently revealed that enhancer of zeste homolog 2 (EZH2) can
bind to the promoter region of B-cell translocation gene
anti-proliferation factor 2 (BTG2) to induce H3K27me3 modification
in GC cells, which inhibits BTG2 transcription. Subsequent rescue
experiments revealed that BTG2 can regulate the proliferation,
migration and invasion of GC cells induced by MNX1-AS1. These
results suggested that lncRNA MNX1-AS1 can suppress the expression
of BTG2 in GC through EZH2-mediated H3K27me3 to promote GC
progression.
Direct interactions alter the
half-life of mRNAs
AS-lncRNAs can regulate the stability of sense RNAs
by forming RNA duplexes with them (28). Pan and Xie (29) previously observed through
subcellular fractionation and fluorescence in situ
hybridization assays that the lncRNA forkhead box C2 (FOXC2)-AS1 is
mainly located in the nucleus where its expression is increased in
CRC tissues, which indicates poor patient prognosis. FOXC2 is the
closest gene to FOXC2-AS1, where their sequences overlap. The
expression of FOXC2 and FOXC2-AS1 in CRC tissues was also found to
be positively correlated. FOXC2-AS1 was subsequently found to
positively regulate the expression of FOXC2. Reverse
transcription-quantitative PCR (RT-qPCR) assay results suggested
that FOXC2-AS1 can enhance the stability of FOXC2 mRNA. According
to the site and gene structure, exon 1 of FOXC2-AS1 was completely
complementary to FOXC2. RNase protection assay then revealed that
FOXC2-AS1 and FOXC2 formed a double-stranded RNA structure, which
was confirmed by RNA pull-down assay. This resulted in the
conclusion that FOXC2-AS1 and FOXC2, and hence AS-lncRNAs and
mRNAs, can form double-stranded structures, which enhanced the
stability of FOXC2 mRNA (27). In
addition, Zhao et al (30)
found that metastasis-associated in colon cancer protein 1
(MACC1)-AS1 expression was increased in GC tissues, indicating a
poor prognosis. MACC1-AS1 was found to promote metabolic plasticity
by regulating MACC1 expression, thereby promoting GC progression.
Through bioinformatics analysis, it was found that MACC1-AS1
contained the binding site of MACC1 mRNA. MACC1-AS1 was
predominantly found to localize in the cytoplasm, allowing for the
regulation of mRNA stability. Subsequent RT-qPCR analysis found
that MACC1-AS1 significantly reduced the degradation of MACC1 mRNA,
whilst affinity pull-down assays confirmed the direct interaction
between MACC1-AS1 and MACC1. These findings suggested that
MACC1-AS1 promoted mRNA stability through direct binding, thereby
promoting GC progression.
Functioning as a ‘scaffold’ to
regulating the half-life of mRNA
AS-lncRNAs can serve as scaffolds to interact with
various proteins to form RNA/protein complexes (31). Zhu et al (32) found that Rho GTPase-activating
protein 5 (ARHGAP5)-AS1 expression was increased in GC cells.
Clinical data from The Cancer Genome Atlas (TCGA) database found
that higher ARHGAP5-AS1 expression levels were associated with
shorter overall survival. m6A modifications were previously found
to regulate mRNA stability through RNA-binding proteins (33). Zhu et al (32) used RMBase and SRAMP analyses to
show that there may be m6A modification in ARHGAP5 mRNA. When
ARHGAP5-AS1 was overexpressed, the binding of both m6A modification
and methyltransferase 3, N6-adenosine-methyltransferase complex
catalytic subunit (METTL3) to ARHGAP5 mRNA were found to be
elevated. Taken together, ARHGAP5-AS1 may recruit METTL3 to promote
the m6A modification of ARHGAP5 mRNA, thereby stabilizing ARHGAP5
mRNA in the cytoplasm and promoting GC chemoresistance.
In summary, AS-lncRNAs can regulate gene expression
through a variety of mechanisms, including ceRNA mechanisms,
interaction with promoters and direct interaction or indirect
interaction with mRNAs by serving as ‘scaffolds’ to alter its
half-life.
Interacting with 5′-UTRs
Tran et al (34) reported an AS-lncRNA, AS-putative
RNA-binding protein 15 (RBM15). The sequences of RBM15 and
RBM15-AS1 both were first analyzed, which found that exon 1 of
RBM15-AS1 coincided with the 5′-UTR of RBM15. The main functional
region of lncRNA was then searched further, before full-length and
multiple truncated versions were constructed. It was found that the
5-end of RBM15 was the main functional region, which could promote
differentiation. The relationship between RBM15-AS1 and RBM15 was
then verified experimentally, revealing that the shortest
functional region exon 1 of RBM15-AS 1 could promote the
translation of RBM15 without significantly affecting its mRNA
levels. By constructing a dual-luciferase reporter system with the
5′-UTR of RBM15 and constructs encoding the different truncated
versions of the lncRNAs, it was found that the region overlapping
the 5′-UTR of AS-RBM15 and RBM15 was responsible for the
upregulation of RBM15 protein translation.
Exerting regulatory effects on sense
mRNAs
Alternative splicing is a common phenomenon during
post-transcriptional regulation, which produces different mRNA
variants and unique isoforms of the same protein (35,36).
Yuan et al (37) previously
identified an oncofetal splicing factor, muscleblind-like protein 3
(MBNL3), which promoted HCC tumorigenesis and suggested poor
prognosis in patients. Transcriptomic analysis revealed that MBNL3
induced lncRNA PXN-AS1 exon 4 encapsulation. Transcripts of lncRNA
PXN-AS1 with exon 4 deleted were found to bind to the coding
sequence of PXN mRNA, leading to translation elongation factor
separation from the PXN mRNA to inhibit PXN translation. By
contrast, exon 4-containing transcripts preferentially bind to the
3′ UTRs of PXN mRNA to protect the PXN mRNA from miR-24-ago2
complex-induced degradation, which in turn increases PXN
expression. Through this mechanism, MBNL3 was concluded to
upregulate PXN expression by promoting exon 4 inclusion, where PXN
mediated the pro-tumor effects of MBNL3 in HCC.
Positive feedback loop of
‘AS-lncRNA/protein/gene of interest’ model
AS-lncRNA/protein/gene of interest can form stable
complexes to continuously promote the expression of AS-lncRNAs and
sense genes (38). Luo et
al (39) previously found that
zinc finger E-box binding homeobox 1 (ZEB1)-AS1 and ZEB1 expression
is increased in triple-negative breast cancer (TNBC), both of which
demonstrated direct binding to embryonic lethal, abnormal vision,
drosophila, homolog-like 1 (ELAVL1) according to pull-down silver
staining and RIP experiments. Specifically, ZEB1-AS1 directed
ELAVL1 to the ZEB1 mRNA to protect it from degradation. ChIP
experiments then demonstrated that ZEB1 exhibited potent binding
affinity to part 1 of the ZEB1-AS1 promoter. ZEB1 could bind to the
promoter of ZEB1-AS1 to promote the activation of ZEB1-AS1
expression, which was also verified by luciferase reporter assays.
This suggested that ZEB1-AS1 interacted with ZEB1, which then
formed a positive feedback loop with ELAVL1 to amplify the
promotion of TNBC.
Transcriptional regulation of
AS-lncRNAs in the nucleus
AS-lncRNAs can regulate gene expression on a
post-transcriptional level by interacting with RNA-binding proteins
to regulate mRNA stability (25).
Wang et al (40) previously
found that EGFR-AS1 expression was significantly increased in renal
cell carcinoma tissues. Based on literature search results and
sequence complementarity between EGFR and EGFR-AS1, the
aforementioned study then confirmed that EGFR-AS1 can directly bind
to EGFR mRNA to inhibit its degradation. In addition, subsequent
RNA pull-down experiments and mass spectrometry analysis revealed
that EGFR-AS1 interacted with human antigen R (HuR), which was the
reason for the stability of EGFR mRNA. In summary, EGFR-AS1
regulated gene expression on the post-transcriptional level in
renal cell carcinoma by enhancing the stability of EGFR mRNA in a
HuR dependent manner.
In conclusion, this section mainly summarized the
mechanism by which AS-lncRNAs can regulate gene expression.
Effect of AS-lncRNAs on tumors and
underlying mechanism
Accumulating evidence has revealed that AS-lncRNAs
are involved in tumorigenesis and development, where they serve
important roles as either tumor suppressors or oncogenes (Table I). They can mediate a variety of
processes, including cell proliferation, migration, invasion,
epithelial-mesenchymal transition (EMT), apoptosis, drug resistance
and immune escape (Fig. 1)
(41). lncRNA MBNL1-AS1 and
ZNF667-AS1 have been documented to be tumor suppressors (42,43),
whilst lncRNA homeobox A11 (HOXA11)-AS, integrin subunit β1-AS1
were reported to be oncogenes (44,45).
 | Table I.AS-lncRNAs play a role as tumor
suppressor or oncogene. |
Table I.
AS-lncRNAs play a role as tumor
suppressor or oncogene.
| Tumor | AS-lncRNA | Upregulated | Downregulated |
|---|
| Colorectal
cancer | ZNF561-AS1 | √ |
|
| Colorectal
cancer | FOXC2-AS1 | √ |
|
| Colorectal
cancer | SLCO4A1-AS1 | √ |
|
| Colorectal
cancer | RAD51-AS1 |
| √ |
| Colorectal
cancer | DNAJC3-AS1 | √ |
|
| Colorectal
cancer | SATB2-AS1 |
| √ |
| Colorectal
cancer | STEAP3-AS1 | √ |
|
| Colorectal
cancer | LDLRAD4-AS1 | √ |
|
| Colorectal
cancer | ZNF667-AS1 |
| √ |
| Colorectal
cancer | SLCO4A1-AS1 | √ |
|
| Colorectal
cancer | ZEB1-AS1 | √ |
|
| Colorectal
cancer | LOXL1-AS1 | √ |
|
| Colorectal
cancer | DLGAP1-AS1 | √ |
|
| Colorectal
cancer | OIP5-AS1 | √ |
|
| Colorectal
cancer | PGM5-AS1 |
| √ |
| Colorectal
cancer | ELFN1-AS1 | √ |
|
| Colorectal
cancer | TTN-AS1 | √ |
|
| Colorectal
cancer | LBX2-AS1 | √ |
|
| Colorectal
cancer | MFI2-AS1 | √ |
|
| Colorectal
cancer | FOXD3-AS1 | √ |
|
| Colorectal
cancer | FGD5-AS1 | √ |
|
| Gastric cancer | MNX1-AS1 | √ |
|
| Gastric cancer | MACC1-AS1 | √ |
|
| Gastric cancer | ARHGAP5-AS1 | √ |
|
| Gastric cancer | OIP5-AS1 | √ |
|
| Gastric cancer | ADAMTS9-AS2 |
| √ |
| Gastric cancer | BDNF-AS | √ |
|
| Gastric cancer | MACC1-AS1 | √ |
|
| Gastric cancer | MSC-AS1 | √ |
|
| Gastric cancer | ELF3-AS1 |
| √ |
| Gastric cancer | MBNL1-AS1 |
| √ |
| Gastric cancer | NKX2-1-AS1 | √ |
|
| Gastric cancer | HNF1A-AS1 | √ |
|
| Gastric cancer | SAMD12-AS1 | √ |
|
| Gastric
adenocarcinoma | HAND2-AS1 |
| √ |
| Gastric
adenocarcinoma | PGM5-AS1 |
| √ |
| Gastric cancer | CCDC144NL-AS1 | √ |
|
| Gastric cancer | NUTM2A-AS1 | √ |
|
| Gastric cancer | VCAN-AS1 | √ |
|
| Gastric cancer | DLX6-AS1 | √ |
|
| Gastric cancer | NKX2-1-AS1 | √ |
|
| Gastric cancer | GATA6-AS |
| √ |
| Hepatocellular
carcinoma | PXN-AS1 | √ |
|
| Hepatocellular
carcinoma | IGF2-AS |
| √ |
| Hepatocellular
carcinoma | GABPB1-AS1 |
| √ |
| Hepatocellular
carcinoma | RAB11B-AS1 |
| √ |
| Hepatocellular
carcinoma | HMMR-AS1 | √ |
|
| Hepatocellular
carcinoma | KDM4A-AS1 | √ |
|
| Hepatocellular
carcinoma | CCDC144NL-AS1 | √ |
|
| Hepatocellular
carcinoma | MCM3AP-AS1 | √ |
|
| Hepatocellular
carcinoma | CDKN2B-AS1 | √ |
|
| Hepatocellular
carcinoma | BZRAP1-AS1 |
| √ |
| Hepatocellular
carcinoma | F11-AS1 |
| √ |
| Hepatocellular
carcinoma | ZFPM2-AS1 | √ |
|
| Hepatocellular
carcinoma | SBF2-AS1 | √ |
|
| Hepatocellular
carcinoma | SLC2A1-AS1 |
| √ |
| Hepatocellular
carcinoma | GAS6-AS1 | √ |
|
| Hepatocellular
carcinoma | UPK1A-AS1 | √ |
|
| Hepatocellular
carcinoma | KTN1-AS1 | √ |
|
| Hepatocellular
carcinoma | CACNA1G-AS1 | √ |
|
| Hepatocellular
carcinoma | AGAP2-AS1 | √ |
|
| Hepatocellular
carcinoma | FOXD2-AS1 | √ |
|
| Hepatocellular
carcinoma | FEZF1-AS1 | √ |
|
| Renal cell
carcinoma | EGFR-AS1 | √ |
|
| Renal cell
carcinoma | ENTPD3-AS1 |
| √ |
| Renal cell
carcinoma | LOXL1-AS1 | √ |
|
| Renal cell
carcinoma | ZFPM2-AS1 | √ |
|
| Renal cell
carcinoma | FGD5-AS1 | √ |
|
| Renal cell
carcinoma | ZNF582-AS1 |
| √ |
| Renal cell
carcinoma | DARS-AS1 | √ |
|
| Renal cell
carcinoma | NNT-AS1 | √ |
|
| Renal cell
carcinoma | DNAJC3-AS1 | √ |
|
| Renal cell
carcinoma | CDKN2B-AS1 | √ |
|
| Renal cell
carcinoma | ITGB2-AS1 | √ |
|
| Renal cell
carcinoma | MSC-AS1 | √ |
|
| Renal cell
carcinoma | ARAP1-AS1 | √ |
|
| Renal cell
carcinoma | TP73-AS1 | √ |
|
| Renal cell
carcinoma | OTUD6B-AS1 |
| √ |
| Renal cell
carcinoma | DLX6-AS1 | √ |
|
| Renal cell
carcinoma | ADAMTS9-AS2 |
| √ |
| Renal cell
carcinoma | PCED1B-AS1 | √ |
|
| Triple negative
breast cancer | ZEB1-AS1 | √ |
|
| Triple negative
breast cancer | HAND2-AS1 |
| √ |
| Triple negative
breast cancer | DLX6-AS1 | √ |
|
| Triple negative
breast cancer | WEE2-AS1 | √ |
|
| Triple negative
breast cancer | HLA-F-AS1 | √ |
|
| Triple negative
breast cancer | FAM83H-AS1 | √ |
|
| Triple negative
breast cancer | LRP11-AS1 | √ |
|
| Triple negative
breast cancer | AFAP1-AS1 | √ |
|
| Breast cancer | VCAN-AS1 | √ |
|
| Breast cancer | A1BG-AS1 | √ |
|
| Breast cancer | CERS6-AS1 | √ |
|
| Breast cancer | ZBED3-AS1 |
| √ |
| Breast cancer | HNF1A-AS1 | √ |
|
| Breast cancer | MIF-AS1 | √ |
|
| Breast cancer | DSCAM-AS1 | √ |
|
| Breast cancer | ADPGK-AS1 | √ |
|
| Breast cancer | MCM3AP-AS1 | √ |
|
| Breast cancer | MBNL1-AS1 |
| √ |
| Breast cancer | OIP5-AS1 | √ |
|
| Breast cancer | HOXD-AS1 | √ |
|
| Breast cancer | FBXL19-AS1 | √ |
|
| Breast cancer | TFAP2A-AS1 |
| √ |
| Breast cancer | MAFG-AS1 | √ |
|
| Breast cancer | CDKN2B-AS1 | √ |
|
| Leukemia | AS-RBM15 | √ |
|
| Leukemia | USP30-AS1 | √ |
|
| Leukemia | SATB1-AS1 | √ |
|
| Leukemia | CD27-AS1 | √ |
|
| Leukemia | SBF2-AS1 | √ |
|
| Leukemia | FBXL19-AS1 | √ |
|
| Leukemia | GAS6-AS1 | √ |
|
| Leukemia | MBNL1-AS1 |
| √ |
| Leukemia | TP73-AS1 | √ |
|
| Leukemia | PRR34-AS1 | √ |
|
| Leukemia | ITGB2-AS1 | √ |
|
| Leukemia | LEF1-AS1 |
| √ |
| Leukemia | ZEB2-AS1 | √ |
|
| Leukemia | LAMP5-AS1 | √ |
|
| Leukemia | GAS6-AS1 | √ |
|
| Leukemia | DNAJC3-AS1 | √ |
|
| Glioma | GATA6-AS | √ |
|
| Glioma | FAM181A-AS1 | √ |
|
| Glioma | PCED1B-AS1 | √ |
|
| Glioma | FOXD2-AS1 | √ |
|
| Glioma | DLX6-AS1 | √ |
|
| Glioma | FLVCR1-AS1 | √ |
|
| Glioma | GAS5-AS1 |
| √ |
| Glioma | AGAP2-AS1 | √ |
|
| Glioma | TP73-AS1 | √ |
|
| Glioma | PSMB1-AS1 | √ |
|
| Glioma | OIP7-AS5 | √ |
|
| Glioma | MATN1-AS1 | √ |
|
| Glioma | HOXD-AS1 | √ |
|
| Glioma | HOXA11-AS | √ |
|
| Glioma | SOX21-AS1 | √ |
|
| Glioma | ZEB1-AS1 | √ |
|
| Glioma | TPT1-AS1 | √ |
|
| Glioma | EGFR-AS1 | √ |
|
| Glioma | FOXD1-AS1 | √ |
|
| Glioblastoma | WEE2-AS1 | √ |
|
| Glioblastoma | UBA6-AS1 |
|
|
| Glioblastoma | TP73-AS1 | √ |
|
| Squamous cell
carcinoma of the hypopharynx | HOXA11-AS1 | √ |
|
| Squamous cell
carcinoma of the hypopharynx | LEF1-AS1 | √ |
|
| Squamous cell
carcinoma of the hypopharynx | MNX1-AS1 | √ |
|
| Nasopharyngeal
carcinoma | DLX6-AS1 | √ |
|
| Nasopharyngeal
carcinoma | SMAD5-AS1 | √ |
|
| Nasopharyngeal
carcinoma | HOXC13-AS | √ |
|
| Nasopharyngeal
carcinoma | PTPRG-AS1 | √ |
|
| Nasopharyngeal
carcinoma | AFAP1-AS1 | √ |
|
| Nasopharyngeal
carcinoma | TP73-AS1 | √ |
|
| Nasopharyngeal
carcinoma | DLX6-AS1 | √ |
|
| Nasopharyngeal
carcinoma | OIP5-AS1 | √ |
|
| Nasopharyngeal
carcinoma | HOXC-AS1 | √ |
|
| Nasopharyngeal
carcinoma | FEZF1-AS1 | √ |
|
| Nasopharyngeal
carcinoma | FOXP4-AS1 | √ |
|
| Nasopharyngeal
carcinoma | FOXD2-AS1 | √ |
|
| Osteosarcoma | OIP5-AS1 | √ |
|
| Osteosarcoma | CCDC144NL-AS1 | √ |
|
| Osteosarcoma | PCED1B-AS1 | √ |
|
| Osteosarcoma | PGM5-AS1 | √ |
|
| Osteosarcoma | TTN-AS1 | √ |
|
| Osteosarcoma | RHPN1-AS1 | √ |
|
| Osteosarcoma | NR2F1-AS1 | √ |
|
| Osteosarcoma | DLX6-AS1 | √ |
|
| Osteosarcoma | C5ORF66-AS1 | √ |
|
| Osteosarcoma | ASB16-AS1 | √ |
|
| Osteosarcoma | ADPGK-AS1 | √ |
|
| Osteosarcoma | AFAP1-AS1 | √ |
|
| Osteosarcoma | HNF1A-AS1 | √ |
|
| Osteosarcoma | RUSC1-AS1 | √ |
|
| Osteosarcoma | FBXL19-AS1 | √ |
|
| Osteosarcoma | MSC-AS1 | √ |
|
| Osteosarcoma | LBX2-AS1 | √ |
|
| Osteosarcoma | FOXD2-AS1 | √ |
|
| Osteosarcoma | NNT-AS1 | √ |
|
| Osteosarcoma | DSCAM-AS1 | √ |
|
Regulation tumor cell death and
proliferation
Programmed cell death mainly includes apoptosis,
autophagy, pyroptosis and ferroptosis pathways (46,47).
Results from several studies have shown that AS-lncRNAs can
regulate tumor cell death and proliferation through the
aforementioned pathways (48–51).
Apoptosis
AS-lncRNAs can reduce cell cycle arrest and
apoptosis and promote cell proliferation (48). Opa-interacting protein 5 (OIP5)-AS1
overexpression was previously found to downregulate
nucleotide-binding oligomerization domain, leucine rich repeat and
pyrin domain-containing (NLRP)6 by interacting with EZH2 to promote
GC cell proliferation whilst suppressing apoptosis (52). Wang et al (53) also reported that OIP5-AS1 can
promote cell proliferation by accelerating cell cycle progression
in GC cells through sponging miR-641 to increase the expression of
cyclin D1 and activation of AKT (53). In a different study, OIP5-AS1 was
found to promote GC cell proliferation but inhibited apoptosis by
regulating the miR-367-3p/high mobility group AT-hook 2 axis and
activating the PI3K/AKT and Wnt/β-catenin pathways (54). In glioma, Liao et al
(55) found that the expression of
GATA6-AS was significantly upregulated, where it promoted
proliferation and inhibited apoptosis through taurine upregulated
1.
Autophagy
Autophagy serves a dual role in the physiology of a
wide variety of cancer types (56,57).
In cancer cells, autophagy can inhibit tumorigenesis by preventing
cancer cell survival and inducing cell death (58), but it can also promote
tumorigenesis by promoting cancer cell proliferation and tumor
growth (59). Wang and Jin
(49) previously detected that
solute carrier organic anion transporter family member 4A1
(SLCO4A1)-AS1 expression levels in patients with CRC were
positively associated with Par-3 family cell polarity regulator
(PARD3) expression. PARD3 protein is a key molecule that can
trigger autophagy initiation. SLCO4A1-AS1 was also found to promote
autophagy and the proliferation of CRC cells both in vitro
and in vivo. Overexpression and knockdown experiments
revealed that SLCO4A1-AS1 functioned as a ceRNA to induce autophagy
and promote the proliferation of CRC cells by sponging endogenous
miR-508-3p, which promoted PARD3 protein expression (47).
Pyroptosis
Pyroptosis is a relatively recently discovered form
of programmed cell death mode that is mediated by the gasdermin
protein family (60), which also
serves a duplicitous role in tumor progression (61). Previous studies have shown that
pyroptosis is involved in the growth, differentiation, invasion and
late metastasis of cancer (62).
Components of pyroptosis have been reported to serve an important
carcinogenic role in GC progression (63). Previously, AS-lncRNAs have been
found to be key regulators of apoptosis (50). Wang et al (64) found that heart and neural crest
derivatives-expressed 2-AS1 and phosphoglucomutases (PGM5)-AS1 were
independently associated with pyroptosis in GC, where they appeared
to participate in the occurrence and development of GC according to
co-expression analysis and Kaplan-Meier survival analysis (64). In addition, Ren et al
(65) found that the
overexpression of lncRNA A disintegrin and metalloproteinase with
thrombospondin motifs (ADAMTS)9-AS2 inhibited GC progression by
regulating the miR-223-3p/NLRP3 axis-mediated pyroptosis.
Ferroptosis
Ferroptosis is another recently discovered form of
cell death that is different from apoptosis, necrosis and autophagy
mechanistically (66). In
particular, epigenetic mechanisms, such as DNA methylation, histone
modifications and AS-lncRNAs, have been reported to serve roles in
ferroptosis (67,68). Huang et al (51) previously found that reactive oxygen
species (ROS), Fe2+ and total iron concentrations were
decreased after overexpressing brain-derived neurotrophic factor
(BDNF)-AS following RT-PCR detection. By contrast, ROS,
Fe2+ and total iron concentrations were revealed to be
increased after BDNF-AS expression was knocked down. A ferroptosis
model was then established using erastin and Ras-selective lethal 3
(RSL3), where it was found that the changes in the level of
ferroptosis markers induced by erastin were more significant
compared with those induced by RSL3. A different previous study
also found that erastin can induce ferroptosis by binding to
voltage-dependent anion-selective channel protein (VDAC)2/VDAC3
(68). The protein expression
profile in GC tissues studied by Huang et al (51) revealed that VDAC2/VDAC3 was
abnormally expressed in GC samples. However, VDAC3 protein
expression was found to be significantly increased when BDNF-AS was
overexpressed but was decreased when BDNF-AS expression was knocked
down, compared with VDAC2 (49).
These results suggested that BDNF-AS can regulate ferroptosis in GC
cells through VDAC3. In conclusion, the aforementioned results from
previous studies suggested that when BDNF-AS was overexpressed, the
degree of ferroptosis in GC cells was reduced, which favored GC
invasion and metastasis.
Regulation of energy metabolism
reprogramming in tumor cells
In cancer cells, glucose metabolism is reprogrammed
to adapt to the increased energy requirements. Unlike in normal
cells, the main metabolic process in cancer cells is glycolysis
(69). The predominance of aerobic
glycolysis is a key metabolic feature of the Warburg phenotype,
which is caused by the active metabolic reprogramming required to
support the sustained cancer cell proliferation and malignant
progression (70). Since the
German scientist Otto Warburg put forward the ‘Warburg effect’
(71), the relationship between
aerobic glycolysis and tumorigenesis has become a topic of intense
scientific research (72). The
mechanism of glucose metabolism in cancer cells can be summarized
mainly as follows (70): i)
Overexpression of glucose transporters and key glycolytic enzymes,
which accelerates the glycolytic flux, followed by the accumulation
and transfer of glycolytic intermediates for cancer biomass
synthesis; ii) high-speed ATP production to meet the energy demand;
and iii) accumulation of lactic acid, which promotes tumor
progression, greatly contributing to tumor acidosis. Therefore, the
Warburg effect is one of the core factors in underlying the
mechanism of cancer progression. Zhao et al (30) previously found that in GC cells
MACC1-AS1 significantly upregulated the expression of glycolysis
components glucose transporter 1 (GLUT1), hexokinase 2 (HK2),
glucose-6-phosphate dehydrogenase and lactate transporter
monocarboxylate transporter 1 on the mRNA level, whilst
significantly upregulating the expression of GLUT1, HK2 and lactate
dehydrogenase (LDH) on the protein level. In addition, following
2-NBDG assays, MACC1-AS1 was found to accelerate glucose uptake in
GC cells. Under either normal conditions or those of glucose
deprivation, MACC1-AS1 also increased the production of ATP and
lactic acid, presumably by increasing the activity of key
glycolytic enzymes HK2 and LDHA. These results suggested that a
potential role of MACC1-AS1 is to promote glycolysis by
upregulating the activity and expression of glycolytic enzymes in
GC cells. This metabolic transformation may serve to improve cell
viability and resistance to apoptosis, which in turn promotes the
progression of GC. In a previous study, Li et al (73) found that the overexpression of
RAD51-AS1 could reduce glucose uptake by CRC cells, which led to a
reduction in lactic acid concentrations in CRC cells. Subsequently,
through western blotting, it was found that the RAD51-AS1
overexpression led to the reduction in HK2 and GLUT1 levels,
suggesting that RAD51-AS1 can inhibit the glycolytic process in CRC
cells. In conclusion, these results strongly suggested that
RAD51-AS1 serves an important role in maintaining the glycolysis
balance, which may affect the proliferation of CRC.
Lipid metabolism reprogramming has also been
proposed to be a marker of cancer (74). In rapidly proliferating tumor
cells, the re-synthesis of fatty acids is enhanced, such that the
expression of key fatty acid synthases, including acetyl-CoA
carboxylase (ACC) and fatty acid synthase (FASN), is increased.
This subsequently results in the accumulation of a variety of fatty
acids (75). Accumulating lipids
can promote cell proliferation and tumor progression in a complex
tumor microenvironment (76).
Previous studies have demonstrated that fibroblasts in the tumor
microenvironment express FASN highly (76), which mediates lipid metabolism
reprogramming, resulting in the accumulation of various fatty acids
to promote the migration of CRC cells (76). EGFR/PI3K/AKT signaling has been
shown to activate sterol regulatory element-binding protein-1
cleavage and upregulate the expression of ACC1 and FASN, resulting
in increased lipid metabolism (77,78).
Tang et al (79) previously
found that interferon-induced, double-stranded RNA-activated
protein kinase inhibitor-AS1 regulated fatty acid synthase through
the EGFR/PI3K/AKT/NF-κB signal pathway to promote the progression
of CRC. Fatty acid is an important energy source through fatty acid
oxidation (FAO), which has been shown to be necessary for the
proliferation and survival of cancer cells (80). Inhibition of FAO can inhibit cell
stemness and alleviate tumor growth (81). Mesenchymal stem cells (MSCs) have
been revealed to promote the dryness of GC cells and predict the
poor prognosis of GC. He et al (82) found that inhibition of FAO could
reverse the MSC-mediated stemness of GC cells, suggesting that FAO
is involved in the self-renewal of GC stem cells induced by MSCs,
whilst having little effect on non-stem cells.
Regulation of inflammation in
cancer
A large number of studies have shown that
inflammatory immune cells can serve diverse but key roles in
promoting tumorigenesis (83).
Tumor-promoting inflammatory cells mainly include macrophages, mast
cells, neutrophils, T and B lymphocytes (84). Nie et al (85) employed the Gene Set Enrichment
Analysis (GSEA) pre-ranking of tumor characteristics to find that
there was a decrease in memory B cells but an increase in naive and
effector CD8T cells in the high-risk group, suggesting
immune-related regulation. Tumor-infiltrating inflammatory cells
have been demonstrated to induce and then maintain tumor
angiogenesis (86), stimulate
cancer cell proliferation, promote foreign tissue invasion and
support cell migration (84,87).
AS-lncRNAs have correspondingly been detected to regulate
inflammation in tumors and cancer transformation, by regulating the
expression of genes associated with the immune response. This was
proposed to be exerted through the synthesis of inflammatory
factors and inflammatory signaling pathways (88). In addition, AS-lncRNAs have been
reported to alter the profile of immune cell infiltration in the
tumor microenvironment, mediating a profound impact on tumor
invasiveness, progression and prognosis (89). Xu et al (90) found that the expression of the CRC
tissue-specific lncRNA special AT-rich sequence-binding protein 2
(SATB2)-AS1 was downregulated in CRC tissues, which predicted a
favourable prognosis. Transwell and wound healing assays
demonstrated that SATB2-AS1 knockdown significantly increased CRC
cell migration and invasion ability. The results of GSEA also
revealed that the expression of SATB2-AS1 was associated with
various immune responses. In addition, it was found that when
SATB2-AS1 was negatively associated with the expression of Th1
chemokines C-X-C motif chemokine ligand (CXCL)9 and CXCL10, these
chemokines could be induced by IFN-γ and mediated effector T cell
transport. In conclusion, these results suggested that SATB2-AS1 is
a mediator of the CRC immune response by regulating immune cell
density in CRC to inhibit CRC cell metastasis, slowing the
progression of CRC.
Regulation of EMT, invasion and
migration in tumor cells
One of the key mechanisms for cancer cells to
enhance their invasive ability is by detaching from their
intercellular adhesion, followed by the acquisition of a more
dynamic mesenchymal phenotype as part of the EMT process (91). EMT has been previously considered
to be a binary process with two distinct cell groups, namely
epithelial cells and mesenchymal cells, characterized by the loss
of the epithelial marker E-cadherin and the increased expression of
the mesenchymal marker vimentin (92,93).
During EMT, epithelial cells lose apical-basal polarity and
E-cadherin expression, whilst upregulating the expression of
mesenchymal biomarkers, including twist, slug, snail, vimentin and
N-cadherin (94,95). EMT has been associated with a
number of tumor functions, including tumor initiation, malignant
progression, tumor stemness, cell migration and metastasis
(92). In addition, EMT is
considered to be a key factor on worsening patient prognosis by
accelerating the invasion and migration of cancer cells (96).
Yu et al (97) previously observed that lncRNA
SLCO4A1-AS1 expression was significantly increased in CRC tissues,
which was in turn associated with poor patient prognosis and CRC
tumor metastasis. By knocking down SLCO4A1-AS1 expression, it was
then detected that SLCO4A1-AS1 inhibited the migration, invasion
and EMT of CRC cells in vitro. In a different study, Zhou
et al (98) found that
SLCO4A1-AS1 activated Wnt/β-catenin signaling in CRC cells, which
promoted cell migration and invasion. The authors explored TCGA
database for antisense lncRNAs involved in CRC under hypoxic
conditions. Among the candidate genes found, lncRNA
six-transmembrane epithelial antigen of prostate 3 (STEAP3)-AS1 was
reported to be aberrantly transcribed under hypoxic conditions in
clinical CRC tissues, where it was positively associated with poor
patient prognosis (96).
Furthermore, it was revealed that lncRNA STEAP3-AS1 interacted with
YTH N6-methyladenosine RNA-binding protein 2 to stabilize STEAP3
mRNA, which increased the expression of the STEAP3 protein. The
STEAP3 protein then activated Wnt/β-catenin signaling in an
iron-dependent manner to promote cell invasion and migration, in
turn accelerating CRC progression (96).
A previous independent study of RNA-sequencing data
found that in A549 and HeLa cells, downregulation of E74-like ETS
transcription factor 3 (ELF3)-AS1 led to the upregulation of snail
family transcriptional repressor (SNAI)2 mRNA expression,
suggesting that the negative regulation of SNAI2 expression by
ELF3-AS1 may be widespread in cancer (99). In addition, Li et al
(26) identified 123 lncRNAs
regulated by SNAI2 in GC by RNA sequencing, where both the
ELF3 gene and ELF3-AS1 were found to be transcriptionally
repressed by EMT-associated transcription factors SNAI2 or SNAI1.
In addition, the gene expression profiles produced by SNAI2
overexpression and ELF3-AS1 knockdown were revealed to be highly
very similar. These results suggested that knocking down ELF3-AS1
expression cannot only upregulate the expression of SNAI2, but can
also activate the downstream signal pathways of SNAI2 in GC. Cell
proliferation, Transwell and wound healing experiments revealed
that downregulation of ELF3-AS1 promoted the proliferation,
migration and invasion of GC cells, indicating poor patient GC
prognosis. In conclusion, ELF3-AS1 was suggested to inhibit EMT
mainly by inhibiting the SNAI2-mediated transduction of
GC-associated transcription factors. Mo et al (100) previously found that the higher
expression levels of lncRNA low density lipoprotein receptor class
A domain-containing 4 (LDLRAD4)-AS1 promoted the progression of
CRC, which is associated with poor patient prognosis. Subsequent
western blotting and immunohistochemistry results revealed that
lncRNA LDLRAD4-AS1 could promote EMT both in vitro and in
vivo. lncRNA LDLRAD4-AS1 was then observed to promote EMT by
reducing the expression of LDLRAD4, promoting the process of
GC.
The TGF-β/EMT pathway is closely associated with
cancer progression (101). In
particular, Smad7 is activated by all members of the TGF-β
superfamily and has a negative regulatory effect on the TGF-β
signal pathway (102). Su et
al (42) previously reported
that MBNL1-AS1 knockdown can increase the proliferation, migration
and invasion of GC cells according to in vitro experimental
results and animal models. MBNL1-AS1 expression was demonstrated to
be downregulated in GC tissues and cells, which played a positive
role in inhibiting the growth of the GC tumor. Western blotting
data demonstrated that MBNL1-AS1 affected the TGF-β/SMAD pathway by
regulating the miR-424-5p/Smad7 axis, which inhibited the
proliferation of GC.
Collectively, the aforementioned observations
strongly suggested that AS-lncRNAs can regulate EMT through the
Wnt, SNAI2 and TGF-β signal pathways, serving an important role in
the occurrence and development of various tumors.
Regulation of tumor
neovascularization
The plasminogen activator system, which includes
urokinase type plasminogen activator (uPA), uPA cell receptor and
its specific inhibitor plasminogen activator inhibitor-1 (PAI-1),
has been reported to serve an important role in tumor progression
and angiogenesis (103). The role
of PAI-1 in tumor angiogenesis has been extensively studied, the
expression of which is essential for tumor angiogenesis (104). Teng et al (105) found that the overexpression of
NK2 homeobox 1 (NKX2)-1-AS1 and PAI-1 was associated with GC
progression and prognosis. Specifically, NKX2-1-AS1 served as a
ceRNA of miR-145-5p and promoted tumor progression and
angiogenesis, by activating VEGFR2 signaling through PAI-1.
Previous studies have shown that the PI3K/AKT signaling pathway is
involved in cancer cell invasion and angiogenesis (106,107). Liu et al (108) previously detected that hepatic
nuclear factor 1 α (HNF1A)-AS1 can promote GC invasion, metastasis
and angiogenesis both in vitro and in vivo. In
particular, the high degree of PI3K/AKT signaling pathway
activation was found to promote the progression of GC. Results from
Transwell migration and matrix-based capillary formation assays
revealed that overexpression of HNF1A-AS1 can promote GC
angiogenesis in vitro. ELISA and rescue experiments
confirmed that HNF1A-AS1 exerted its biological function via the
PI3K/AKT signaling pathway. The activation of PI3K/AKT signaling in
tumor cells could increase the secretion of VEGFA, which serves a
role in promoting angiogenesis in human cancers. In conclusion,
these aforementioned findings suggested that lncRNA-HNF1A-AS1 can
function as a ceRNA of miR-30b-3p to activate PI3K/AKT signaling to
increase the secretion of VEGF-A, which promotes angiogenesis and
therefore the progression of GC.
AS-lncRNAs in the diagnosis, prognosis and
treatment of tumors
AS-lncRNAs have the potential to
assist in tumor diagnosis and prognosis
A number of previous studies have demonstrated that
AS-lncRNAs have significant potential as a biomarker for both
cancer diagnosis and prognosis (10). Guo et al (109) previously found that
overexpression of muskelin 1, intracellular mediator containing
Kelch motifs-AS enhanced the stability of Yes-associated protein 1
mRNA, which promoted carcinogenic effects. This was proposed to
facilitate the diagnosis and prognosis of HCC. In a different
previous study, El-Ashmawy et al (110) revealed that family with sequence
similarity 83-member H (FAM83H)-AS1 was involved in the progression
of various malignant tumors. Coincidentally, it was found to be
particularly dysregulated in cancers, potentially serving a crucial
role in their diagnosis and prognosis (110). Supporting this, Da et al
(111) demonstrated that
FAM83H-AS1 may serve a prognostic and diagnostic role in GC.
AS-lncRNAs in the treatment of
cancer
AS-lncRNAs have been documented to not only have the
potential for cancer diagnosis and prognosis, but may also
facilitate the design of treatment strategies for patients with
cancer (41). AS-lncRNAs are
increasingly becoming biomarkers for the treatment of tumors
(45,64,112,113).
Activation of tumor suppressors
Tumor suppressors refer to a large number of
molecules that can reliably control cell division, promote
apoptosis and inhibit metastasis (114). The loss of tumor suppressor
functions may lead to uncontrolled cell division malignant
transformation (115). Tumor
suppressors mainly exert their roles through the following four
main mechanisms: i) Inhibition of cell division; ii) induction of
apoptosis; iii) repair of DNA damage; and iv) inhibition of
metastasis (116). Lu et
al (113) previously observed
that the expression of sterile α motif domain-containing 12
(SAMD12)-AS1 was increased in human GC tissues and cell lines.
SAMD12-AS1 was then detected to exert its biological role in GC
through direct interactions with DNA methyltransferase 1 (DNMT1),
which inhibits p53 signaling to promote the malignant
transformation of GC cells. Silencing SAMD12-AS1 expression
restored p53 signaling to reverse the progression of GC, suggesting
that SAMD12-AS1 may serve as a potential diagnostic and therapeutic
target for GC. Liu et al (117) previously reported that the
expression of HNF1A-AS1 was significantly upregulated in GC
tissues. Similar to the function of HNF1A-AS1, the overexpression
of early growth response 1 (EGR1) was reported to enhance cell
proliferation and promote GC cell cycle progression. The RT-qPCR
assay showed that EGR1 promotes the expression of HNF1A-AS1. EGR1
and HNF1A-AS also inhibited the expression of the antibiotic growth
factor p21 by promoting cell division cycle 34-mediated
ubiquitination and p21 degradation. It was then suggested that the
inhibition of EGR1-activated HNF1A-AS1 and the subsequent
upregulation of anti-growth factors could inhibit the development
of GC. In another study, Zhuang et al (43) previously observed that the lower
expression levels of ZNF667-AS1 predicted poorer prognosis and were
associated with the progression of CRC. Upregulation of ankyrin 2
by the overexpression of ZNF667-AS1 was then found to inhibit the
proliferation, migration and invasion of CRC cells, which hindered
the development of CRC progression.
In conclusion, AS-lncRNA could regulate tumor
development by activating tumor suppressor factors, which may be
exploited as a target for tumor therapy.
Inhibition of oncogene expression
SLCO4A1-AS1 is highly expressed in CRC cells and
tissues, which is associated with poor patient prognosis (118). Zhang et al (118) previously documented that
SLCO4A1-AS1 increased the expression of CDK2 by enhancing the
interaction between CDK2 and heat shock protein 90 (HSP90) and
activating c-MYC signaling. This increase in CDK2 expression
downstream of SLCO4A1-AS enhanced tumor growth by promoting the
phosphorylation of c-Myc at the Ser6 site, activating this protein
(115). Downregulation of
SLCO4A1-AS1 inhibited the proliferation of CRC cells in vivo
and in vitro. Ni et al (119) reported that the expression of
ZEB1-AS1 in CRC was significantly upregulated, which was associated
with poor prognosis in patients with colon adenocarcinoma.
Specifically, ZEB1-AS1 promoted the expression of p21 Protein
(Cdc42/Rac)-activated kinase 2 (PAK2) by sponging miR-455-3p, which
advanced the proliferation and metastasis of colon adenocarcinoma
cells. Knocking down ZEB1-AS expression was then observed to
increase the expression of miR-455-3p and decreased the expression
of PAK2, which suppressed the proliferation and metastasis of
cancer cells. It would be of significance to identify novel
therapeutic targets for patients with colon adenocarcinoma. Wu
et al (120) previously
found that the expression of lysyl oxidase homolog 1 (LOXL1)-AS1
was upregulated in CRC, which then increased the expression of
target gene CD44 by sponging miR708-5p. This in turn
promoted the expression of EGF to facilitate CRC progression.
Knocking down LOXL1-AS1 expression was found to reverse this
upregulated CD44/EGFR signaling pathway to inhibit the progression
of CRC, providing another potential novel pathway to explore the
treatment strategy of CRC.
In conclusion, these previous findings
aforementioned suggested that knocking down AS-lncRNAs can regulate
tumor development by inhibiting the expression of oncogenes, which
may become a novel method for tumor therapy.
Activation of antitumor immunity
Possibly the biggest natural antagonist of cancer
development is the host immune system, especially T cells, which
can initiate an antitumor response by expressing specific receptors
of tumor antigens (121). To
prevent immune hyperactivation, effector lymphocytes express immune
checkpoints, which are mainly receptor proteins expressed on the
cell surface (122). Several
tumors, including liver cancer, exploited this by expressing
corresponding ligands to these checkpoints (123). After ligand binding, the activity
of effector lymphocytes becomes impaired. Programmed cell death
protein 1 (PD-1) was one of the main such co-inhibitory receptors
(124), whereas programmed death
ligand (PD-L)1 is one of the ligands for PD-1 (125). As a co-suppressor molecule during
cancer development, PD-L1 is expressed on T lymphocytes and
promotes immune escape (126).
Fan et al (127)
previously found that PC-esterase domain-containing 1B (PCED1B)-AS1
enhanced the expression and function of PD-L1 and PD-L2 by sponging
miR-194-5p, which induced immunosuppression by HCC. Therefore,
PCED1B-AS1 was a potential therapeutic target for HCC. In cancer,
the PD-1/PD-L1 pathway is responsible for T cell activation,
proliferation and cytotoxic secretion, leading to the degeneration
of the antitumor immune response (128). Zhou et al (129) reported that in hypopharyngeal
squamous cell carcinoma there was a positive regulatory
relationship between HOXA11-AS1 and PD-L1. In particular,
downregulation of HOXA11-AS1 inhibited PD-L1-mediated immune escape
and metastasis by reducing the association between polypyrimidine
tract binding protein 1 and FOS-like 1 in hypopharyngeal squamous
cell carcinoma. Results from the aforementioned study provided a
preliminary basis for applying HOXA11-AS1 as a potential target for
the immunotherapy of hypopharyngeal squamous cell carcinoma. In
addition, AS-lncRNAs can regulate the expression of genes
associated with immune responses, changing the status of immune
cell infiltration in the tumor microenvironment and profoundly
dictate tumor invasiveness, progression and prognosis (88,130). Nie et al (85) performed an integrative analysis
using publicly available gene expression datasets from TCGA project
and the Genotype-Tissue Expression project from various GC
patients' cohorts and a novel pairing algorithm to identify and
validate 13 immune-related lncRNA pair signatures. This may provide
novel insights into the role of AS-lncRNAs in tumor immunity and to
facilitate the development of antitumor immunotherapeutic
methods.
In conclusion, AS-lncRNAs can regulate gene
expression related to immune response, thereby regulating antitumor
immunity to regulate tumor development, and it may become a novel
target for tumor therapy.
To sum up, tumor gene therapy is currently a topic
of intense research, with main fields including the activation of
tumor suppressor factors, inhibiting oncogene expression and
inducing antitumor immune activation, which can all be regulated by
AS-lncRNAs according to the aforementioned previous findings.
Enhancing chemoradiotherapy and
chemotherapy by interfering with tumor drug resistance
Radiotherapy and chemotherapy form the core of all
treatment strategies for cancer; however, the majority of patients
will develop drug resistance, which is a major obstacle to the
long-term efficacy of radiotherapy and chemotherapy (131,132). The mechanisms underlying drug
resistance are complex, which can involve a wide array of factors,
such as drug efflux, DNA damage repair, apoptosis and target
mutations (133,134). Since AS-lncRNAs have been
reported to be aberrantly expressed in tumors, they regulate
chemoradiotherapy resistance (135–138).
5-Fluorouracil resistance
In a previous study, Zhou et al (135) found the lncRNA transmembrane
protein 44 (TMEM44)-AS1 to be associated with resistance to 5-FU by
GC cells. TMEM44-AS1 was observed to function as a ceRNA, which
upregulated protein phosphatase 1 regulatory subunit 13-like
expression and inhibited the p53 pathway, finally promoting
resistance to 5-FU. To combat this, Zhou et al (129) developed the nano-carrier
chitosan-gelatin epigallocatechin gallate to deliver small
interfering RNA-TMEM44-AS1 to knock down TMEM44-AS1 expression,
which effectively reversed 5-FU resistance, specifically by
observing a significant enhancement in the therapeutic effects of
5-FU in xenograft mouse models of GC. Gui et al (137) recently observed that FEZ family
zinc finger (FEZF1)-AS1 was upregulated in 5-FU chemically
resistant GC tissues. RIP then revealed that FEZF1-AS1 increased
the chemical resistance of GC cells through directly targeting
autophagy-related 5 ganglion autophagy. By contrast, knocking down
FEZF1-AS1 expression was documented to increase the sensitivity to
5-FU in GC cells in vivo. In a recent study, Qu et al
(136) revealed that lncRNA discs
large homolog associated protein 1 (DLGAP1)-AS1 promoted the
progression of CRC. Subsequently, functional assays revealed that
silencing DLGAP1-AS1 expression enhanced the chemosensitivity of
cells to 5-FU, suggesting that silencing DLGAP1-AS1 expression may
be a promising therapeutic target for CRC.
Cisplatin resistance
Ren et al (65) found that lncRNA ADAMTS9-AS2
overexpression inhibited the progression of GC and promoted
cisplatin chemosensitivity by regulating the pyroptosis process
mediated by the miR-223-3p/NLRP3 axis. This molecular mechanism
underlying the drug resistance in GC to cisplatin provided a novel
therapeutic target for the clinical treatment of GC. Song et
al (139) previously reported
that OIP5-AS1 was highly expressed in cisplatin-resistant (CR)
osteosarcoma cells. Silencing OIP5-AS1 expression promoted the
apoptosis in CR osteosarcoma cells, suggesting that the
downregulation of OIP5-AS1 can significantly reverse cisplatin
resistance in osteosarcoma cells. These results deepened the
understanding into the role of OIP5-AS1 in cisplatin resistance in
osteosarcoma, possibly providing a novel therapeutic target for
patients with CR osteosarcoma. Wu et al (140) previously revealed that
upregulation of FOXD1-AS1 increased the resistance of GC cells to
cisplatin. FOXD1-AS1 promoted FOXD1 translocation through
PIK3CA/PI3K/AKT/mTOR signaling, which increased the progression of
GC and chemotherapy resistance. These findings provided a novel
target for the treatment of patients with CR GC.
Oxaliplatin resistance
Recent studies have demonstrated that lncRNAs are
involved in the regulation of oxaliplatin resistance (141–143). Liang et al (144) previously found that OIP5-AS1
expression was upregulated in CRC tissues, whilst that of miR-137
was downregulated. Therefore, there was a negative association
between the expression of OIP5-AS1 and miR-137. OIP5-AS1 was
subsequently found to directly target miR-137, such that silencing
OIP5-AS1 expression reversed the resistance of CRC cells to
oxaliplatin by promoting the expression of miR-137. Furthermore,
Hui et al (145) reported
that overexpression of PGM5-AS1 inhibited the proliferation of
oxaliplatin-resistant colon cancer cells by using colony formation
assays, Cell Counting Kit-8 analysis and EdU assays. Transwell
analysis revealed that compared with those in the control group,
the overexpression of PGM5-AS1 inhibited the invasion and migration
of oxaliplatin-resistant colon cancer cells. These results
suggested that PGM5-AS1 overexpression can render colon cancer
cells more sensitive to oxaliplatin. Hui et al (136) also found that PGM5-AS1 and
oxaliplatin could enter colon cancer cells through engineered
exosomes to effectively reverse drug resistance. Li et al
(143) previously observed that
extracellular leucine rich repeat and fibronectin type III
domain-containing 1 (ELFN1)-AS1 interacted with the promoter region
of meis homeobox 1 (MEIS1) to inhibit the expression of MEIS1. By
contrast, MEIS1 can enhance the sensitivity of CRC cells to
oxaliplatin. This resulted in the conclusion that the combination
of oxaliplatin and ELFN1-AS1 ASO can reverse oxaliplatin resistance
in CRC, highlighting the potential of targeting ELFN1-AS1 as a
treatment method oxaliplatin resistance.
Radiotherapy resistance
Zou et al (138) previously found that lncRNA
OIP5-AS1 and dual specificity tyrosine-phosphorylation-regulated
kinase 1A (DYRK1A) were downregulated in radiation-resistant CRC
cell lines. Specially, overexpression of OIP5-AS1 can regulate
DYRK1A expression through miR-369-3p, damage the survival of cell
clones and promote apoptosis after radiotherapy, suggestive of the
reversal of radiation resistance in CRC cells.
Conclusion
Malignant tumors are one of the main causes of
mortality worldwide. As the second leading cause of mortality in
China, cancer seriously threatens the lives of the population and
restricts social and economic development. lncRNAs are RNAs that
contain >200 nucleotides. Although the majority of lncRNAs do
not encode proteins, they can mediate a variety of important
physiological functions. AS-lncRNAs are part of the antisense
chains of coding genes that are transcribed from the opposite
strands of protein or non-protein coding genes. Accumulating
evidence has shown that AS-lncRNAs can serve an important role in
tumorigenesis, metastasis, prognosis and drug resistance.
AS-lncRNAs can regulate gene expression in tumors through
endogenous competition mechanisms, promoter interactions, direct
interactions with mRNA, acting as ‘scaffolds’ to regulate the
half-life of mRNA, interactions with 5′-UTR, regulation of mRNAs of
interest, positive feedback loop of ‘AS-lncRNA/mediator
protein/gene of interest’ model and transcriptional regulation in
the nucleus. Over the past decade, AS-lncRNAs have been found to
mediate a variety of molecular functions, such as the regulation of
cell proliferation, invasion, migration and apoptosis. In addition,
they have been shown to regulate tumor energy metabolism,
inflammation and tumor neovascularization. Although the discovery
of AS-lncRNAs has only been relatively recent, their possible
application in tumor treatment has been widely studied. AS-lncRNAs
can potentially serve as markers of tumor therapy, as they may
activate tumor suppressor factors, inhibit oncogene expression and
induce antitumor immune activation. In addition, there is evidence
that AS-lncRNAs can regulate the mechanism underlying tumor
treatment and drug resistance. To conclude, AS-lncRNAs are likely
to serve a role in tumor gene expression, the mechanisms regulating
tumors physiology and tumor response to treatment. Treatment of
abnormally expressed AS-lncRNAs in tumors is a promising method.
The study into AS-lncRNAs has opened up a new era for the
exploration, diagnosis and treatment of tumors.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82174379), the Special Project of
Jiangsu Provincial Traditional Chinese Medicine Hospital Innovation
and Development Fund (grant no. Y2020CX38) and the National
Clinical Research Base of Traditional Chinese Medicine in Jiangsu
(grant no. JD2023SZ04).
Availability of data and materials
Not applicable.
Authors' contributions
YS wrote the manuscript, searched the literature
and prepared the table. YD, YL and YC was involved in the design of
the study, and revised the manuscript. YL prepared the figure. JZ,
ZL, CC and XY provided article ideas, modified the tables and
revised the manuscript. QL, SZ, WT, ZC, YW, LH, ZQ, KW and ZM
performed literature research and collected relevant articles. Data
authentication is not applicable. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
GBD 2019 Diseases and Injuries
Collaborators, . Global burden of 369 diseases and injuries in 204
countries and territories, 1990–2019: A systematic analysis for the
Global Burden of Disease Study 2019. Lancet. 396:1204–1222. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qiu H, Cao S and Xu R: Cancer incidence,
mortality, and burden in China: A time-trend analysis and
comparison with the United States and United Kingdom based on the
global epidemiological data released in 2020. Cancer Commun (Lond).
41:1037–1048. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miller KD, Nogueira L, Devasia T, Mariotto
AB, Yabroff KR, Jemal A, Kramer J and Siegel RL: Cancer treatment
and survivorship statistics, 2022. CA Cancer J Clin. 72:409–436.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Agirre E and Eyras E: Databases and
resources for human small non-coding RNAs. Hum Genomics. 5:192–199.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takahashi A, Tsutsumi R, Kikuchi I, Obuse
C, Saito Y, Seidi A, Karisch R, Fernandez M, Cho T, Ohnishi N, et
al: SHP2 tyrosine phosphatase converts parafibromin/Cdc73 from a
tumor suppressor to an oncogenic driver. Mol Cell. 43:45–56. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Bio. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hua Q, Mi B, Xu F, Wen J, Zhao L, Liu J
and Huang G: Hypoxia-induced lncRNA-AC020978 promotes proliferation
and glycolytic metabolism of non-small cell lung cancer by
regulating PKM2/HIF-1α axis. Theranostics. 10:4762–4778. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y,
Xue C, Ren F, Ren Z, Li J, et al: Long non-coding RNA PVT1 promotes
tumor progression by regulating the miR-143/HK2 axis in gallbladder
cancer. Mol Cancer. 18:332019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Devaux Y, Zangrando J, Schroen B, Creemers
EE, Pedrazzini T, Chang CP, Dorn GN, Thum T II and Heymans S;
Cardiolinc network, : Long noncoding RNAs in cardiac development
and ageing. Nat Rev Cardiol. 12:415–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Magistri M, Faghihi MA, St Laurent G III
and Wahlestedt C: Regulation of chromatin structure by long
noncoding RNAs: Focus on natural antisense transcripts. Trends
Genet. 28:389–396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cui XY, Zhan JK and Liu YS: Roles and
functions of antisense lncRNA in vascular aging. Ageing Res Rev.
72:1014802021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bian Z, Zhang J, Li M, Feng Y, Wang X,
Zhang J, Yao S, Jin G, Du J, Han W, et al: LncRNA-FEZF1-AS1
promotes tumor proliferation and metastasis in colorectal cancer by
regulating PKM2 Signaling. Clin Cancer Res. 24:4808–4819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang MH, Zhao L, Wang L, Ou-Yang W, Hu SS,
Li WL, Ai ML, Wang YQ, Han Y, Li TT, et al: Nuclear lncRNA HOXD-AS1
suppresses colorectal carcinoma growth and metastasis via
inhibiting HOXD3-induced integrin β3 transcriptional activating and
MAPK/AKT signalling. Mol Cancer. 18:312019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Braga EA, Fridman MV, Moscovtsev AA,
Filippova EA, Dmitriev AA and Kushlinskii NE: LncRNAs in ovarian
cancer progression, metastasis, and main pathways: ceRNA and
alternative mechanisms. Int J Mol Sci. 21:88552020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pan H, Ding Y, Jiang Y, Wang X, Rao J,
Zhang X, Yu H, Hou Q and Li T: LncRNA LIFR-AS1 promotes
proliferation and invasion of gastric cancer cell via
miR-29a-3p/COL1A2 axis. Cancer Cell Int. 21:72021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Si Z, Yu L, Jing H, Wu L and Wang X:
Oncogenic lncRNA ZNF561-AS1 is essential for colorectal cancer
proliferation and survival through regulation of
miR-26a-3p/miR-128-5p-SRSF6 axis. J Exp Clin Canc Res. 40:782021.
View Article : Google Scholar
|
|
22
|
Shuai Y, Ma Z, Liu W, Yu T, Yan C, Jiang
H, Tian S, Xu T and Shu Y: TEAD4 modulated LncRNA MNX1-AS1
contributes to gastric cancer progression partly through
suppressing BTG2 and activating BCL2. Mol Cancer. 19:62020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yao W, Du X, Zhang J, Wang Y, Wang M, Pan
Z and Li Q: SMAD4-induced knockdown of the antisense long noncoding
RNA BRE-AS contributes to granulosa cell apoptosis. Mol Ther
Nucleic Acids. 25:251–263. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang B, Yuan Y, Yi T and Dang W: The
roles of antisense long noncoding RNAs in tumorigenesis and
development through Cis-Regulation of neighbouring genes.
Biomolecules. 13:6842023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu B, Xiang W and Liu J, Tang J, Wang J,
Liu B, Long Z, Wang L, Yin G and Liu J: The regulatory role of
antisense lncRNAs in cancer. Cancer Cell Int. 21:4592021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li D, Shen L, Zhang X, Chen Z, Huang P,
Huang C and Qin S: LncRNA ELF3-AS1 inhibits gastric cancer by
forming a negative feedback loop with SNAI2 and regulates ELF3 mRNA
stability via interacting with ILF2/ILF3 complex. J Exp Clin Cancer
Res. 41:3322022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bartl J, Zanini M, Bernardi F, Forget A,
Blumel L, Talbot J, Picard D, Qin N, Cancila G, Gao Q, et al: The
HHIP-AS1 lncRNA promotes tumorigenicity through stabilization of
dynein complex 1 in human SHH-driven tumors. Nat Commun.
13:40612022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jadaliha M, Gholamalamdari O, Tang W,
Zhang Y, Petracovici A, Hao Q, Tariq A, Kim TG, Holton SE, Singh
DK, et al: A natural antisense lncRNA controls breast cancer
progression by promoting tumor suppressor gene mRNA stability. PLoS
Genet. 14:e10078022018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pan K and Xie Y: LncRNA FOXC2-AS1 enhances
FOXC2 mRNA stability to promote colorectal cancer progression via
activation of Ca(2+)-FAK signal pathway. Cell Death Dis.
11:4342020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang
S, Dong S, Wen Z, Rao J, Liao W and Shi M: The lncRNA MACC1-AS1
promotes gastric cancer cell metabolic plasticity via AMPK/Lin28
mediated mRNA stability of MACC1. Mol Cancer. 17:692018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu L, Zhu Y, Han S, Chen M, Song P, Dai
D, Xu W, Jiang T, Feng L, Shin VY, et al: Impaired autophagic
degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in
gastric cancer. Cell Death Dis. 10:3832019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Visvanathan A, Patil V, Arora A, Hegde AS,
Arivazhagan A, Santosh V and Somasundaram K: Essential role of
METTL3-mediated m(6)A modification in glioma stem-like cells
maintenance and radioresistance. Oncogene. 37:522–533. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tran NT, Su H, Khodadadi Jamayran A, Lin
S, Zhang L, Zhou D, Pawlik KM, Townes TM, Chen Y, Mulloy JC and
Zhao X: The AS-RBM15 lncRNA enhances RBM15 protein translation
during megakaryocyte differentiation. EMBO Rep. 17:887–900. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang J and Manley JL: Misregulation of
pre-mRNA alternative splicing in cancer. Cancer Discov.
3:1228–1237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Oltean S and Bates DO: Hallmarks of
alternative splicing in cancer. Oncogene. 33:5311–5318. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yuan JH, Liu XN, Wang TT, Pan W, Tao QF,
Zhou WP, Wang F and Sun SH: The MBNL3 splicing factor promotes
hepatocellular carcinoma by increasing PXN expression through the
alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 19:820–832.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou M, Guo X, Wang M and Qin R: The
patterns of antisense long non-coding RNAs regulating corresponding
sense genes in human cancers. J Cancer. 12:1499–1506. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Luo N, Zhang K, Li X and Hu Y: ZEB1
induced-upregulation of long noncoding RNA ZEB1-AS1 facilitates the
progression of triple negative breast cancer by binding with ELAVL1
to maintain the stability of ZEB1 mRNA. J Cell Biochem.
121:4176–4187. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi
J, Liu B, Sun S, Yang F, Wang L and Qu L: Long noncoding RNA
EGFR-AS1 promotes cell growth and metastasis via affecting HuR
mediated mRNA stability of EGFR in renal cancer. Cell Death Dis.
10:1542019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gong YQ, Lu TL, Hou FT and Chen CW:
Antisense long non-coding RNAs in gastric cancer. Clin Chim Acta.
534:128–137. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Su J, Chen D, Ruan Y, Tian Y, Lv K, Zhou
X, Ying D and Lu Y: LncRNA MBNL1-AS1 represses gastric cancer
progression via the TGF-β pathway by modulating miR-424-5p/Smad7
axis. Bioengineered. 13:6978–6995. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhuang L, Ding W, Ding W, Zhang Q, Xu X
and Xi D: lncRNA ZNF667-AS1 (NR_036521.1) inhibits the progression
of colorectal cancer via regulating ANK2/JAK2 expression. J Cell
Physiol. 236:2178–2193. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: LncRNA HOXA11-AS promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin X, Zhuang S, Chen X, Du J, Zhong L,
Ding J, Wang L, Yi J, Hu G, Tang G, et al: lncRNA ITGB8-AS1
functions as a ceRNA to promote colorectal cancer growth and
migration through integrin-mediated focal adhesion signaling. Mol
Ther. 30:688–702. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bedoui S, Herold MJ and Strasser A:
Emerging connectivity of programmed cell death pathways and its
physiological implications. Nat Rev Mol Cell Bio. 21:678–695. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu J, Hong M, Li Y, Chen D, Wu Y and Hu
Y: Programmed cell death tunes tumor immunity. Front Immunol.
13:8473452022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zheng C, Chu M, Chen Q, Chen C, Wang ZW
and Chen X: The role of lncRNA OIP5-AS1 in cancer development and
progression. Apoptosis. 27:311–321. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Z and Jin J: LncRNA SLCO4A1-AS1
promotes colorectal cancer cell proliferation by enhancing
autophagy via miR-508-3p/PARD3 axis. Aging (Albany NY).
11:4876–4889. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xia X, Wang X, Cheng Z, Qin W, Lei L,
Jiang J and Hu J: The role of pyroptosis in cancer: Pro-cancer or
pro-‘host’? Cell Death Dis. 10:6502019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang G, Xiang Z, Wu H, He Q, Dou R, Lin
Z, Yang C, Huang S, Song J, Di Z, et al: The lncRNA
BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer
peritoneal metastasis by regulating VDAC3 ubiquitination. Int J
Biol Sci. 18:1415–1433. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bai Y and Li S: Long noncoding RNA
OIP5-AS1 aggravates cell proliferation, migration in gastric cancer
by epigenetically silencing NLRP6 expression via binding EZH2. J
Cell Biochem. 121:353–362. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang LW, Li XB, Liu Z, Zhao LH, Wang Y and
Yue L: Long non-coding RNA OIP5-AS1 promotes proliferation of
gastric cancer cells by targeting miR-641. Eur Rev Med Pharmacol
Sci. 23:10776–10784. 2019.PubMed/NCBI
|
|
54
|
Tao Y, Wan X, Fan Q, Wang Y, Sun H, Ma L,
Sun C and Wu Y: Long non-coding RNA OIP5-AS1 promotes the growth of
gastric cancer through the miR-367-3p/HMGA2 axis. Dig Liver Dis.
52:773–779. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liao Y, Zhang B, Zhang T, Zhang Y and Wang
F: LncRNA GATA6-AS promotes cancer cell proliferation and inhibits
apoptosis in glioma by downregulating lncRNA TUG1. Cancer Biother
Radiopharm. 34:660–665. 2019.PubMed/NCBI
|
|
56
|
Russell RC and Guan KL: The multifaceted
role of autophagy in cancer. EMBO J. 41:e1100312022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li J, Zhan H, Ren Y, Feng M, Wang Q, Jiao
Q, Wang Y, Liu X, Zhang S, Du L, et al: Sirtuin 4 activates
autophagy and inhibits tumorigenesis by upregulating the p53
signaling pathway. Cell Death Differ. 30:313–326. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Feng X, Zhang H, Meng L, Song H, Zhou Q,
Qu C, Zhao P, Li Q, Zou C, Liu X and Zhang Z: Hypoxia-induced
acetylation of PAK1 enhances autophagy and promotes brain
tumorigenesis via phosphorylating ATG5. Autophagy. 17:723–742.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Elias EE, Lyons B and Muruve DA:
Gasdermins and pyroptosis in the kidney. Nat Rev Nephrol.
19:337–350. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wei X, Xie F, Zhou X, Wu Y, Yan H, Liu T,
Huang J, Wang F, Zhou F and Zhang L: Role of pyroptosis in
inflammation and cancer. Cell Mol Immunol. 19:971–992. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tang R, Xu J, Zhang B, Liu J, Liang C, Hua
J, Meng Q, Yu X and Shi S: Ferroptosis, necroptosis, and pyroptosis
in anticancer immunity. J Hematol Oncol. 13:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yang P, Yang W, Wei Z, Li Y, Yang Y and
Wang J: Novel targets for gastric cancer: The tumor
microenvironment (TME), N6-methyladenosine (m6A), pyroptosis,
autophagy, ferroptosis and cuproptosis. Biomed Pharmacother.
163:1148832023. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang Z, Cao L, Zhou S, Lyu J, Gao Y and
Yang R: Construction and validation of a novel pyroptosis-related
Four-lncRNA prognostic signature related to gastric cancer and
immune infiltration. Front Immunol. 13:8547852022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ren N, Jiang T, Wang C, Xie S, Xing Y,
Piao D, Zhang T and Zhu Y: LncRNA ADAMTS9-AS2 inhibits gastric
cancer (GC) development and sensitizes chemoresistant GC cells to
cisplatin by regulating miR-223-3p/NLRP3 axis. Aging (Albany NY).
12:11025–11041. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Tang D, Chen X, Kang R and Kroemer G:
Ferroptosis: Molecular mechanisms and health implications. Cell
Res. 31:107–125. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wu Y, Zhang S, Gong X, Tam S, Xiao D, Liu
S and Tao Y: The epigenetic regulators and metabolic changes in
ferroptosis-associated cancer progression. Mol Cancer. 19:392020.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Katada S, Imhof A and Sassone-Corsi P:
Connecting threads: Epigenetics and metabolism. Cell. 148:24–28.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ganapathy-Kanniappan S and Geschwind JF:
Tumor glycolysis as a target for cancer therapy: Progress and
prospects. Mol Cancer. 12:1522013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Vaupel P, Schmidberger H and Mayer A: The
Warburg effect: Essential part of metabolic reprogramming and
central contributor to cancer progression. Int J Radiat Biol.
95:912–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Koppenol WH, Bounds PL and Dang CV: Otto
Warburg's contributions to current concepts of cancer metabolism.
Nat Rev Cancer. 11:325–337. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liberti MV and Locasale JW: The warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li C, Wang P, Du J, Chen J, Liu W and Ye
K: LncRNA RAD51-AS1/miR-29b/c-3p/NDRG2 crosstalk repressed
proliferation, invasion and glycolysis of colorectal cancer. IUBMB
life. 73:286–298. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Cheng X, Li J and Guo D: SCAP/SREBPs are
central players in lipid metabolism and novel metabolic targets in
cancer therapy. Curr Top Med Chem. 18:484–493. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li C, Zhang L, Qiu Z, Deng W and Wang W:
Key molecules of fatty acid metabolism in gastric cancer.
Biomolecules. 12:7062022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gong J, Lin Y, Zhang H, Liu C, Cheng Z,
Yang X, Zhang J, Xiao Y, Sang N, Qian X, et al: Reprogramming of
lipid metabolism in cancer-associated fibroblasts potentiates
migration of colorectal cancer cells. Cell Death Dis. 11:2672020.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Guo D, Reinitz F, Youssef M, Hong C,
Nathanson D, Akhavan D, Kuga D, Amzajerdi AN, Soto H, Zhu S, et al:
An LXR agonist promotes glioblastoma cell death through inhibition
of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov.
1:442–456. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang J, Song F, Zhao X, Jiang H, Wu X,
Wang B, Zhou M, Tian M, Shi B, Wang H, et al: EGFR modulates
monounsaturated fatty acid synthesis through phosphorylation of
SCD1 in lung cancer. Mol Cancer. 16:1272017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Tang Y, Tang R, Tang M, Huang P, Liao Z,
Zhou J, Zhou L, Su M, Chen P, Jiang J, et al: LncRNA DNAJC3-AS1
Regulates fatty acid synthase via the EGFR pathway to promote the
progression of colorectal cancer. Front Oncol. 10:6045342020.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Currie E, Schulze A, Zechner R, Walther TC
and Farese RJ: Cellular fatty acid metabolism and cancer. Cell
Metab. 18:153–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Carracedo A, Cantley LC and Pandolfi PP:
Cancer metabolism: Fatty acid oxidation in the limelight. Nat Rev
Cancer. 13:227–232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
He W, Liang B, Wang C, Li S, Zhao Y, Huang
Q, Liu Z, Yao Z, Wu Q, Liao W, et al: MSC-regulated lncRNA
MACC1-AS1 promotes stemness and chemoresistance through fatty acid
oxidation in gastric cancer. Oncogene. 38:4637–4654. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Greten FR and Grivennikov SI: Inflammation
and cancer: Triggers, mechanisms, and consequences. Immunity.
51:27–41. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Nie C, Zhai J, Wang Q, Zhu X, Xiang G, Liu
C, Liu T, Wang W, Wang Y, Zhao Y, et al: Comprehensive analysis of
an individualized Immune-Related lncRNA pair signature in gastric
cancer. Front Cell Dev Biol. 10:8056232022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Cui X, Morales RT, Qian W, Wang H, Gagner
JP, Dolgalev I, Placantonakis D, Zagzag D, Cimmino L, Snuderl M, et
al: Hacking macrophage-associated immunosuppression for regulating
glioblastoma angiogenesis. Biomaterials. 161:164–178. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wang H, Tian T and Zhang J:
Tumor-Associated Macrophages (TAMs) in Colorectal Cancer (CRC):
From mechanism to therapy and prognosis. Int J Mol Sci.
22:84702021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lazăr DC, Avram MF, Romoșan I, Cornianu M,
Tăban S and Goldiș A: Prognostic significance of tumor immune
microenvironment and immunotherapy: Novel insights and future
perspectives in gastric cancer. World J Gastroenterol.
24:3583–3616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Denaro N, Merlano MC and Lo Nigro C: Long
noncoding RNAs as regulators of cancer immunity. Mol Oncol.
13:61–73. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K,
Liu X, Xu T, Sun L, Qin J, et al: LncRNA SATB2-AS1 inhibits tumor
metastasis and affects the tumor immune cell microenvironment in
colorectal cancer by regulating SATB2. Mol Cancer. 18:1352019.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Sanchez-Tillo E, Liu Y, de Barrios O,
Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A
and Postigo A: EMT-activating transcription factors in cancer:
Beyond EMT and tumor invasiveness. Cell Mol Life Sci. 69:3429–3456.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Bio. 20:69–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Song Y, Lin M, Liu Y, Wang ZW and Zhu X:
Emerging role of F-box proteins in the regulation of
epithelial-mesenchymal transition and stem cells in human cancers.
Stem Cell Res Ther. 10:1242019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Brabletz S, Schuhwerk H, Brabletz T and
Stemmler MP: Dynamic EMT: A multi-tool for tumor progression. EMBO
J. 40:e1086472021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lin J, Shi Z, Yu Z and He Z: LncRNA
HIF1A-AS2 positively affects the progression and EMT formation of
colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed
Pharmacother. 98:433–439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Yu J, Han Z, Sun Z, Wang Y, Zheng M and
Song C: LncRNA SLCO4A1-AS1 facilitates growth and metastasis of
colorectal cancer through β-catenin-dependent Wnt pathway. J Exp
Clin Canc Res. 37:2222018. View Article : Google Scholar
|
|
98
|
Zhou L, Jiang J, Huang Z, Jin P, Peng L,
Luo M, Zhang Z, Chen Y, Xie N, Gao W, et al: Hypoxia-induced lncRNA
STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal
cancer progression by preventing m(6)A-mediated degradation of
STEAP3 mRNA. Mol Cancer. 21:1682022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ali MM, Akhade VS, Kosalai ST, Subhash S,
Statello L, Meryet-Figuiere M, Abrahamsson J, Mondal T and Kanduri
C: PAN-cancer analysis of S-phase enriched lncRNAs identifies
oncogenic drivers and biomarkers. Nat Commun. 9:8832018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Mo S, Zhang L, Dai W, Han L, Wang R, Xiang
W, Wang Z, Li Q, Yu J, Yuan J, et al: Antisense lncRNA LDLRAD4-AS1
promotes metastasis by decreasing the expression of LDLRAD4 and
predicts a poor prognosis in colorectal cancer. Cell Death Dis.
11:1552020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Slattery ML, Herrick J, Curtin K, Samowitz
W, Wolff RK, Caan BJ, Duggan D, Potter JD and Peters U: Increased
risk of colon cancer associated with a genetic polymorphism of
SMAD7. Cancer Res. 70:1479–1485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Rakic JM, Maillard C, Jost M, Bajou K,
Masson V, Devy L, Lambert V, Foidart JM and Noel A: Role of
plasminogen activator-plasmin system in tumor angiogenesis. Cell
Mol Life Sci. 60:463–473. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Bajou K, Maillard C, Jost M, Lijnen RH,
Gils A, Declerck P, Carmeliet P, Foidart JM and Noel A:
Host-derived plasminogen activator inhibitor-1 (PAI-1)
concentration is critical for in vivo tumoral angiogenesis and
growth. Oncogene. 23:6986–6990. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Teng F, Zhang JX, Chen Y, Shen XD, Su C,
Guo YJ, Wang PH, Shi CC, Lei M, Cao YO and Liu SQ: LncRNA
NKX2-1-AS1 promotes tumor progression and angiogenesis via
upregulation of SERPINE1 expression and activation of the VEGFR-2
signaling pathway in gastric cancer. Mol Oncol. 15:1234–1255. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Duan S, Huang W, Liu X, Liu X, Chen N, Xu
Q, Hu Y, Song W and Zhou J: IMPDH2 promotes colorectal cancer
progression through activation of the PI3K/AKT/mTOR and
PI3K/AKT/FOXO1 signaling pathways. J Exp Clin Canc Res. 37:3042018.
View Article : Google Scholar
|
|
107
|
Peng Y, Wang Y, Zhou C, Mei W and Zeng C:
PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we
making headway? Front Oncol. 12:8191282022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Liu HT, Ma RR, Lv BB, Zhang H, Shi DB, Guo
XY, Zhang GH and Gao P: LncRNA-HNF1A-AS1 functions as a competing
endogenous RNA to activate PI3K/AKT signalling pathway by sponging
miR-30b-3p in gastric cancer. Br J Cancer. 122:1825–1836. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Guo C, Zhou S, Yi W, Yang P, Li O, Liu J
and Peng C: Long non-coding RNA muskelin 1 antisense RNA (MKLN1-AS)
is a potential diagnostic and prognostic biomarker and therapeutic
target for hepatocellular carcinoma. Exp Mol Pathol.
120:1046382021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
El-Ashmawy NE, Al-Ashmawy GM and Hamouda
SM: Long non-coding RNA FAM83H-AS1 as an emerging marker for
diagnosis, prognosis and therapeutic targeting of cancer. Cell
Biochem Funct. 39:350–356. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Da J, Liu P, Wang R and Bu L: Upregulation
of the long non-coding RNA FAM83H-AS1 in gastric cancer and its
clinical significance. Pathol Res Pract. 215:1526162019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Chen Q, Zhou L, Ma D, Hou J, Lin Y, Wu J
and Tao M: LncRNA GAS6-AS1 facilitates tumorigenesis and metastasis
of colorectal cancer by regulating TRIM14 through
miR-370-3p/miR-1296-5p and FUS. J Transl Med. 20:3562022.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Lu GH, Zhao HM, Liu ZY, Cao Q, Shao RD and
Sun G: LncRNA SAMD12-AS1 promotes the progression of gastric cancer
via DNMT1/p53 Axis. Arch Med Res. 52:683–691. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Macleod K: Tumor suppressor genes. Curr
Opin Genet Dev. 10:81–93. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Sherr CJ: Principles of tumor suppression.
Cell. 116:235–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Sun W and Yang J: Functional mechanisms
for human tumor suppressors. J Cancer. 1:136–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Liu HT, Liu S, Liu L, Ma RR and Gao P:
EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 promotes cell-cycle
progression in gastric cancer. Cancer Res. 78:5877–5890. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhang J, Cui K, Huang L, Yang F, Sun S,
Bian Z, Wang X, Li C, Yin Y, Huang S, et al: SLCO4A1-AS1 promotes
colorectal tumourigenesis by regulating Cdk2/c-Myc signalling. J
Biomed Sci. 29:42022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Ni X, Ding Y, Yuan H, Shao J, Yan Y, Guo
R, Luan W and Xu M: Long non-coding RNA ZEB1-AS1 promotes colon
adenocarcinoma malignant progression via miR-455-3p/PAK2 axis. Cell
Prolif. 53:e127232020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wu X, Cui F, Chen Y, Zhu Y and Liu F: Long
Non-Coding RNA LOXL1-AS1 enhances colorectal cancer proliferation,
migration and invasion through miR-708-5p/CD44-EGFR Axis. Onco
Targets Ther. 13:7615–7627. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Flecken T, Schmidt N, Hild S, Gostick E,
Drognitz O, Zeiser R, Schemmer P, Bruns H, Eiermann T, Price DA, et
al: Immunodominance and functional alterations of tumor-associated
antigen-specific CD8+ T-cell responses in hepatocellular carcinoma.
Hepatology. 59:1415–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Chow A, Perica K, Klebanoff CA and Wolchok
JD: Clinical implications of T cell exhaustion for cancer
immunotherapy. Nat Rev Clin Oncol. 19:775–790. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Pardoll DM: The blockade of immune
checkpoints in cancer immunotherapy. Nat Rev Cancer. 12:252–264.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
He X and Xu C: Immune checkpoint signaling
and cancer immunotherapy. Cell Res. 30:660–669. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Pu Y and Ji Q: Tumor-Associated
macrophages regulate PD-1/PD-L1 immunosuppression. Front Immunol.
13:8745892022. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Topalian SL, Drake CG and Pardoll DM:
Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor
immunity. Curr opin immunol. 24:207–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Fan F, Chen K, Lu X, Li A, Liu C and Wu B:
Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging
hsa-miR-194-5p induces immunosuppression in hepatocellular
carcinoma. Hepatol Int. 15:444–458. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Han Y, Liu D and Li L: PD-1/PD-L1 pathway:
Current researches in cancer. Am J Cancer Res. 10:727–742.
2020.PubMed/NCBI
|
|
129
|
Zhou Z, Liu Q, Zhang G, Mohammed D, Amadou
S, Tan G and Zhang X: HOXA11-AS1 Promotes PD-L1-Mediated immune
escape and metastasis of hypopharyngeal carcinoma by facilitating
PTBP1 and FOSL1 Association. Cancers (Basel). 14:36942022.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Chen YG, Satpathy AT and Chang HY: Gene
regulation in the immune system by long noncoding RNAs. Nat
immunol. 18:962–972. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wu Y, Song Y, Wang R and Wang T: Molecular
mechanisms of tumor resistance to radiotherapy. Mol Cancer.
22:962023. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Hou J, Zhang G, Wang X, Wang Y and Wang K:
Functions and mechanisms of lncRNA MALAT1 in cancer chemotherapy
resistance. Biomark Res. 11:232023. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chen QN, Wei CC, Wang ZX and Sun M: Long
non-coding RNAs in anti-cancer drug resistance. Oncotarget.
8:1925–1936. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Ni J, Bucci J, Malouf D, Knox M, Graham P
and Li Y: Exosomes in cancer radioresistance. Front Oncol.
9:8692019. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhou M, Dong J, Huang J, Ye W, Zheng Z,
Huang K, Pan Y, Cen J, Liang Y, Shu G, et al: Chitosan-Gelatin-EGCG
Nanoparticle-Meditated LncRNA TMEM44-AS1 Silencing to Activate the
P53 signaling pathway for the synergistic reversal of 5-FU
resistance in gastric cancer. Adv Sci (Weinh). 9:e21050772022.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Qu L, Chen Y, Zhang F and He L: The lncRNA
DLGAP1-AS1/miR-149-5p/TGFB2 axis contributes to colorectal cancer
progression and 5-FU resistance by regulating smad2 pathway. Mol
Ther Oncolytics. 20:607–624. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Gui Z, Zhao Z, Sun Q, Shao G, Huang J,
Zhao W and Kuang Y: LncRNA FEZF1-AS1 promotes multi-drug resistance
of gastric cancer cells via upregulating ATG5. Front Cell Dev Biol.
9:7491292021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zou Y, Yao S, Chen X, Liu D, Wang J, Yuan
X, Rao J, Xiong H, Yu S, Yuan X, et al: LncRNA OIP5-AS1 regulates
radioresistance by targeting DYRK1A through miR-369-3p in
colorectal cancer cells. Eur J Cell Biol. 97:369–378. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Song L, Zhou Z, Gan Y, Li P, Xu Y, Zhang
Z, Luo F, Xu J, Zhou Q and Dai F: Long noncoding RNA OIP5-AS1
causes cisplatin resistance in osteosarcoma through inducing the
LPAATβ/PI3K/AKT/mTOR signaling pathway by sponging the miR-340-5p.
J Cell Biochem. 120:9656–9666. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Wu Q, Ma J, Wei J, Meng W, Wang Y and Shi
M: FOXD1-AS1 regulates FOXD1 translation and promotes gastric
cancer progression and chemoresistance by activating the
PI3K/AKT/mTOR pathway. Mol Oncol. 15:299–316. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Qi FF, Yang Y, Zhang H and Chen H: Long
non-coding RNAs: Key regulators in oxaliplatin resistance of
colorectal cancer. Biomed Pharmacother. 128:1103292020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Shi CJ, Xue ZH, Zeng WQ, Deng LQ, Pang FX,
Zhang FW, Fu WM and Zhang JF: LncRNA-NEF suppressed oxaliplatin
resistance and epithelial-mesenchymal transition in colorectal
cancer through epigenetically inactivating MEK/ERK signaling.
Cancer Gene Ther. 30:855–865. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Li Y, Gan Y, Liu J, Li J, Zhou Z, Tian R,
Sun R, Liu J, Xiao Q, Li Y, et al: Downregulation of MEIS1 mediated
by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and
oxaliplatin resistance in colorectal cancer. Signal Transduct
Target. 7:872022. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Liang J, Tian XF and Yang W: Effects of
long non-coding RNA Opa-interacting protein 5 antisense RNA 1 on
colon cancer cell resistance to oxaliplatin and its regulation of
microRNA-137. World J Gastroenterol. 26:1474–1489. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Hui B, Lu C, Wang J, Xu Y, Yang Y, Ji H,
Li X, Xu L, Wang J, Tang W, et al: Engineered exosomes for
co-delivery of PGM5-AS1 and oxaliplatin to reverse drug resistance
in colon cancer. J Cell Physiol. 237:911–933. 2022. View Article : Google Scholar : PubMed/NCBI
|















