Introduction
Breast cancer remains the most commonly diagnosed
type of cancer in women, accounting for one in eight cancer
diagnoses globally, with >2,000,000 new breast cancer cases
diagnosed and 685,000 breast cancer-associated deaths recorded in
2020 (1). Due to population growth
and aging alone, the global burden of breast cancer is expected to
increase to >3,000,000 new cases and >1,000,000 deaths by
2040 (2). Notably, there has been
a significant decrease in the breast cancer-related death rate over
the years, with an overall decline of 43% in 2020 compared with in
1989 (1); however, this decline
has slowed slightly in recent years. This decrease has been due to
advances in treatment and earlier disease detection through
screening. An improved understanding of the biological
heterogeneity of breast cancer has also led to advances in the
effective and personalized approach of molecular targeted therapies
(2).
Based on the expression of hormone receptors (HRs)
and growth factor receptors, breast cancer is categorized into
distinct molecular subtypes: Luminal A [HR+ and human
epidermal growth factor receptor 2 (HER2−)]; luminal B
(HR+/HER2+); HER2+/HR−;
and triple-negative breast cancer (TNBC;
HR−/HER2−) (3). Globally, of patients with breast
cancer, 83% have HR+ subtypes; of which 71% are luminal
A and 12% are luminal B (4). The
luminal A subgroup is less aggressive, more responsive to hormonal
interventions and has a better prognosis compared with the luminal
B subgroup, which is further defined by its high expression of the
proliferation marker antigen Ki67 and HER2+. The
HER2+ subtype has high expression of the HER2 oncogene;
thus, anti-HER2 therapies may be used for treatment. Globally, the
HER2+ subgroup accounts for only 5% of patients with
breast cancer, while the remaining 12% of the total patient
population has the TNBC subtype (4). Due to the absence of molecular
targets, such as overexpression of hormone receptors and HER2, TNBC
is more difficult to treat than other subtypes of breast cancer
(5).
Clinical classification of breast cancer informs the
choice of treatment. In women with advanced or metastatic estrogen
receptor (ER)+ and HER2− breast cancer,
palbociclib and abemaciclib were recently approved by the United
States Food and Drug Administration for their use in combination
with hormone therapy. In addition, alspelisib has been approved as
combination therapy with hormonal therapy for the treatment of
HR+ and HER2− breast cancer with
phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit α
(PIK3CA) gene mutations (6). Trastuzumab and pertuzumab are target
therapies selected for the treatment of HER2+ breast
cancer. Additionally, trastuzumab has been used in the prevention
of relapsed early-stage HER2+ breast cancer.
Chemotherapy is the main treatment for TNBC due to its lack of
expression of both HRs and HER2; therefore, patients with this type
of cancer do not exhibit a response to targeted therapies (7). Pembrolizumab and atezolizumab are
some of the immunotherapy drugs recommended for use in patients
with metastatic TNBC with tumors expressing programmed death ligand
1 (PD-L1). PARP inhibitor drugs, including olaparib and
talazoparib, have been used as targeted therapies for TNBC caused
by inherited breast cancer gene (BRCA) alterations (8).
Several biomarkers for the diagnosis, prognostic
prediction and treatment selection of breast cancer have been
developed. These include HRs, HER2 and multigene tests, such as
Oncotype DX, EndoPredict, MammaPrint and Prosigna (9). Breast cancer biomarkers are
constantly evolving from the conventional biomarkers used in
immunohistochemistry, such as ER, progesterone receptor (PR) and
HER2, to genetic biomarkers with therapeutic implications, such as
BRCA1/2, ER, HER2, PIK3CA and neurotrophic tyrosine receptor
kinase gene mutations, and microsatellite instability (10). Additionally, immunomarkers, such as
PD-L1, are being employed. While these biomarkers have been
developed, they are still limited in their specificity and clinical
efficacy (11). Therefore, it is
crucial to develop targeted molecular therapies with high accuracy
and easily measurable companion diagnostics.
In a preliminary study, target screening for the
diagnosis and treatment of cancer was performed by gene expression
profile analyses, followed by tissue microarray analyses of solid
breast cancer and normal tissues (Fig. S1). Several oncoantigens that were
related to carcinogenesis and cancer development have previously
been isolated using this screening process (12–32).
Following target screening shown in Fig. S1, the present study identified
kinesin family member 20B [KIF20B, also known as M-phase
phosphoprotein 1 (MPHOSPH1)] as a candidate molecular target that
was frequently and highly expressed in breast cancer. KIF20B
belongs to the kinesin superfamily, which possesses a highly
conserved motor domain, the ATPase activity of which enables it to
undergo microtubule-dependent plus-end movement. As a
microtubule-associated protein, KIF20B is important for cytokinesis
(33). There has been increasing
evidence of high expression levels of kinesin family proteins in
cancer, suggesting that, in addition to their normal physiological
cell function, their upregulation may have oncogenic potential in
some types of cancer. Previous reports have suggested that
KIF20B is highly expressed in some types of human cancer,
including hepatocellular carcinoma, bladder, colorectal, pancreatic
and tongue cancer (34–38); however, to the best of our
knowledge, there has been no detailed investigation on the function
of KIF20B in tumorigenesis, and its value as a therapeutic and
diagnostic target of various subtypes of breast cancer. This
prompted an investigation into the possible role of KIF20B in the
development of breast cancer. Therefore, the present study reveals
the functional role of KIF20B in breast cancer development, and
demonstrates its value as a therapeutic target and prognostic
biomarker.
Materials and methods
Breast cancer cell lines and clinical
tissue samples
The present study used eight breast cancer cell
lines (T-47D, ZR-75-1, MCF7, AU565, SK-BR-3, HCC1599, HCC1937 and
MDA-MB-468), which were commercially purchased. Normal primary
human mammary epithelial cells (HMECs) were also commercially
purchased and the use of primary human cells was approved by the
ethics committee. The features of all cells are described in
Table I. RPMI-1640 (FUJIFILM Wako
Pure Chemical Corporation), EMEM (FUJIFILM Wako Pure Chemical
Corporation), McCoy's 5A (Gibco; Thermo Fisher Scientific, Inc.)
and Leibovitz's L-15 (Gibco; Thermo Fisher Scientific, Inc.) media
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) were prepared and the cells were cultured in
monolayers. The cells were incubated at 37°C in an incubator
containing 5% CO2, with the exception of the MDA-MB-468
cells, which required 0% CO2 incubation. HMECs were
maintained in Mammary Epithelial Cell Basal Medium (cat. no.
PCS-600-030) supplemented with Mammary Epithelial Cell Growth Kit
(cat. no. PCS-600-040) (both from American Type Culture
Collection).
 | Table I.Human breast cancer cell lines and
normal breast cells. |
Table I.
Human breast cancer cell lines and
normal breast cells.
| Cell line | Histology | Subtype | Medium | Catalog no. |
|---|
| T-47D | Ductal
carcinoma | Luminal A | RPMI-1640 | HTB-133 |
| ZR-75-1 | Ductal
carcinoma | Luminal B | RPMI-1640 | CRL-1500 |
| MCF7 | Adenocarcinoma | Luminal A | EMEM | HTB-22 |
| AU565 | Adenocarcinoma |
HER2+ | RPMI-1640 | CRL-2351 |
| SK-BR-3 | Adenocarcinoma |
HER2+ | McCoy's 5A | HTB-30 |
| HCC1599 | Ductal
carcinoma | TNBC | RPMI-1640 | CRL-2331 |
| HCC1937 | Primary ductal
carcinoma | TNBC | RPMI-1640 | CRL-2336 |
| MDA-MB-468 | Adenocarcinoma | TNBC | Leibovitz's
L-15 | HTB-132 |
| HMECs | Normal breast | N/A | Mammary epithelial
cell basal medium | PCS-600-030 |
For reverse transcription-quantitative PCR
(RT-qPCR), 15 breast tumor samples and three normal breast tissue
samples were collected between June 2005 and April 2010 from
patients who provided written informed consent at Kanagawa Cancer
Center (Yokohama, Japan) (Table
SI). The pathologist confirmed that the obtained normal tissues
were non-neoplastic and were not contaminated with tumor cells.
The use of the tissues was approved by the Ethics
Committee of Kanagawa Cancer Center (approval date: June 8, 2005).
In addition, 251 formalin-fixed primary breast cancer tissues and
adjacent healthy tissues were obtained from Japanese female
patients (median age, 65 years; range, 28–89 years) from Kanagawa
Cancer Center for tissue microarray generation. The clinical stage
of the breast cancer samples was determined according to the Union
for International Cancer Control TNM classification 7th Edition
(39). The project to establish
tumor tissue microarrays from archival formalin-fixed and
paraffin-embedded, surgically obtained tissues and to use the
tissue microarrays for later researches was approved by the
Kanagawa Cancer Center Ethics Committee (approval no. Rin-177;
Yokohama, Japan). The patients whose tissues were used to generate
the breast cancer tissue microarrays received surgery at Kanagawa
Cancer Center Hospital between January 2004 and March 2006. Written
informed consent was obtained from the patients for the use of
their clinical information and for specimens remaining after
clinical examinations, such as archival formalin-fixed and
paraffin-embedded specimens. The ethics committee at Shiga
University of Medical Science (approval no. 21-163) approved the
present study including the use of all aforementioned clinical
materials.
RT-qPCR
Maxwell® 16 LEV simplyRNA Cells and
Tissue kits (Promega Corporation) were used to extract total RNA
from the cultured cells and clinical tissues according to the
manufacturer's protocol. PrimeScript RT Master Mix (Takara Bio,
Inc.) was used to synthesize cDNA from total RNA according to the
manufacturer's instructions. Samples were incubated at 37°C for 15
min and 85°C for 5 sec. mRNA expression was quantified by qPCR
analysis using TaqMan® Fast Universal PCR Master Mix
(Thermo Fisher Scientific, Inc.) and QuantStudio™ 3
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. For the
assays, KIF20B (assay ID Hs01027505_m1) primers were used,
and β-actin (ACTB; assay ID Hs01060665_g1) (both from Thermo
Fisher Scientific, Inc.) was used as a control. All experiments
were performed in triplicate. The thermal cycling conditions were
as follows: Initial denaturation for 20 sec at 95°C, followed by 40
cycles at 95°C for 1 sec and 60°C for 20 sec. Using the
2−ΔΔCq method (40) and
with ACTB mRNA expression as a reference, KIF20B mRNA
expression levels were quantified.
Western blot analysis
Cold phosphate-buffered saline (PBS) was used to
wash the cells, and RIPA buffer (Pierce; Thermo Fisher Scientific,
Inc.) containing a 1% protease inhibitor cocktail was used to lyse
the cells. After homogenization, lysates were incubated at 4°C for
30 min, and the supernatant was centrifuged at 20,400 × g for 15
min at 4°C to separate proteins from debris. The DC Protein assay
kit (Thermo Fisher Scientific, Inc.) was used to measure total
protein concentration. A sample buffer containing SDS was then
added to the proteins, boiled for 5 min at 100°C and then incubated
at room temperature for 5 min. Using Mini-Protean® TGX
7.5% precast gels (Bio-Rad Laboratories, Inc.), proteins were
electrophoresed and transferred onto Trans-Blot® Turbo
0.2 µm polyvinylidene difluoride membranes (cat. no. 1704156;
Bio-Rad Laboratories, Inc.). The membranes were blocked in 5%
nonfat dried milk in 1X TBS-0.05% Tween 20 (cat. no. 9997; Cell
Signaling Technology, Inc.) overnight at 4°C, after which they were
incubated with rabbit polyclonal anti-KIF20B antibody (1:200; cat.
no. ab122165; Abcam) or anti-ACTB antibody (1:2,000; cat. no.
12262S; Cell Signaling Technology, Inc.) overnight at 4°C.
Membranes were then incubated with anti-rabbit horseradish
peroxidase (HRP)-conjugated secondary antibodies (1:2,000; cat. no.
NA934; Cytiva) for 1 h at room temperature. A Fusion Solo S system
(Vilber Lourmat) was used to visualize protein bands with
chemiluminescence western blotting detection reagents (cat no.
RPN2232; Cytiva).
Immunocytochemistry
Cells (5.0×103 cells/well) were seeded
onto an 8-well Lab-Tek II chamber slides (Nalge Nunc
International]. The cells were fixed in 4% formaldehyde solution
for 15 min at room temperature, washed and then permeabilized using
PBS (−) containing 0.2% Triton X-100 for 2 min at room temperature.
Thereafter, they were blocked in 3% bovine serum albumin (BSA;
FUJIFILM Wako Pure Chemical Corporation) in PBS (−) for 30 min at
4°C. The cells were then incubated with the rabbit polyclonal
anti-KIF20B antibody (1:100; cat. no. ab122165; Abcam) in PBS (−)
containing 1% BSA and 0.1% Tween-20 at 4°C overnight. After washing
with PBS (−), the cells were incubated with Alexa Fluor®
488-conjugated goat anti-rabbit IgG secondary antibody (1:800; cat.
no. A11008; Thermo Fisher Scientific, Inc.) in a wet chamber for 90
min at room temperature. Nuclei were stained with VECTASHIELD
Antifade Mounting Medium containing DAPI (Vector Laboratories,
Inc.) and visualized with a confocal laser scanning microscope
(Leica TCS SP8 X; Leica Microsystems, Inc.).
Immunohistochemistry and tissue
microarray analysis
Tumor tissue microarrays were constructed according
to a previously published process (13). Formalin-fixed paraffin-embedded
breast cancer tissues and normal tissues were obtained from the
Kanagawa Cancer Center. A visually aligned section of hematoxylin-
and eosin-stained tissue was used to select the areas for sampling.
Slides for tissue microarrays were deparaffinized in xylene and
rehydrated in graded concentrations of ethanol. Target Retrieval
Solution (pH 9) (Dako; Agilent Technologies, Inc.) was used in a
microwave oven for heat-induced antigen retrieval. After blocking
endogenous peroxidase activity with a 0.3% hydrogen
peroxide/methanol mixture and nonspecific protein binding sites
with protein blocking buffer (cat. no. X0909; Dako; Agilent
Technologies, Inc.), the sections were incubated with anti-KIF20B
antibody (1:200; cat. no. ab122165; Abcam) overnight at 4°C. After
primary antibody incubation, sections were incubated at room
temperature for 30 min with polymer anti-rabbit secondary
antibodies labeled with EnVision+System-HRP (cat. no. K4003; Dako;
Agilent Technologies, Inc.). Specimens were counterstained at room
temperature with hematoxylin for 1 min, after being incubated with
3,3′-DAB chromogen and DAB substrate buffer. Since the staining
intensity within each tumor tissue core was mostly homogenous, the
semi-quantitative evaluation of KIF20B staining intensity was
performed by three independent investigators. The expression
patterns of KIF20B in tissue arrays were classified from
absent/weak to strong. Images of the immunostained samples were
acquired with a NanoZoomer whole slide scanner (Hamamatsu Photonics
K.K.) and positivity of the KIF20B protein was semi-quantitatively
analyzed (30,32) by three independent investigators
without prior knowledge of the clinicopathological data. Since the
intensity of staining within each tumor tissue core was mostly
homogeneous, the staining intensity for KIF20B was recorded as
strong positive, weak positive or negative. The staining patterns
were defined as strong positive if all of the three reviewers
independently classified them as such.
RNA interference assay
To examine the biological functions of KIF20B, small
interfering RNAs (siRNAs, 100 µM; Sigma-Aldrich; Merck KGaA) were
transfected into T-47D, HCC1937 and SK-BR-3 breast cancer cells
(0.5×106 cells/dish in 10-cm culture dishes) at 37°C for
5 h using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Each gene was targeted using the following sequences:
si-KIF20B-#1, sense 5′-GAUGUAUCACUAGACAGUA-3′, antisense
5′-UACUGUCUAGUGAUACAUC-3′; si-KIF20B-#2, sense
5-CAACGAAUUUCAGAACCUA-3, antisense 5′-UAGGUUCUGAAAUUCGUUG-3′;
si-LUC control 1, sense 5′-CGUACGCGGAAUACUUCGA-3′, antisense
5′-UCGAAGUAUUCCGCGUACG-3′; and si-EGFP control 2, sense
5′-GAAGCAGCACGACUUCUUC-3′, antisense 5′-GAAGAAGUCGUGCUGCUUC-3′
(Sigma-Aldrich; Merck KGaA). Western blot analysis was performed 72
h post-transfection, with antibodies against KIF20B to confirm
inhibition of KIF20B expression by siRNAs.
For the cell viability assay, breast cancer cells
transfected with siRNAs against KIF20B or si-controls were plated
in 6-well plates at a density of 0.5×105 cells/well. On
day 7 post-transfection, the Cell Counting Kit-8 (CCK-8) solution
(Dojindo Laboratories, Inc.) was used to measure cell viability.
The cells were incubated with CCK-8 reagent for 4 h and the
absorbance was measured at 450 nm using a multi-label plate reader
(ARVO X4 system; PerkinElmer, Inc.).
Colony formation assay
A total of 0.5×106 cells/dish were seeded
in 10-cm culture dishes and transfected with siRNAs against KIF20B
or si-LUC at 37°C for 5 h. After 7 days, the cells were washed with
PBS (−) and fixed with 4% paraformaldehyde phosphate-buffered
solution (FUJIFILM Wako Pure Chemical Corporation) at 4°C for 30
min. The cells were then stained with Giemsa at room temperature
for 1 h. A Canon PIXUS-MP990 multifunction device was used to
capture the images and colonies, defined as ≥50 cells, were
counted.
Annexin V fluorescein isothiocyanate
(FITC)-propidium iodide (PI) assay
Breast cancer cells that had been transfected with
siRNAs against KIF20B or si-LUC at 37°C for 5 h were plated in
10-cm dishes at a density of 0.5×106 cells/dish. After
48 h, the cells were trypsinized, washed twice with PBS and then
resuspended in 1X binding buffer at a concentration of
1×106 cells/ml. Apoptotic cells were stained using the
FITC Annexin V Apoptosis Detection Kit I (BD Pharmingen; BD
Biosciences) according to manufacturer's instructions. Control
assays were performed to set up compensation and quadrants. The
PI-A and Annexin V FITC-A plots were used for gating cells and
identifying changes in the scatter properties of the cells. The
data were analyzed and the plots were automatically generated using
the BD FACSCanto™ II flow cytometer (Ver1.2) and BD
FACSDiva™ software (Ver7.0), (BD Biosciences).
Caspase 3/7 expression assay
Caspase 3/7 activities were analyzed using the
CellEvent Caspase-3/7 Green Detection Reagent (cat. no. C10423;
Invitrogen; Thermo Fisher Scientific, Inc.), according to
manufacturer's instructions. Breast cancer cells transfected with
si-KIF20B or si-LUC (0.5×106 cells/dish) were seeded
into 6-well plates at a density of 0.5×105 cells/well in
an appropriate culture medium containing 10% FBS. The cells were
then incubated with CellEvent Caspase-3/7 Green Detection Reagent
at a final concentration of 4 µM for 30 min at 37°C, and then
monitored by live cell imaging at 15 min interval for 6 h using the
Evos M7000 Auto Imaging System (Thermo Fisher Scientific,
Inc.).
Flow cytometric analysis
The BD FACSCanto™ II flow cytometer
(Ver1.2), BD FACSDiva™ software (Ver7.0) and the
CycleTEST PLUS DNA Reagent Kit (cat. no. 340242), (all BD
Biosciences) were used to carry out flow cytometric analysis of
cell cycle progression following the manufacturer's instructions.
The breast cancer cell lines were transfected with si-KIF20B-#2 or
si-LUC siRNA oligonucleotides at 37°C for 5 h. To assess DNA
ploidy, 1×106 cells/ml were harvested 72 h
post-transfection. Briefly, 250 µl solution A (trypsin buffer) was
added to the cells and incubated at room temperature for 10 min.
Subsequently, 200 µl solution B (trypsin inhibitor and RNase
buffer) was then added to the cells, gently mixed by tapping the
tube and incubated at room temperature for 10 min. After
incubation, 200 µl cold solution C (PI staining solution) was added
to the cells and incubated in the dark at 4°C for 10 min. A 70-µm
nylon mesh was used to filter the samples and DNA content was
analyzed within 3 h in 20,000 ungated cells.
Live cell imaging
The Evos M7000 Auto Imaging System (Thermo Fisher
Scientific, Inc.) was used to monitor cytokinetics. Briefly, breast
cancer cells (0.5×106 cells/dish) transfected with
si-KIF20B or si-LUC were seeded into 6-well plates at a density of
0.5×105 cells/well in an appropriate culture medium
containing 10% FBS. Cell morphology and dynamics were monitored
using the Evos M7000 Auto Cell Imaging System. Images were captured
every 15 min over a period of 24 h up to 120 h.
Matrigel invasion assay
Cells transfected with siRNA against KIF20B or with
si-LUC were grown to 60% confluence in culture medium containing
10% FBS. After trypsinization, cells were washed with PBS (−) and
suspended in medium without serum or protease inhibitor. Matrigel
invasion assay was conducted according to the manufacturer's
recommendation using Matrigel matrix (BD Biosciences) as described
previously (30).
Statistical analysis
Statistical analyses were performed using the Stat
View 5.0 Statistical Program (SAS Institute, Inc.). The unpaired
Student's t-test was used to analyze the difference between two
groups in cell-based assays. Multiple comparisons were conducted
using one-way ANOVA followed by Tukey's post hoc test to compare
means of each group with those of the other groups. The results are
presented as the mean ± standard deviation, and each experiment was
performed in triplicate. An analysis of the association between
KIF20B expression and clinical variables, including age,
histological type, ER, PR and HER2 status, pathological T (pT) and
N (pN) stages was conducted using Fisher's exact test. An overall
survival (OS) curve was calculated based on the date of surgery and
the date of breast cancer-related death or last follow-up. Breast
tumors were analyzed for KIF20B expression and Kaplan-Meier curves
were calculated for each relevant variable. Log-rank tests were
used to analyze differences in survival duration between patient
subgroups. To determine whether clinicopathological factors and
cancer-related mortality were associated, a Cox proportional hazard
regression model was used for univariate and multivariate analyses.
In the first step, individual associations between death and
possible prognostic factors were examined, including age, ER
status, PR status, HER2 status, pT classification and pN
classification. Using backward (stepwise) procedures, a
multivariate analysis was conducted incorporating KIF20B expression
into the model, along with all the variables statistically
significant (P<0.05) and independent of the other variables.
P<0.05 was considered to indicate a statistically significant
difference.
Database analysis
BioGPS (http://biogps.org/#goto=welcome), GTex Dataset
(https://gtexportal.org/home/) and UALCAN
(http://ualcan.path.uab.edu/) databases
were used to examine the expression of KIF20B in tumor and normal
tissues and organs. cBioportal for Cancer Genomics (http://www.cbioportal.org/) was used to investigate
the mutational status of the KIF20B gene. The PrognoScan
(http://www.prognoscan.org/) database was
used to assess the association between KIF20B expression and the OS
of patients with breast cancer.
Results
KIF20B expression in breast cancer
cell lines and tissues
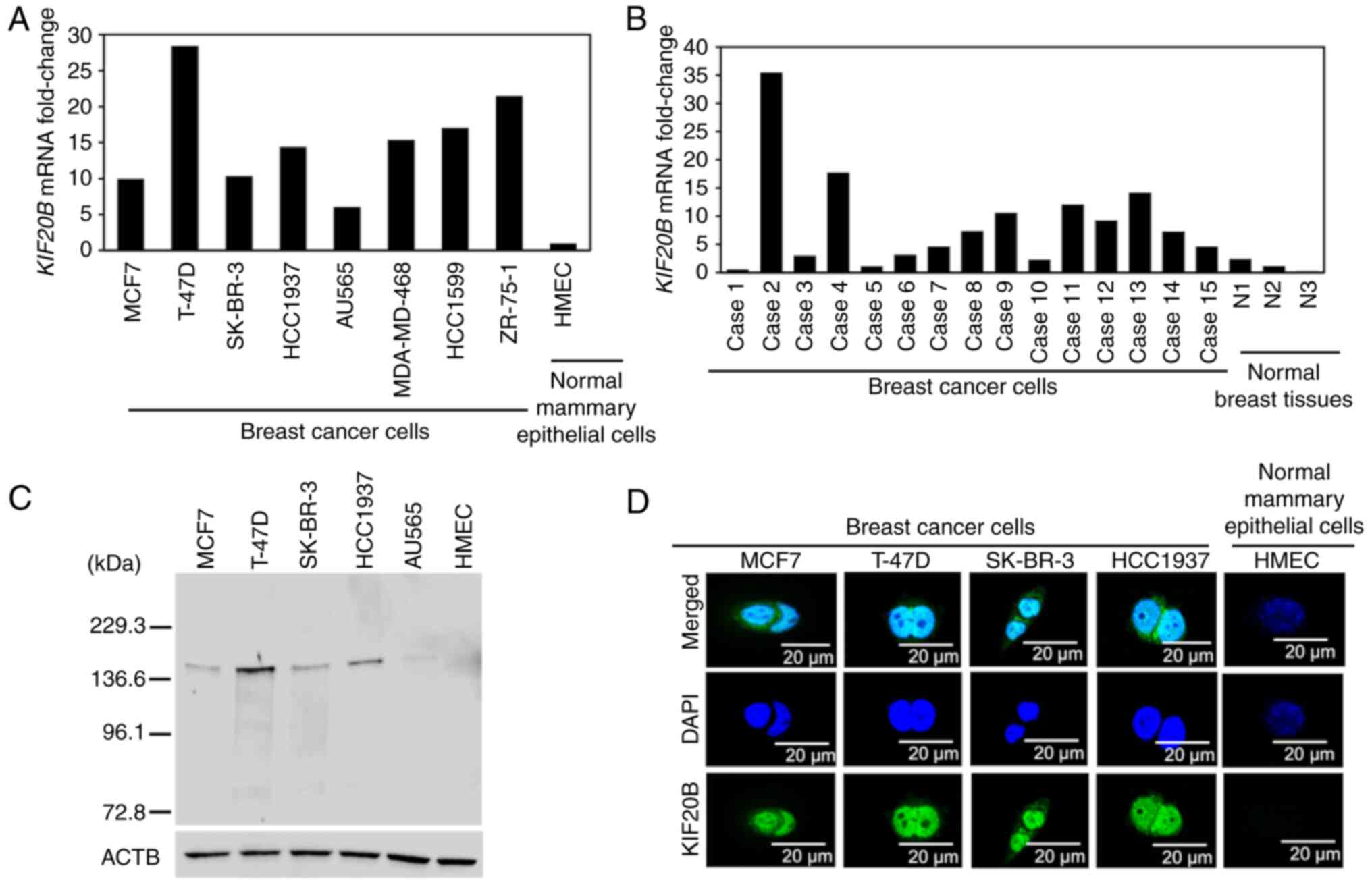
Using RT-qPCR, KIF20B mRNA expression was
detected in all of the breast cancer cell lines and tissues;
however, scarce expression was observed in normal breast epithelial
cells (HMECs) and tissues (Fig. 1A and
B). Cell lines representing each of the three breast cancer
subtypes were selected for western blotting and immunocytochemistry
experiments in the present study. KIF20B protein was found to be
highly expressed in breast cancer cells when compared with HMECs as
detected by western blotting (Fig.
1C). Furthermore, immunocytochemical analysis showed that
KIF20B protein was mainly localized in the cytoplasm and nuclei of
KIF20B-positive breast cancer cells (Fig. 1D).
Association of KIF20B expression with
poor prognosis in patients with breast cancer
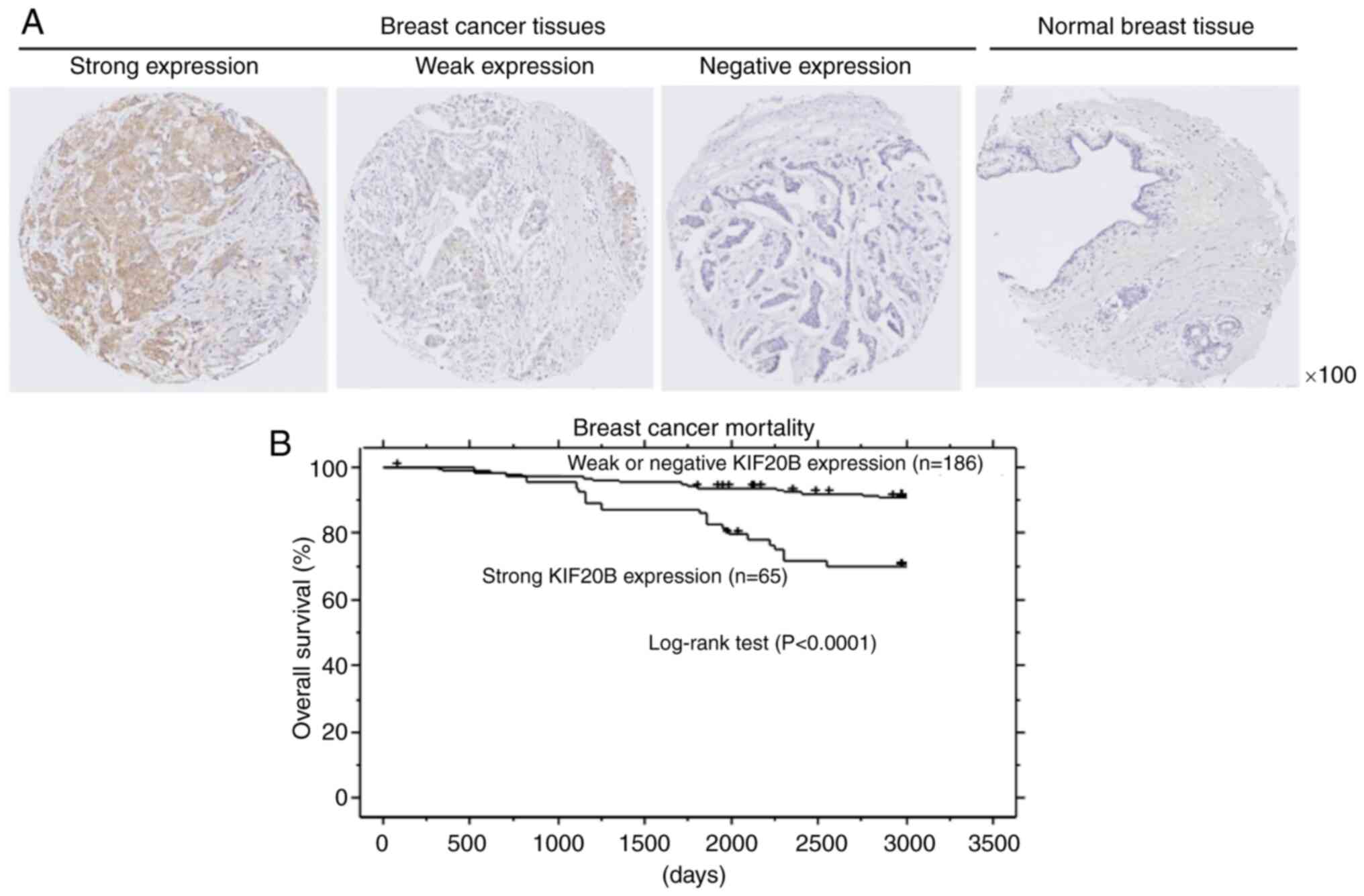
Immunohistochemical analysis detected KIF20B
expression in 145 (57.8%) out of the 251 breast cancer tissues
(Fig. 2A and Table II). In addition,
clinicopathological parameters related to KIF20B protein expression
were evaluated (Table II). Strong
KIF20B expression was significantly related to advanced pN stage.
Kaplan-Meier analysis showed that strong KIF20B protein expression
was associated with a poorer prognosis in patients with breast
cancer (Fig. 2B). Furthermore,
univariate analysis was conducted to determine the relationship
between patient prognosis and variables, such as KIF20B expression
status (strong positive vs. weak positive and negative), age (≥65
vs. <65 years), Luminal type (negative vs. positive), HER2
status (positive vs. negative), pT classification (T3-4 vs. T1-2)
and pN classification (N1-2 vs. N0). A strong positive KIF20B
expression, an advanced pT stage and an advanced pN stage were
significantly associated with a worse prognosis. Furthermore, in
multivariate analysis, both strong positive KIF20B expression and
advanced pN stage were independent prognostic factors (Table III).
 | Table II.Association of KIF20B protein
expression in breast cancer tissues with patient characteristics
(n=251). |
Table II.
Association of KIF20B protein
expression in breast cancer tissues with patient characteristics
(n=251).
| Parameter | All patients | Strong positive
KIF20B expression | Weak positive
KIF20B expression | Negative KIF20B
expression |
P-valuea |
|---|
| Total | 251 | 65 | 80 | 106 |
|
| Age, years |
|
|
|
| 0.2419 |
|
<65 | 188 | 45 | 59 | 84 |
|
|
≥65 | 63 | 20 | 21 | 22 |
|
| Luminal |
|
|
|
| 0.2687 |
|
Positive | 177 | 42 | 59 | 76 |
|
|
Negative | 74 | 23 | 21 | 30 |
|
| HER2 |
|
|
|
| 0.3145 |
|
Positive | 61 | 19 | 22 | 20 |
|
|
Negative | 190 | 46 | 58 | 86 |
|
| pT stage |
|
|
|
| 0.2331 |
|
T1-2 | 86 | 16 | 28 | 42 |
|
|
T3-4 | 165 | 49 | 52 | 64 |
|
| pN stage |
|
|
|
| 0.0134b |
| N0 | 142 | 28 | 45 | 69 |
|
|
N1-2 | 109 | 37 | 35 | 37 |
|
 | Table III.Cox's proportional hazards model
analysis of prognostic factors in patients with breast cancer. |
Table III.
Cox's proportional hazards model
analysis of prognostic factors in patients with breast cancer.
| Variable | Hazard ratio | 95% CI | P-value |
|---|
| Univariate
analysis |
|
|
|
| KIF20B
expression (strong positive vs. weak positive and negative) | 3.509 | 1.823–6.755 | 0.0002a |
| Age
(≥65 vs. <65 years) | 1.382 | 0.68–2.808 | 0.3717 |
| Luminal
type (negative vs. positive) | 1.834 | 0.945–3.558 | 0.0729 |
| HER2
(positive vs. negative) | 2.157 | 0.725–2.997 | 0.2835 |
| pT
stage (T3-4 vs. T1-2) | 2.886 | 1.201–6.936 | 0.0178a |
| pN
stage (N1-2 vs. N0) | 5.998 | 2.627–13.695 |
<0.0001a |
| Multivariate
analysis |
|
|
|
| KIF20B
expression (strong positive vs. weak positive and negative) | 2.674 | 1.375–5.198 | 0.0002a |
| pT
stage (T3-4 vs. T1-2) | 1.76 | 0.714–4.339 | 0.2198 |
| pN
stage (N1-2 vs. N0) | 4.554 | 1.94–10.69 | 0.0005a |
KIF20B knockdown inhibits breast
cancer cell proliferation
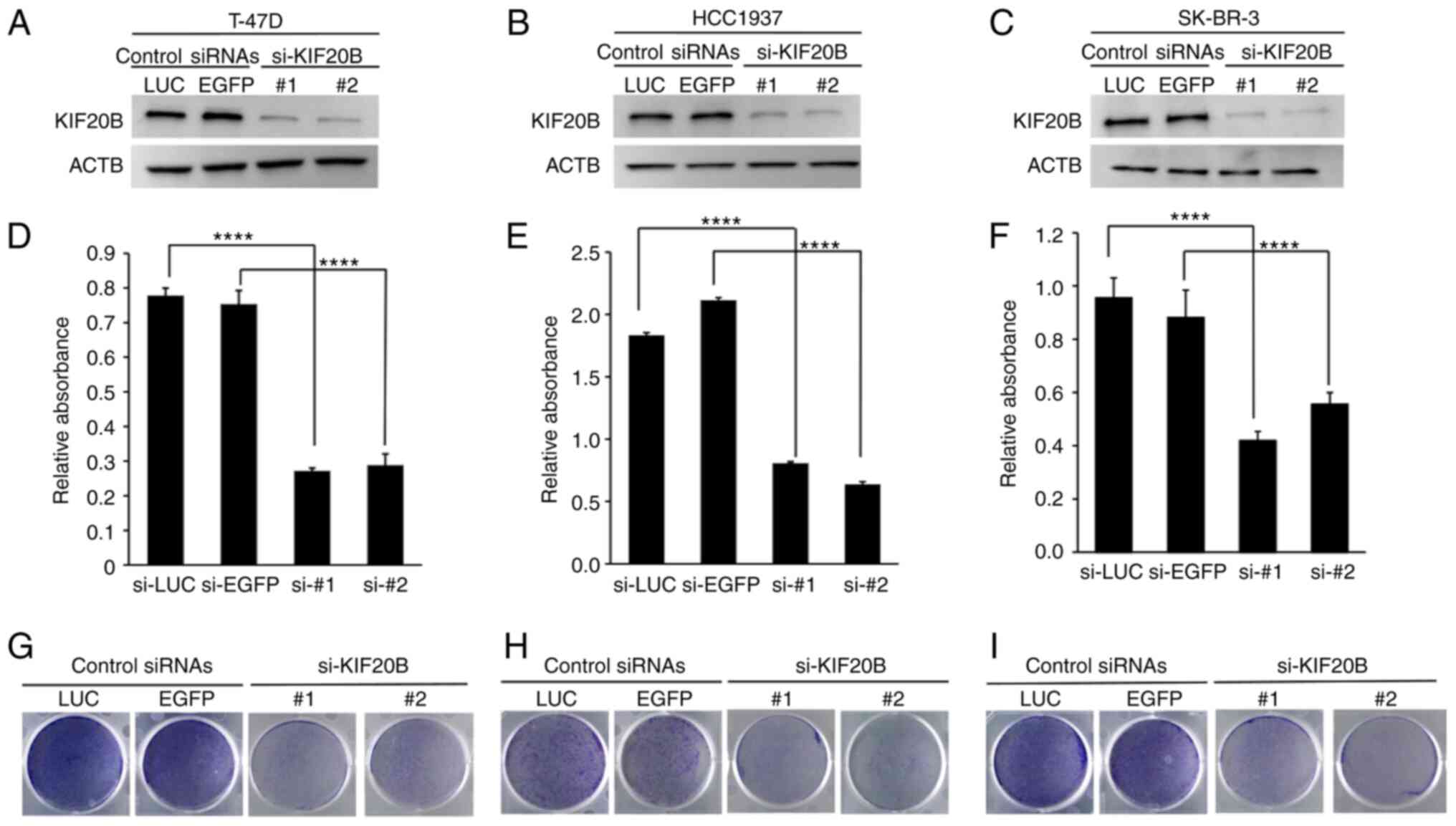
The expression of KIF20B was suppressed using
si-KIF20Bs in T-47D (luminal A), HCC1937 (TNBC) and SK-BR-3
(HER2/neu-positive) breast cancer cell lines. Compared with control
siRNAs, si-KIF20B decreased KIF20B protein expression levels in
breast cancer cells (Fig. 3A-C).
Additionally, MTT assay revealed that si-KIF20B significantly
inhibited breast cancer cell viability compared with control siRNAs
(Fig. 3D-F). Moreover, colony
formation assay showed a marked reduction in breast cancer cell
proliferation in response to KIF20B knockdown (Fig. 3G-I).
KIF20B knockdown induces dysregulation
of cell division, cell cycle arrest and apoptosis in breast cancer
cells
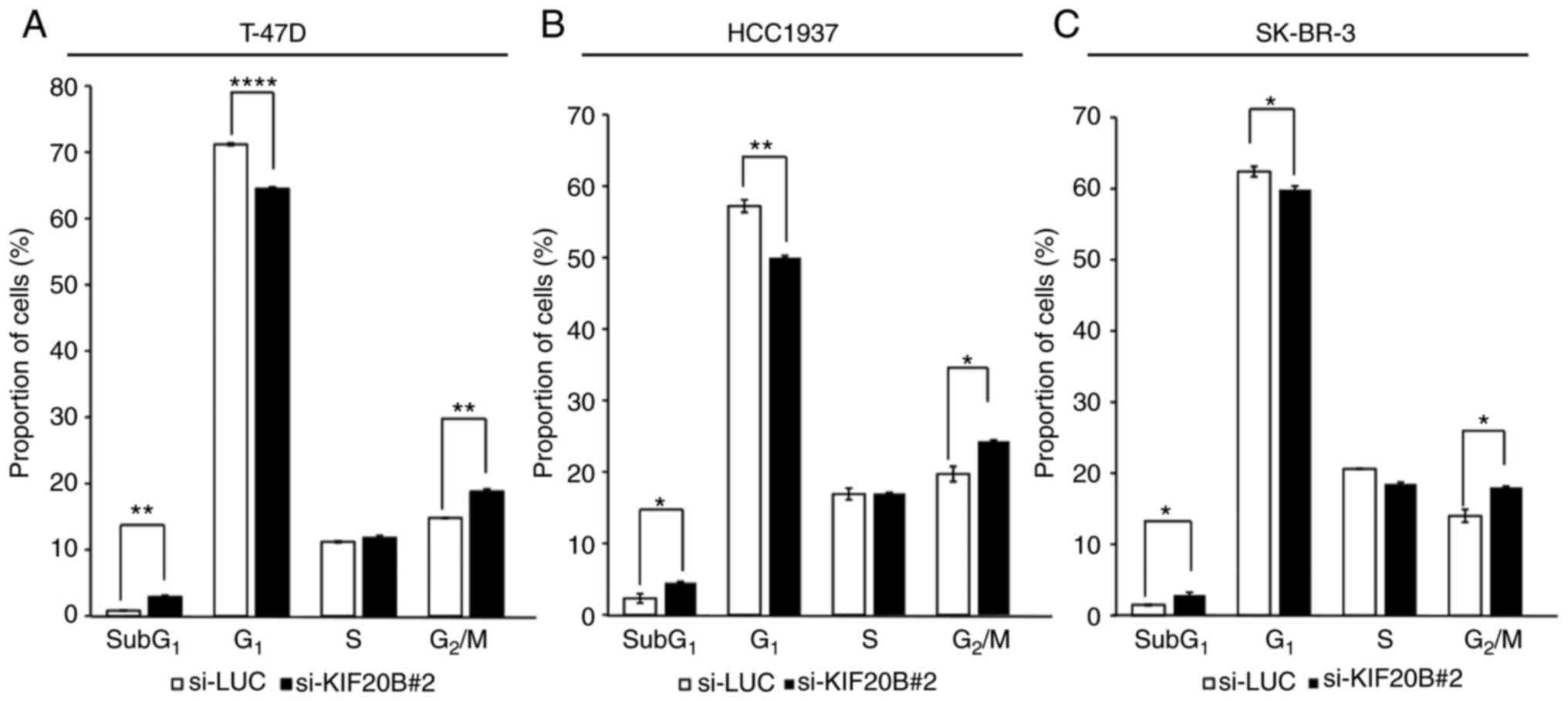
To further examine the mechanism of breast cancer
cell proliferation controlled by KIF20B, flow cytometric analysis
of cell cycle progression was performed after siRNA transfection.
si-LUC was selected for further experiments due to its stability
and lower toxicity to breast cancer cells, whereas si-KIF20B-#2 was
selected for further experiments because it showed better knockdown
effect on KIF20B expression. Compared with si-LUC, at 72 h
post-transfection, si-KIF20B#2 significantly increased the
proportion of cells at the sub-G1 and G2/M
phases (Figs. 4A-C and S2). In addition, the number of apoptotic
cells was significantly increased after siRNA-mediated KIF20B
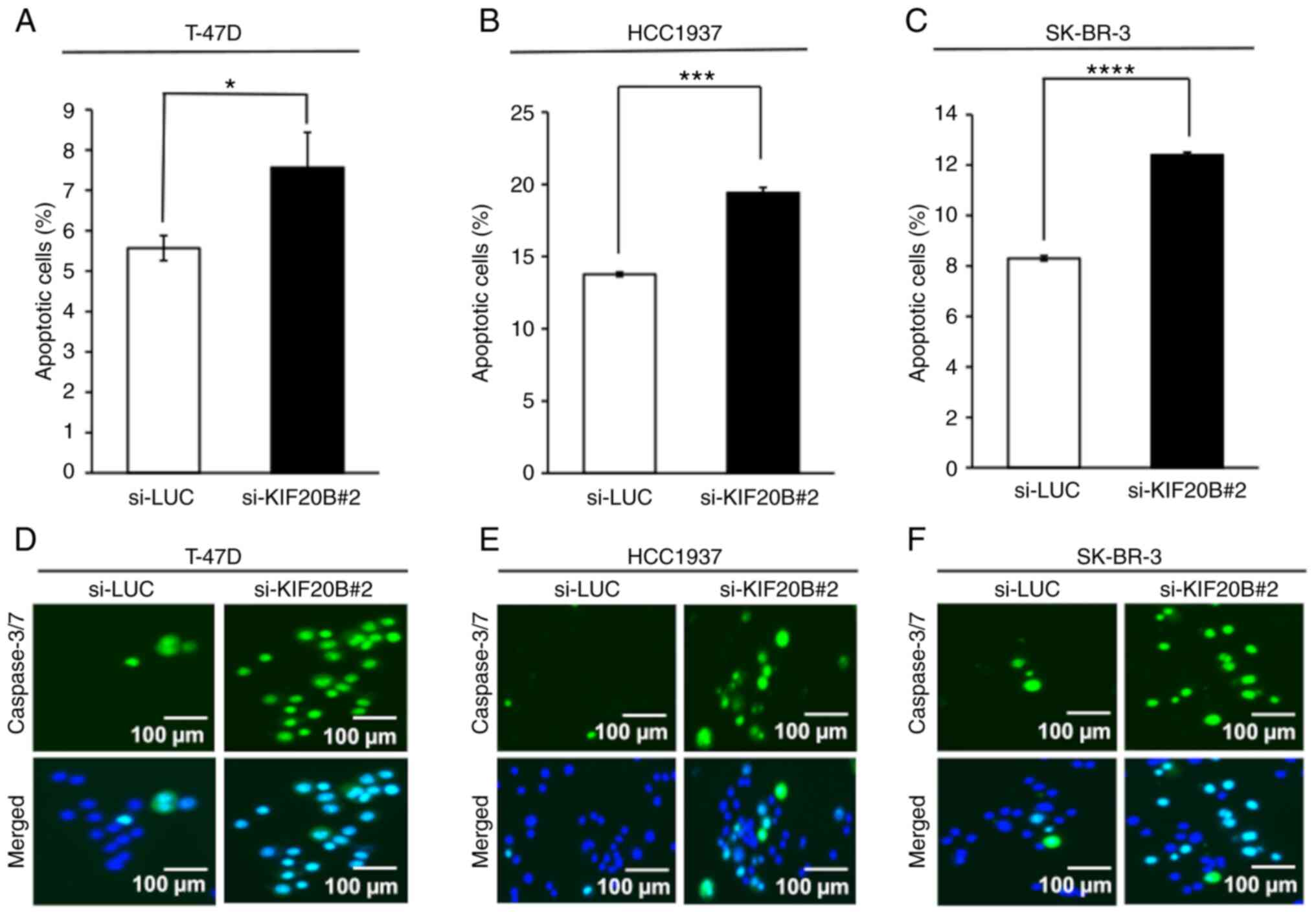
knockdown compared with that in the control group (Figs. 5A-C and S3). Additionally, the caspase 3/7 assay
demonstrated increased activation of caspase 3/7 in breast cancer
cells transfected with si-KIF20B#2 compared with the control siRNA
(Fig. 5D-F).
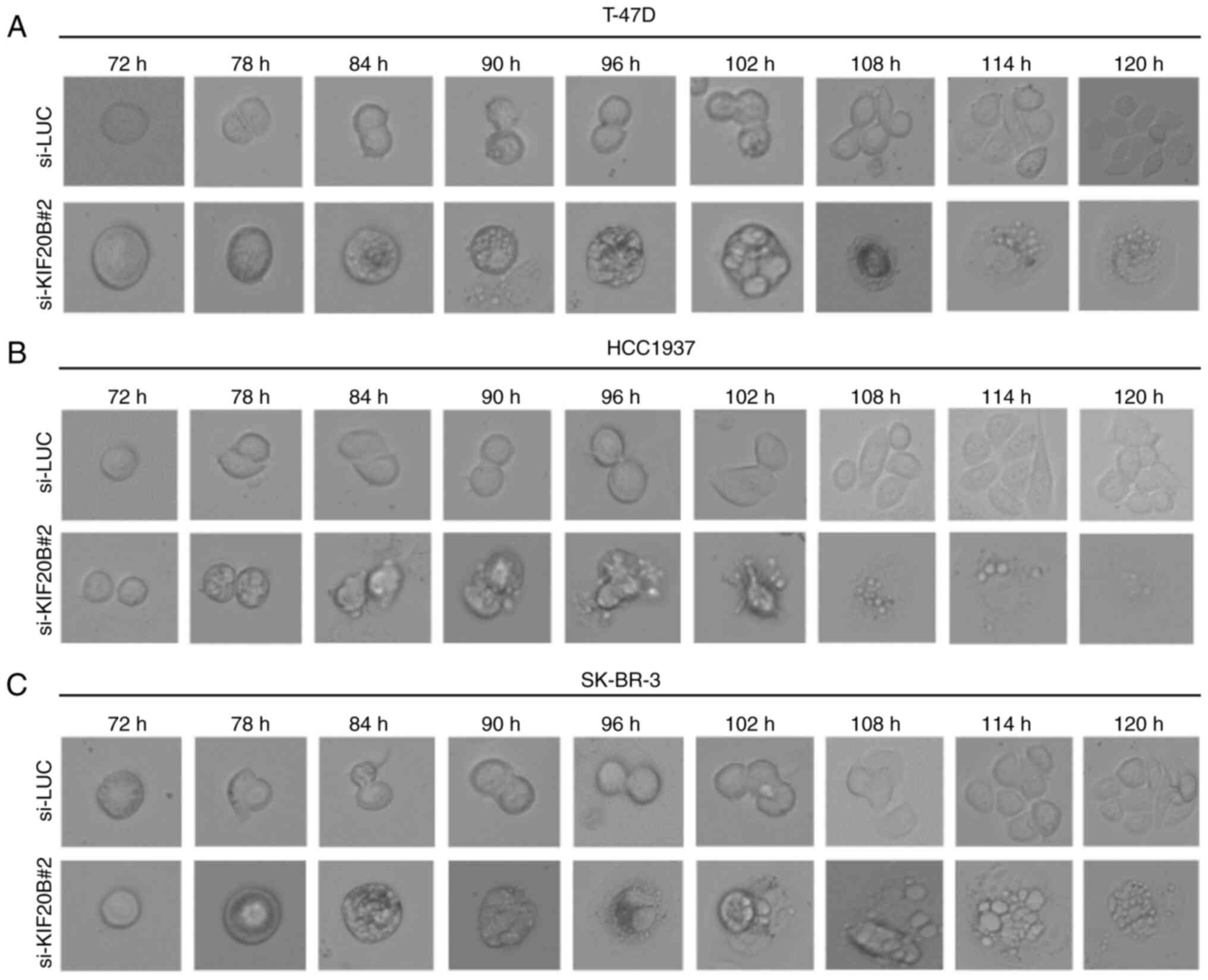
Morphological changes in T-47D, HCC1937 and SK-BR-3
cells transfected with si-KIF20B#2 were further monitored by live
cell imaging (Fig. 6A-C). Cells
transfected with control siRNAs divided regularly, whereas cells
transfected with si-KIF20B#2 did not properly divide and exhibited
increased cell death, suggesting that knockdown of KIF20B induces
G2/M arrest and subsequent cell death.
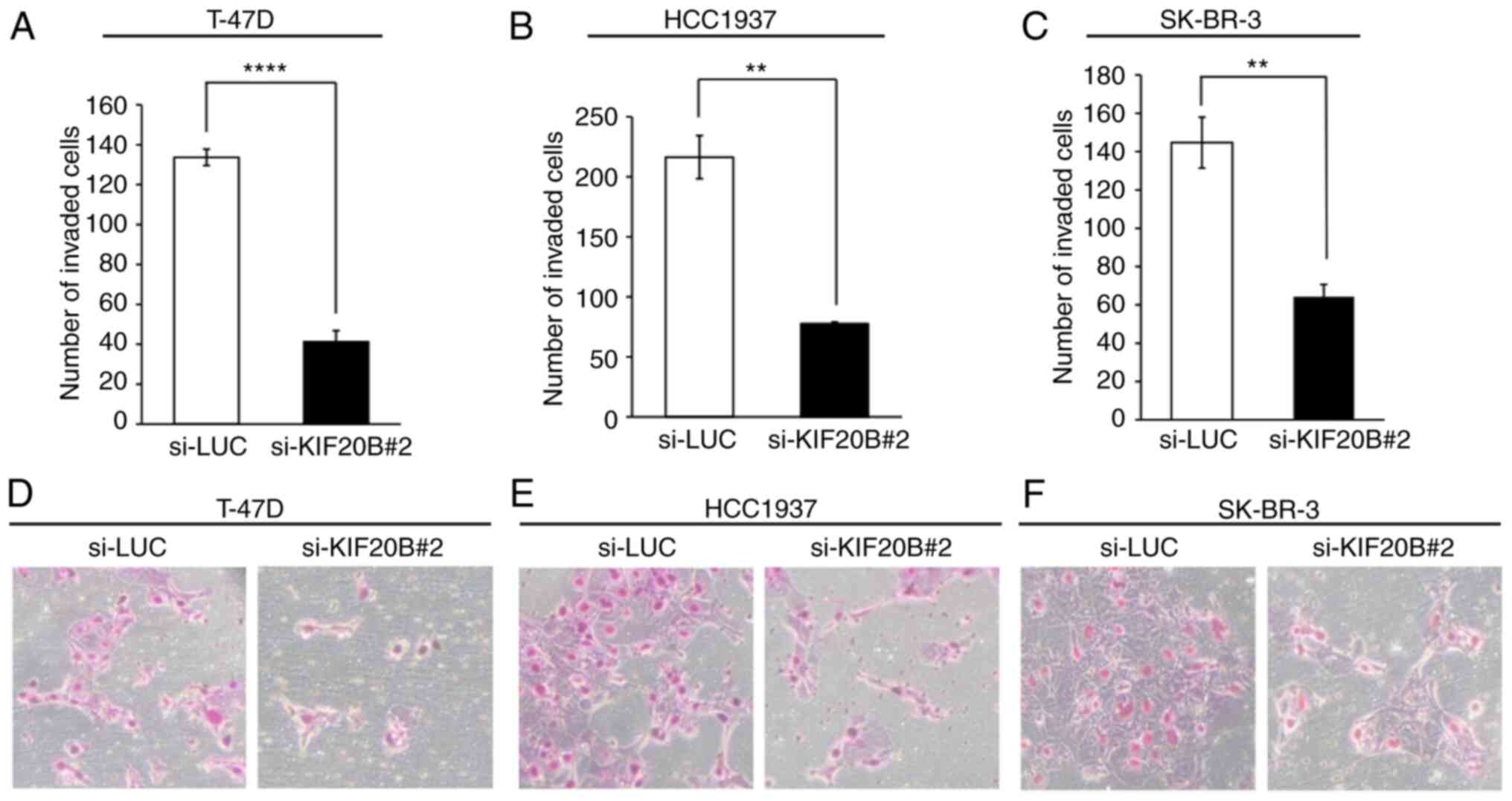
KIF20B knockdown inhibits the invasive
phenotype of breast cancer cells
To further elucidate the mechanism underlying the
effects of strong KIF20B expression on advanced pN stage, as
detected by tissue microarray analysis, Matrigel assays were
performed to assess the potential role of KIF20B in the invasion of
breast cancer cells. The invasive activity of T-47D, HCC1937 and
SK-BR-3 breast cancer cells was significantly reduced after
knockdown of KIF20B with si-KIF20B#2 compared with that in the
control group (Fig. 7A-F).
Database analysis of KIF20B
expression, OS and mutations
In order to validate the expression of KIF20B
in breast cancer and normal tissues, we examined KIF20B data
using databases. The BioGPS, GTex Dataset and UALCAN databases
showed scarce expression of KIF20B gene in normal tissues
and organs, and indicated that KIF20B was significantly upregulated
in breast cancer tissues compared to normal tissues (data not
shown). Using PrognoScan, a significant association between high
KIF20B expression and reduced OS of patients with breast
cancer was identified (dataset no. GSE143; P=0.036199). In order to
examine the mechanisms underlying the effects of KIF20B
upregulation in breast cancer, KIF20B genetic alterations were
investigated using cBioportal for cancer genomics. The genetic
aberrations in KIF20B were detected in only 9 of 1,084 breast
invasive carcinoma cases (0.83%) (data not shown). Therefore, it
was hypothesized that the high expression of KIF20B may be due to
several epigenetic mechanisms.
Discussion
Breast cancer is a biologically heterogeneous
disease; therefore, the treatment responses and survival outcomes
of patients vary. Although significant advances in breast cancer
research have been made, it still has high incidence and mortality
rates. A wide range of treatments have been developed; however, the
OS rates remain low and some subtypes, such as TNBC, are difficult
to treat (41). Therefore, the
development of new molecular targeted drugs, as well as biomarkers
that are novel, precise and cost effective, is desired.
In the present study, KIF20B was found to be highly
expressed in breast cancer cells and tissues, but was barely
expressed in normal breast epithelial cells and tissues. In
addition, knockdown of KIF20B markedly inhibited the proliferation
of breast cancer cells and subsequently induced apoptosis. These
data suggested the potential of KIF20B as a diagnostic and
therapeutic target. Moreover, patients with breast cancer and
strong KIF20B expression had a significantly poorer prognosis than
those with weak or negative KIF20B expression. The findings from
the PrognoScan database independently support the current
experimental findings, indicating that KIF20B might be a prognostic
biomarker for breast cancer.
As determined by tissue microarray analysis, strong
KIF20B expression was significantly related to advanced pN stage
(nodal involvement). Furthermore, the results of a Matrigel cell
invasion assay showed that the invasiveness of breast cancer cells
was significantly decreased by KIF20B silencing. These data
potentially highlight the relevance of KIF20B activation in the
malignant potential of breast cancer cells, such as in cell
invasion, which is an important factor and the first step in cancer
metastasis, a hallmark of malignant tumors, which leads to the
dissemination of primary tumor cells to distant organs (42).
KIF20B, a member of the kinesin-6 family, is also
known as membrane palmitoylated protein 1 or MPHOSPH1. It is a
plus-end-directed kinesin-related protein that serves a crucial
role in cytokinesis. In addition, it has been reported to be
phosphorylated during the G2/M transition (43). KIF20B also influences
microtubule-binding and microtubule-bundling properties, and
microtubule-stimulated adenosine triphosphatase activity in
vitro (44). Knockdown of
KIF20B also potentially induced failed cytokinesis, due to the
observed dysregulation of cell division monitored by the live cell
imaging and subsequent apoptotic cell death. The present data
demonstrated that siRNA-mediated knockdown of KIF20B increased the
proportion of breast cancer cells in the sub-G1 and
G2/M phases. These data independently suggested that
KIF20B-mediated cytokinesis and G2/M transition are
important factors in breast cancer cell proliferation and/or
survival, and are potential therapeutic targets (Fig. S4). KIF20B is a mitotic target
regulated by Pin1, as evidenced by an in vitro interaction
of the tail domain of KIF20B with the WW domain of Pin1 (45). Pin1 serves an essential role in the
regulation of mitosis through its interaction with various mitotic
phosphorylated proteins at G2/M phase transition
(46). Furthermore, Pin1 protein
is involved in multiple cellular processes, including suppression
of apoptosis by directly regulating anti-apoptotic proteins
(47). Pin1 has also been reported
to be highly expressed in most types of cancer, including human
breast cancer (48). These reports
indicated the important role of KIF20B in the completion of
cytokinesis and that of Pin1 in the regulation of mitosis. In
addition, the present data demonstrated the increased proportion of
breast cancer cells in the sub-G1 and G2/M
phases, dysregulation of cell division and subsequent cell death
after knockdown of KIF20B. Based on these findings, it was
hypothesized that the dysregulation of cell division in
KIF20B-depleted breast cancer cells may be caused by mitotic
arrest, probably due to failure in cytokinesis, which may lead to
errors in chromosomal segregation and subsequent induction of
apoptotic cell death. Notably, direct elucidation of the role of
KIF20B in cytokinesis, and its relation to microtubules and
chromosomal segregation in breast cancer is required. Future
studies that examine the potential molecular function of the
KIF20B-Pin1 interaction, as well as the oncogenic functions of
KIF20B in breast cancer progression, are also warranted.
In conclusion, the findings of the present study
suggested that KIF20B is an oncoprotein that serves an essential
role in breast cancer cell proliferation and survival, probably
through various unknown oncogenic pathways. Notably, KIF20B was
also shown to serve a pivotal role in breast cancer cell invasion.
Therefore, targeting KIF20B could be an effective approach in the
development of new molecular therapies and cancer biomarkers for
patients with breast cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported in part by a Grant-in-Aid for Scientific
Research (B), Grant-in-Aid for Challenging Research (Exploratory),
and Grant-in-Aid for Scientific Research on Innovative Areas from
the Japan Society for the Promotion of Science (JSPS KAKENHI grant
nos. 15H04761, 19H03559, 21K19444, 16H06277, 22H04923 and
23H02815). YD is a member of Shiga Cancer Treatment Project
supported by Shiga Prefecture (Japan) and the International Joint
Research Project (grant no. FY2016-2024) of the Institute of
Medical Sciences (The University of Tokyo).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding authors.
Authors' contributions
RWM, AT and YD conceived the research concept and
designed the study. RWM, AT and YD developed the study methodology.
TYa, TYo, YM and YD acquired the data, managed the patients and
provided the facilities. RWM, AT, BT and YD analyzed and
interpreted the data (e.g., statistical analysis, biostatistics and
computational analysis) and also confirm the accuracy of all the
raw data. RWM, AT and YD wrote, reviewed, and/or revised the
manuscript. RWM, AT and YD were involved in administrative,
technical, and/or material support (i.e., reporting or organizing
data, constructing databases). YD supervised the study. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The ethics committee at Kanagawa Cancer Center
(approval date: June 8, 2005; approval no. Rin-177) approved the
present research and the use of clinical materials. The project to
establish tumor tissue microarrays from archival formalin-fixed and
paraffin-embedded, surgically obtained tissues and to use the
tissue microarrays for later researches was approved by the
Kanagawa Cancer Center Ethics Committee (approval no. Rin-177;
Yokohama, Japan). Written informed consent was obtained from all
patients from Kanagawa Cancer Center for the use of the clinical
materials in this study. The ethics committee at Shiga University
of Medical Science (approval no. 21-163) approved the present study
including the use of all clinical materials.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
FBS
|
fetal bovine serum
|
|
KIF20B
|
kinesin family member 20B
|
|
HMECs
|
human mammary epithelial cells
|
|
TNBC
|
triple-negative breast cancer
|
References
|
1
|
Giaquinto AN, Sung H, Miller KD, Kramer
JL, Newman LA, Minihan A, Jemal A and Siegel RL: Breast cancer
statistics, 2022. CA Cancer J Clin. 72:524–541. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arnold M, Morgan E, Rumgay H, Mafra A,
Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S
and Soerjomataram I: Current and future burden of breast cancer:
Global statistics for 2020 and 2040. Breast. 66:15–23. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Voduc KD, Cheang MCU, Tyldesley S, Gelmon
K, Nielsen TO and Kennecke H: Breast cancer subtypes and the risk
of local and regional relapse. J Clin Oncol. 28:1684–1691. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tong CWS, Wu M, Cho WCS and To KKW: Recent
advances in the treatment of breast cancer. Front Oncol. 8:2272018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jamdade VS, Sethi N, Mundhe NA, Kumar P,
Lahkar M and Sinha N: Therapeutic targets of triple negative breast
cancer: A review. Br J Pharmacol. 172:4228–4237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raphael A, Salmon-Divon M, Epstein J,
Zahavi T, Sonnenblick A and Shachar SS: Alpelisib efficacy in
hormone receptor-positive HER2-negative PIK3CA-mutant advanced
breast cancer post-everolimus treatment. Genes (Basel).
29:17632022. View Article : Google Scholar
|
|
7
|
Hudis CA and Gianni L: Triple-negative
breast cancer: An unmet medical need. Oncologist. 16 (Suppl
1):S1–S11. 2011. View Article : Google Scholar
|
|
8
|
Robson M, Im SA, Senkus E, Xu B, Domchek
SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, et al:
Olaparib for metastatic breast cancer in patients with a germline
BRCA mutation. N Engl J Med. 377:523–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hou Y, Peng Y and Li Z: Update on
prognostic and predictive biomarkers of breast cancer. Semin Diagn
Pathol. 39:322–332. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chung C, Yeung VTY and Wong KCW:
Prognostic and predictive biomarkers with therapeutic targets in
breast cancer: A 2022 update on current developments, evidence, and
recommendations. J Oncol Pharm Pract. 15:1343–1360. 2023.
View Article : Google Scholar
|
|
11
|
Ovcaricek T, Takac I and Matos E:
Multigene expression signatures in early hormone receptor positive
HER 2 negative breast cancer. Radiol Oncol. 53:285–292. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Daigo Y and Nakamura Y: From cancer
genomics to thoracic oncology: Discovery of new biomarkers and
therapeutic targets for lung and esophageal carcinoma. Gen Thorac
Cardiovasc Surg. 56:43–53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Daigo Y, Takano A, Teramoto K, Chung S and
Nakamura Y: A systematic approach to the development of novel
therapeutics for lung cancer using genomic analyses. Clin Pharmacol
Ther. 94:218–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ishikawa N, Daigo Y, Takano A, Taniwaki M,
Kato T, Hayama S, Murakami H, Takeshima Y, Inai K, Nishimura H, et
al: Increases of amphiregulin and transforming growth factor-alpha
in serum as predictors of poor response to gefitinib among patients
with advanced non-small cell lung cancers. Cancer Res.
65:9176–9184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ishikawa N, Daigo Y, Yasui W, Inai K,
Nishimura H, Tsuchiya E, Kohno N and Nakamura Y: ADAM8 as a novel
serological and histochemical marker for lung cancer. Clin Cancer
Res. 10:8363–8370. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kakiuchi S, Daigo Y, Ishikawa N, Furukawa
C, Tsunoda T, Yano S, Nakagawa K, Tsuruo T, Kohno N, Fukuoka M, et
al: Prediction of sensitivity of advanced non-small cell lung
cancers to gefitinib (Iressa, ZD1839). Hum Mol Genet. 13:3029–3043.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kato T, Daigo Y, Hayama S, Ishikawa N,
Yamabuki T, Ito T, Miyamoto M, Kondo S and Nakamura Y: A novel
human tRNA-dihydrouridine synthase involved in pulmonary
carcinogenesis. Cancer Res. 65:5638–5646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kikuchi T, Daigo Y, Katagiri T, Tsunoda T,
Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi
K, et al: Expression profiles of non-small cell lung cancers on
cDNA microarrays: identification of genes for prediction of
lymph-node metastasis and sensitivity to anticancer drugs.
Oncogene. 22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suzuki C, Daigo Y, Ishikawa N, Kato T,
Hayama S, Ito T, Tsuchiya E and Nakamura Y: ANLN plays a critical
role in human lung carcinogenesis through the activation of RHOA
and by involvement in the phosphoinositide 3-kinase/AKT pathway.
Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kakiuchi S, Daigo Y, Tsunoda T, Yano S,
Sone S and Nakamura Y: Genome-wide analysis of organ-preferential
metastasis of human small cell lung cancer in mice. Mol Cancer Res.
1:485–499. 2003.PubMed/NCBI
|
|
21
|
Taniwaki M, Daigo Y, Ishikawa N, Takano A,
Tsunoda T, Yasui W, Inai K, Kohno N and Nakamura Y: Gene expression
profiles of small-cell lung cancers: Molecular signatures of lung
cancer. Int J Oncol. 29:567–575. 2006.PubMed/NCBI
|
|
22
|
Oshita H, Nishino R, Takano A, Fujitomo T,
Aragaki M, Kato T, Akiyama H, Tsuchiya E, Kohno N, Nakamura Y and
Daigo Y: RASEF is a novel diagnostic biomarker and a therapeutic
target for lung cancer. Mol Cancer Res. 11:937–951. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hayama S, Daigo Y, Yamabuki T, Hirata D,
Kato T, Miyamoto M, Ito T, Tsuchiya E, Kondo S and Nakamura Y:
Phosphorylation and activation of cell division cycle associated 8
by aurora kinase B plays a significant role in human lung
carcinogenesis. Cancer Res. 67:4113–4122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ishikawa N, Daigo Y, Takano A, Taniwaki M,
Kato T, Tanaka S, Yasui W, Takeshima Y, Inai K, Nishimura H, et al:
Characterization of SEZ6L2 cell-surface protein as a novel
prognostic marker for lung cancer. Cancer Sci. 97:737–745. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kato T, Sato N, Hayama S, Yamabuki T, Ito
T, Miyamoto M, Kondo S, Nakamura Y and Daigo Y: Activation of
Holliday junction recognizing protein involved in the chromosomal
stability and immortality of cancer cells. Cancer Res.
67:8544–8553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takano A, Ishikawa N, Nishino R, Masuda K,
Yasui W, Inai K, Nishimura H, Ito H, Nakayama H, Miyagi Y, et al:
Identification of nectin-4 oncoprotein as a diagnostic and
therapeutic target for lung cancer. Cancer Res. 69:6694–6703. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kobayashi Y, Takano A, Miyagi Y, Tsuchiya
E, Sonoda H, Shimizu T, Okabe H, Tani T, Fujiyama Y and Daigo Y:
Cell division cycle-associated protein 1 overexpression is
essential for the malignant potential of colorectal cancers. Int J
Oncol. 44:69–77. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thang PM, Takano A, Yoshitake Y, Shinohara
M, Murakami Y and Daigo Y: Cell division cycle associated 1 as a
novel prognostic biomarker and therapeutic target for oral cancer.
Int J Oncol. 49:1385–1393. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Daigo K, Takano A, Thang PM, Yoshitake Y,
Shinohara M, Tohnai I, Murakami Y, Maegawa J and Daigo Y:
Characterization of KIF11 as a novel prognostic biomarker and
therapeutic target for oral cancer. Int J Oncol. 52:155–165.
2018.PubMed/NCBI
|
|
30
|
Nakamura M, Takano A, Thang PM, Tsevegjav
B, Zhu M, Yokose T, Yamashita T, Miyagi Y and Daigo Y:
Characterization of KIF20A as a prognostic biomarker and
therapeutic target for different subtypes of breast cancer. Int J
Oncol. 57:277–288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsevegjav B, Takano A, Zhu M, Yoshitake Y,
Shinohara M and Daigo Y: Holliday junction recognition protein as a
prognostic biomarker and therapeutic target for oral cancer. Int J
Oncol. 60:262022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu M, Takano A, Tsevegjav B, Yoshitake Y,
Shinohara M and Daigo Y: Characterization of Opa interacting
protein 5 as a new biomarker and therapeutic target for oral
cancer. Int J Oncol. 60:272022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hirokawa N, Noda Y and Okada Y: Kinesin
and dynein superfamily proteins in organelle transport and cell
division. Curr Opin Cell Biol. 10:60–73. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kanehira M, Katagiri T, Shimo A, Takata R,
Shuin T, Miki T, Fujioka T and Nakamura Y: Oncogenic role of
MPHOSPH1, a cancer-testis antigen specific to human bladder cancer.
Cancer Res. 67:3276–3285. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin WF, Lin XL, Fu SW, Yang L, Tang CT,
Gao YJ, Chen HY and Ge ZZ: Pseudopod-associated protein KIF20B
promotes Gli1-induced epithelial-mesenchymal transition modulated
by pseudopodial actin dynamic in human colorectal cancer. Mol
Carcinog. 57:911–925. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X, Zhou Y, Liu X, Peng A, Gong H,
Huang L, Ji K, Petersen RB, Zheng L and Huang K: MPHOSPH1: A
potential therapeutic target for hepatocellular carcinoma. Cancer
Res. 74:6623–6634. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen J, Zhao CC, Chen FR, Feng GW, Luo F
and Jiang T: KIF20B promotes cell proliferation and may be a
potential therapeutic target in pancreatic cancer. J Oncol.
2021:55724022021.PubMed/NCBI
|
|
38
|
Li ZY, Wang ZX and Li CC: Kinesin family
member 20B regulates tongue cancer progression by promoting cell
proliferation. Mol Med Rep. 19:2202–2210. 2019.PubMed/NCBI
|
|
39
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM classification of malignant tumours. 7th edition. John Wiley
& Sons; 2011
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cao L and Niu Y: Triple negative breast
cancer: Special histological types and emerging therapeutic
methods. Cancer Biol Med. 17:293–306. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pearson GW: Control of invasion by
epithelial-to-mesenchymal transition programs during metastasis. J
Clin Med. 8:6462019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Matsumoto-Taniura N, Pirollet F, Monroe R,
Gerace L and Westendorf JM: Identification of novel M phase
phosphoproteins by expression cloning. Mol Biol Cell. 7:1455–1469.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Abaza A, Soleilhac JM, Westendorf J, Piel
M, Crevel I, Roux A and Pirollet F: M phase phosphoprotein 1 is a
human plus-end-directed kinesin-related protein required for
cytokinesis. J Biol Chem. 278:27844–27852. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kamimoto T, Zama T, Aoki R, Muro Y and
Hagiwara M: Identification of a novel kinesin-related protein,
KRMP1, as a target for mitotic peptidyl-prolyl isomerase Pin1. J
Biol Chem. 276:37520–37528. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cheng CW and Tse E: PIN1 in cell cycle
control and cancer. Front Pharmacol. 9:13672018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liou YC, Zhou XZ and Lu KP: Prolyl
isomerase Pin1 as a molecular switch to determine the fate of
phosphoproteins. Trends Biochem Sci. 36:501–514. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T,
Petkova V and Lu KP: Pin1 is overexpressed in breast cancer and
cooperates with Ras signaling in increasing the transcriptional
activity of c-Jun towards cyclin D1. EMBO J. 20:3459–3472. 2001.
View Article : Google Scholar : PubMed/NCBI
|