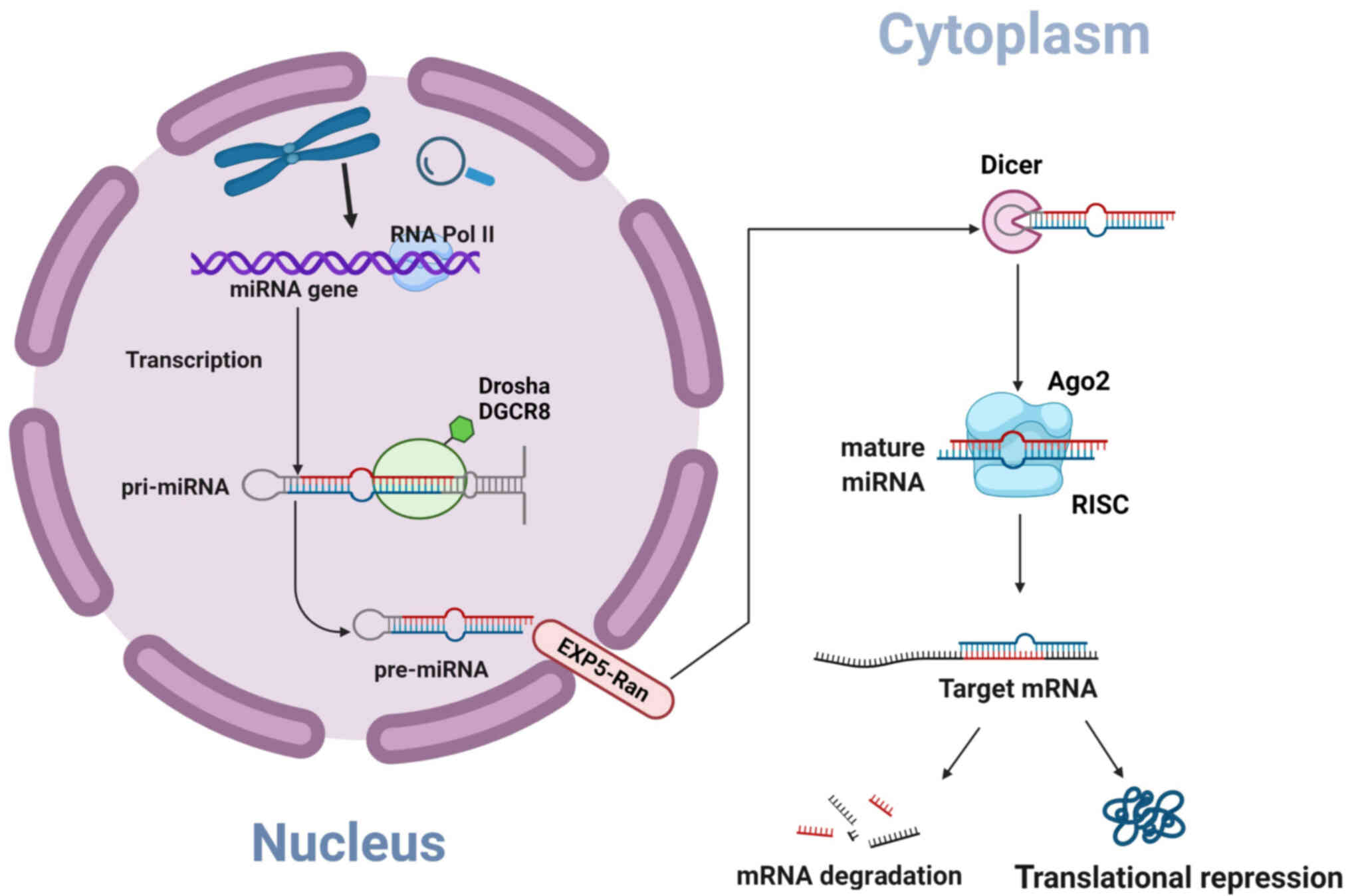
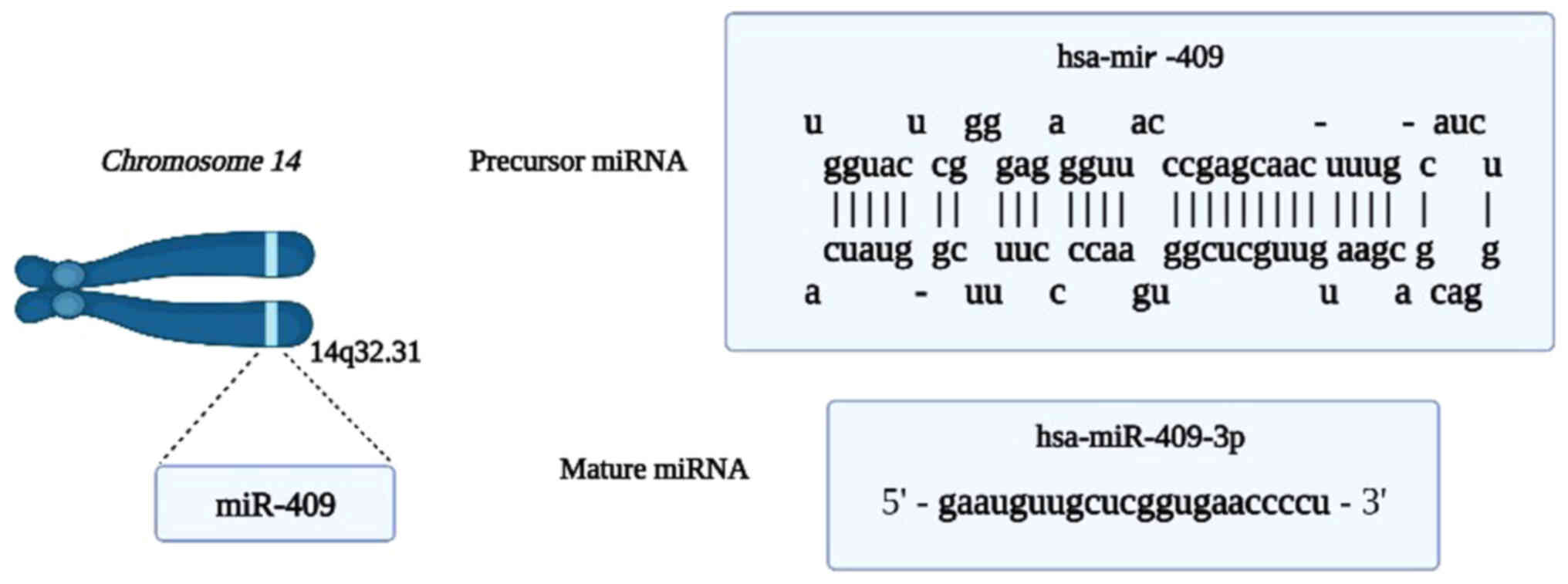
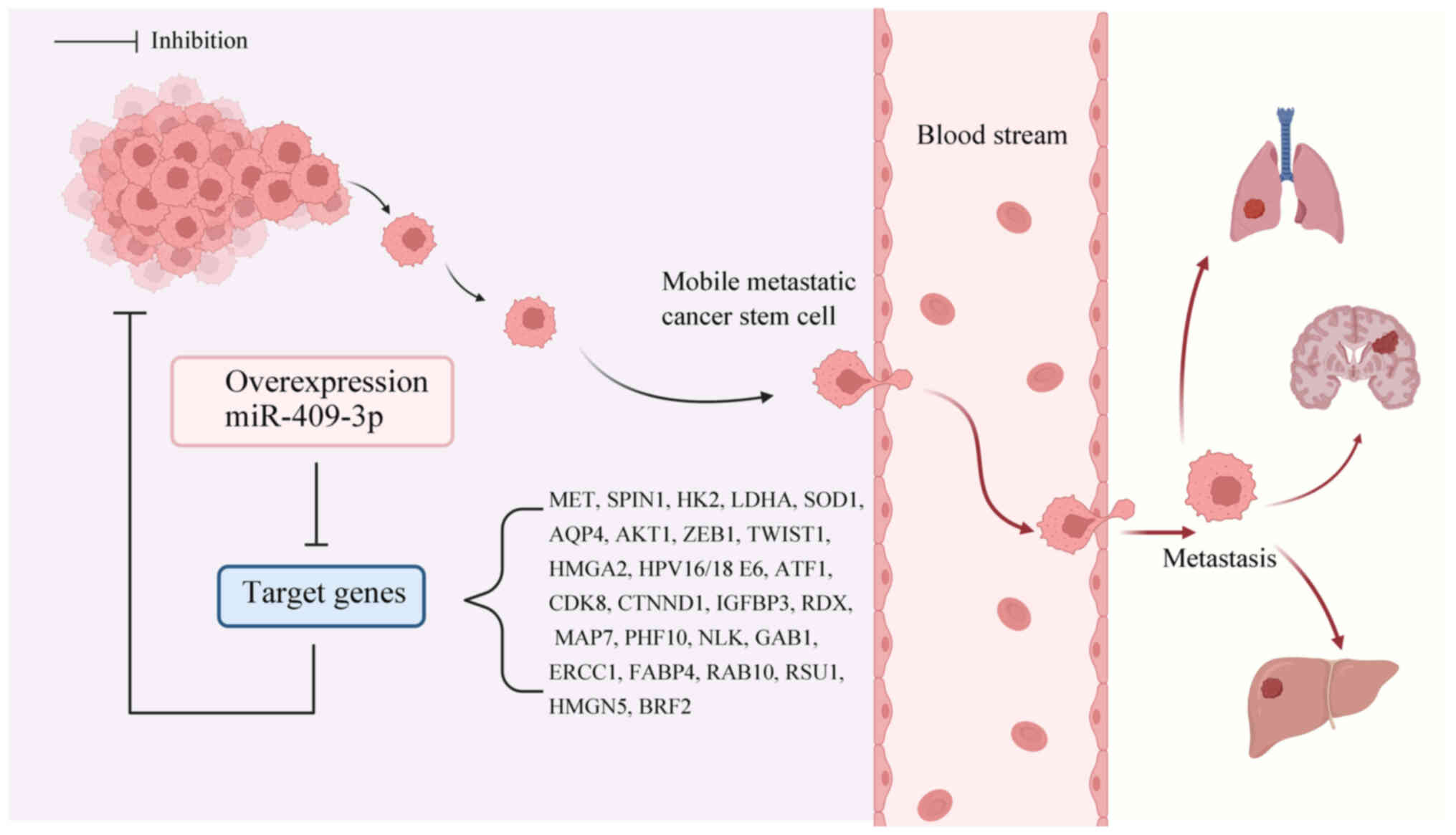
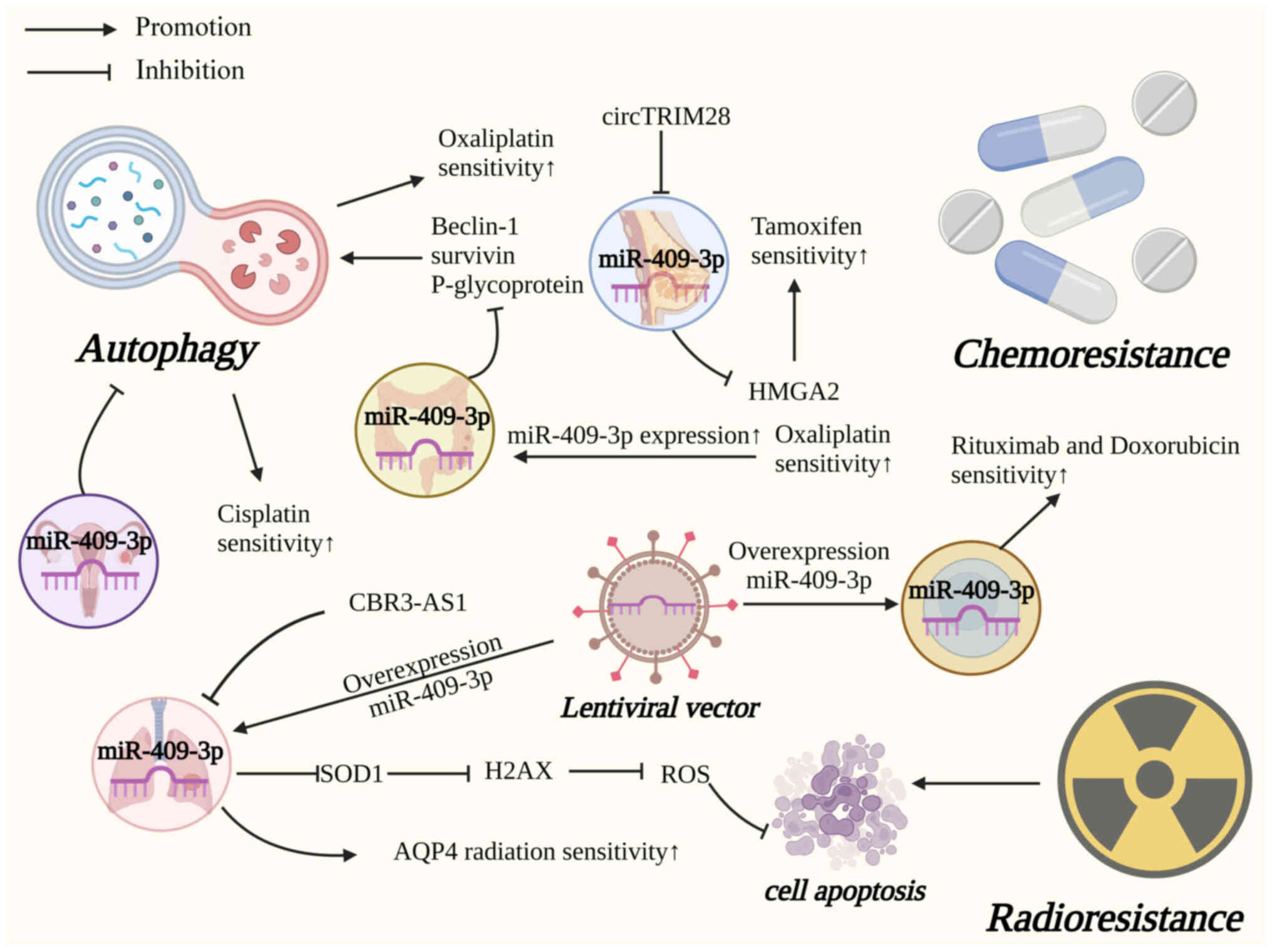
|
1
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar :
|
|
2
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body
fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol.
8:467–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar
|
|
6
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bueno MJ and Malumbres M: MicroRNAs and
the cell cycle. Biochim Biophys Acta. 1812:592–601. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget.
6:8474–8490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma L: MicroRNA and metastasis. Adv Cancer
Res. 132:165–207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tiwari A, Mukherjee B and Dixit M:
MicroRNA key to angiogenesis regulation: MiRNA biology and therapy.
Curr Cancer Drug Targets. 18:266–277. 2018. View Article : Google Scholar
|
|
11
|
Altuvia Y, Landgraf P, Lithwick G, Elefant
N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T and Margalit H:
Clustering and conservation patterns of human microRNAs. Nucleic
Acids Res. 33:2697–2706. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wan L, Zhu L, Xu J, Lu B, Yang Y, Liu F
and Wang Z: MicroRNA-409-3p functions as a tumor suppressor in
human lung adenocarcinoma by targeting c-Met. Cell Physiol Biochem.
34:1273–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R,
Cheng W, Wang F, Qi LW, Chen Y, et al: A six-microRNA panel in
plasma was identified as a potential biomarker for lung
adenocarcinoma diagnosis. Oncotarget. 8:6513–6525. 2017. View Article : Google Scholar :
|
|
14
|
Song Q, Ji Q, Xiao J, Li F, Wang L, Chen
Y, Xu Y and Jiao S: miR-409 inhibits human non-small-cell lung
cancer progression by directly targeting SPIN1. Mol Ther Nucleic
Acids. 13:154–163. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qu R, Chen X and Zhang C: LncRNA
ZEB1-AS1/miR-409-3p/ ZEB1 feedback loop is involved in the
progression of non-small cell lung cancer. Biochem Biophys Res
Commun. 507:450–456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yin D, Hua L, Wang J, Liu Y and Li X: Long
non-coding RNA DUXAP8 facilitates cell viability, migration, and
glycolysis in non-small-cell lung cancer via regulating HK2 and
LDHA by inhibition of miR-409-3p. Onco Targets Ther. 13:7111–7123.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu S, Li B, Xu J, Hu S, Zhan N, Wang H,
Gao C, Li J and Xu X: SOD1 promotes cell proliferation and
metastasis in non-small cell lung cancer via an
miR-409-3p/SOD1/SETDB1 epigenetic regulatory feedforward loop.
Front Cell Dev Biol. 8:2132020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Zhang C, Peng X, Liu K, Zhao L,
Chen X, Yu H and Lai Y: A combination of four serum miRNAs for
screening of lung adenocarcinoma. Hum Cell. 33:830–838. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Wu L and Pang J: Long noncoding
RNA PSMA3-AS1 functions as a microRNA-409-3p sponge to promote the
progression of non-small cell lung carcinoma by targeting spindlin
1. Oncol Rep. 44:1550–1560. 2020.PubMed/NCBI
|
|
20
|
Liu S, Zhan N, Gao C, Xu P, Wang H, Wang
S, Piao S and Jing S: Long noncoding RNA CBR3-AS1 mediates
tumorigenesis and radiosensitivity of non-small cell lung cancer
through redox and DNA repair by CBR3-AS1/miR-409-3p/SOD1 axis.
Cancer Lett. 526:1–11. 2022. View Article : Google Scholar
|
|
21
|
Yang X, Li M, Zhao Y, Tan X, Su J and
Zhong X: Hsa_circ_0079530/AQP4 axis is related to non-small cell
lung cancer development and radiosensitivity. Ann Thorac Cardiovasc
Surg. 28:307–319. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cuk K, Zucknick M, Madhavan D, Schott S,
Golatta M, Heil J, Marmé F, Turchinovich A, Sinn P, Sohn C, et al:
Plasma microRNA panel for minimally invasive detection of breast
cancer. PLoS One. 8:e767292013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cuk K, Zucknick M, Heil J, Madhavan D,
Schott S, Turchinovich A, Arlt D, Rath M, Sohn C, Benner A, et al:
Circulating microRNAs in plasma as early detection markers for
breast cancer. Int J Cancer. 132:1602–1612. 2013. View Article : Google Scholar
|
|
24
|
Li S, Meng H, Zhou F, Zhai L, Zhang L, Gu
F, Fan Y, Lang R, Fu L, Gu L and Qi L: MicroRNA-132 is frequently
down-regulated in ductal carcinoma in situ (DCIS) of breast and
acts as a tumor suppressor by inhibiting cell proliferation. Pathol
Res Pract. 209:179–183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen J, Hu Q, Schrauder M, Yan L, Wang D,
Medico L, Guo Y, Yao S, Zhu Q, Liu B, et al: Circulating miR-148b
and miR-133a as biomarkers for breast cancer detection. Oncotarget.
5:5284–5294. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang G, Liu Z, Xu H and Yang Q:
miR-409-3p suppresses breast cancer cell growth and invasion by
targeting Akt1. Biochem Biophys Res Commun. 469:189–195. 2016.
View Article : Google Scholar
|
|
27
|
Ma Z, Li Y, Xu J, Ren Q, Yao J and Tian X:
MicroRNA-409-3p regulates cell invasion and metastasis by targeting
ZEB1 in breast cancer. IUBMB Life. 68:394–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Venkatadri R, Muni T, Iyer AKV, Yakisich
JS and Azad N: Role of apoptosis-related miRNAs in
resveratrol-induced breast cancer cell death. Cell Death Dis.
7:e21042016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su Q, Shen H, Gu B and Zhu N: Circular RNA
CNOT2 knockdown regulates twist family BHLH transcription factor
via targeting microRNA 409-3p to prevent breast cancer invasion,
migration and epithelial-mesenchymal transition. Bioengineered.
12:9058–9069. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang S, Zou C, Li Y, Yang X, Liu W, Zhang
G and Lu N: Knockdown circTRIM28 enhances tamoxifen sensitivity via
the miR-409-3p/HMGA2 axis in breast cancer. Reprod Biol Endocrinol.
20:1462022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shukla V, Varghese VK, Kabekkodu SP,
Mallya S, Chakrabarty S, Jayaram P, Pandey D, Banerjee S, Sharan K
and Satyamoorthy K: Enumeration of deregulated miRNAs in liquid and
tissue biopsies of cervical cancer. Gynecol Oncol. 155:135–143.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sommerova L, Anton M, Bouchalova P,
Jasickova H, Rak V, Jandakova E, Selingerova I, Bartosik M,
Vojtesek B and Hrstka R: The role of miR-409-3p in regulation of
HPV16/18-E6 mRNA in human cervical high-grade squamous
intraepithelial lesions. Antiviral Res. 163:185–192. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui X, Chen J, Zheng Y and Shen H:
Circ_0000745 promotes the progression of cervical cancer by
regulating miR-409-3p/ATF1 axis. Cancer Biother Radiopharm.
37:766–778. 2022.
|
|
34
|
Zhou B, Li T, Xie R, Zhou J, Liu J, Luo Y
and Zhang X: CircFAT1 facilitates cervical cancer malignant
progression by regulating ERK1/2 and p38 MAPK pathway through
miR-409-3p/CDK8 axis. Drug Dev Res. 82:1131–1143. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu P, Li C, Ye DM, Yu K, Li Y, Tang H, Xu
G, Yi S and Zhang Z: Circular RNA circEPSTI1 accelerates cervical
cancer progression via miR-375/409-3P/515-5p-SLC7A11 axis. Aging
(Albany NY). 13:4663–4673. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu S, Du X, Wu M, Du H, Shi X and Zhang T:
MicroRNA-409-3p inhibits osteosarcoma cell migration and invasion
by targeting catenin-δ1. Gene. 584:83–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang J, Hou W, Jia J, Zhao Y and Zhao B:
MiR-409-3p regulates cell proliferation and tumor growth by
targeting E74-like factor 2 in osteosarcoma. FEBS Open Bio.
7:348–357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu L, Zhang Y, Huang Z, Gu H, Zhou K, Yin
X and Xu J: MiR-409-3p inhibits cell proliferation and invasion of
osteosarcoma by targeting zinc-finger E-box-binding homeobox-1.
Front Pharmacol. 10:1372019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Long Z, Gong F, Li Y, Fan Z and Li J:
Circ_0000285 regulates proliferation, migration, invasion and
apoptosis of osteosarcoma by miR-409-3p/IGFBP3 axis. Cancer Cell
Int. 20:4812020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li C, Nie H, Wang M, Su L, Li J, Yu B, Wei
M, Ju J, Yu Y, Yan M, et al: MicroRNA-409-3p regulates cell
proliferation and apoptosis by targeting PHF10 in gastric cancer.
Cancer Lett. 320:189–197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng B, Liang L, Huang S, Zha R, Liu L,
Jia D, Tian Q, Wang Q, Wang C, Long Z, et al: MicroRNA-409
suppresses tumour cell invasion and metastasis by directly
targeting radixin in gastric cancers. Oncogene. 31:4509–4516. 2012.
View Article : Google Scholar
|
|
42
|
Yu L, Xie J, Liu X, Yu Y and Wang S:
Plasma exosomal CircNEK9 accelerates the progression of gastric
cancer via miR-409-3p/MAP7 axis. Dig Dis Sci. 66:4274–4289. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng J, Li K, Liu G, Feng Y, Shi H and
Zhang X: Precision hyperthermia-induced miRNA-409-3p upregulation
inhibits migration, invasion, and EMT of gastric cancer cells by
targeting KLF17. Biochem Biophys Res Commun. 549:113–119. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Zhang J, Chen X and Gao L:
Circ_0001023 promotes proliferation and metastasis of gastric
cancer cells through miR-409-3p/PHF10 axis. Onco Targets Ther.
13:4533–4544. 2020. View Article : Google Scholar :
|
|
45
|
Liu M, Xu A, Yuan X, Zhang Q, Fang T, Wang
W and Li C: Downregulation of microRNA-409-3p promotes
aggressiveness and metastasis in colorectal cancer: An indication
for personalized medicine. J Transl Med. 13:1952015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bai R, Weng C, Dong H, Li S, Chen G and Xu
Z: MicroRNA-409-3p suppresses colorectal cancer invasion and
metastasis partly by targeting GAB1 expression. Int J Cancer.
137:2310–2322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X,
Yu L, Wang L, Wang J, Wu Y, et al: A plasma microRNA panel for
early detection of colorectal cancer. Int J Cancer. 136:152–161.
2015. View Article : Google Scholar
|
|
48
|
Tan S, Shi H, Ba M, Lin S, Tang H, Zeng X
and Zhang X: miR-409-3p sensitizes colon cancer cells to
oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol
Med. 37:1030–1038. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
López-Rosas I, López-Camarillo C,
Salinas-Vera YM, Hernández-de la Cruz ON, Palma-Flores C,
Chávez-Munguía B, Resendis-Antonio O, Guillen N, Pérez-Plasencia C,
Álvarez-Sánchez ME, et al: Entamoeba histolytica up-regulates
MicroRNA-643 to promote apoptosis by targeting XIAP in human
epithelial colon cells. Front Cell Infect Microbiol. 8:4372019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Han W, Yin H, Ma H, Wang Y, Kong D and Fan
Z: Curcumin regulates ERCC1 expression and enhances oxaliplatin
sensitivity in resistant colorectal cancer cells through its
effects on miR-409-3p. Evid Based Complement Alternat Med.
2020:83945742020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen J, Wang R, Lu E, Song S and Zhu Y:
LINC00630 as a miR-409-3p sponge promotes apoptosis and glycolysis
of colon carcinoma cells via regulating HK2. Am J Transl Res.
14:863–875. 2022.PubMed/NCBI
|
|
52
|
Zhang J, Raju GS, Chang DW, Lin SH, Chen Z
and Wu X: Global and targeted circulating microRNA profiling of
colorectal adenoma and colorectal cancer. Cancer. 124:785–796.
2018. View Article : Google Scholar
|
|
53
|
Gharpure KM, Pradeep S, Sans M, Rupaimoole
R, Ivan C, Wu SY, Bayraktar E, Nagaraja AS, Mangala LS, Zhang X, et
al: FABP4 as a key determinant of metastatic potential of ovarian
cancer. Nat Commun. 9:29232018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cheng Y, Ban R, Liu W, Wang H, Li S, Yue
Z, Zhu G, Zhuan Y and Wang C: MiRNA-409-3p enhances
cisplatin-sensitivity of ovarian cancer cells by blocking the
autophagy mediated by Fip200. Oncol Res. Jan 2–2018.Epub ahead of
print. View Article : Google Scholar
|
|
55
|
Zhang S, Zhang X, Fu X, Li W, Xing S and
Yang Y: Identification of common differentially-expressed miRNAs in
ovarian cancer cells and their exosomes compared with normal
ovarian surface epithelial cell cells. Oncol Lett. 16:2391–2401.
2018.PubMed/NCBI
|
|
56
|
Li Y, Chen L, Zhang B, Ohno Y and Hu H:
miR-409-3p inhibits the proliferation and migration of human
ovarian cancer cells by targeting Rab10. Cell Mol Biol
(Noisy-le-grand). 66:197–201. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhu J, Ma X, Zhang Y, Ni D, Ai Q, Li H and
Zhang X: Establishment of a miRNA-mRNA regulatory network in
metastatic renal cell carcinoma and screening of potential
therapeutic targets. Tumour Biol. Nov 2–2016.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Y, He Y, Bai H, Dang Y, Gao J and Lv
P: Phosphoinositide-dependent kinase 1-associated glycolysis is
regulated by miR-409-3p in clear cell renal cell carcinoma. J Cell
Biochem. 120:126–134. 2019. View Article : Google Scholar
|
|
59
|
Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, Xu
X, Zhu Y, Li S, Zheng X and Xie L: MicroRNA-409-3p inhibits
migration and invasion of bladder cancer cells via targeting c-Met.
Mol Cells. 36:62–68. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu X, Zhu Y, Liang Z, Li S, Xu X, Wang X,
Wu J, Hu Z, Meng S, Liu B, et al: c-Met and CREB1 are involved in
miR-433-mediated inhibition of the epithelial-mesenchymal
transition in bladder cancer by regulating Akt/GSK-3β/Snail
signaling. Cell Death Dis. 7:e20882016. View Article : Google Scholar
|
|
61
|
Lian J, Lin SH, Ye Y, Chang DW, Huang M,
Dinney CP and Wu X: Serum microRNAs as predictors of risk for
non-muscle invasive bladder cancer. Oncotarget. 9:14895–14908.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao Z, Yang F, Liu Y, Fu K and Jing S:
MicroRNA-409-3p suppresses cell proliferation and cell cycle
progression by targeting cyclin D2 in papillary thyroid carcinoma.
Oncol Lett. 16:5237–5242. 2018.PubMed/NCBI
|
|
63
|
Kim K, Yoo D, Lee HS, Lee KJ, Park SB, Kim
C, Jo JH, Jung DE and Song SY: Identification of potential
biomarkers for diagnosis of pancreatic and biliary tract cancers by
sequencing of serum microRNAs. BMC Med Genomics. 12:622019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Leivonen SK, Icay K, Jäntti K, Siren I,
Liu C, Alkodsi A, Cervera A, Ludvigsen M, Hamilton-Dutoit SJ,
d'Amore F, et al: MicroRNAs regulate key cell survival pathways and
mediate chemosensitivity during progression of diffuse large B-cell
lymphoma. Blood Cancer J. 7:6542017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li M, Cui X and Guan H: The expression and
clinical significance of MicroRNA-409-3p in acute myeloid leukemia.
Clin Lab. 66:2020. View Article : Google Scholar
|
|
66
|
Xie W, Wang Z, Guo X and Guan H:
MiR-409-3p regulates the proliferation and apoptosis of THP-1
through targeting Rab10. Leuk Res. 132:1073502023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kumar A, Nayak S, Pathak P, Purkait S,
Malgulawar PB, Sharma MC, Suri V, Mukhopadhyay A, Suri A and Sarkar
C: Identification of miR-379/miR-656 (C14MC) cluster downregulation
and associated epigenetic and transcription regulatory mechanism in
oligodendrogliomas. J Neurooncol. 139:23–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Venza I, Visalli M, Beninati C, Benfatto
S, Teti D and Venza M: IL-10Rα expression is post-transcriptionally
regulated by miR-15a, miR-185, and miR-211 in melanoma. BMC Med
Genomics. 8:812015. View Article : Google Scholar
|
|
69
|
Jiang L, Zhang Y, Li B, Kang M, Yang Z,
Lin C, Hu K, Wei Z, Xu M, Mi J, et al: miRNAs derived from
circulating small extracellular vesicles as diagnostic biomarkers
for nasopharyngeal carcinoma. Cancer Sci. 112:2393–2404. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen H and Dai J: miR-409-3p suppresses
the proliferation, invasion and migration of tongue squamous cell
carcinoma via targeting RDX. Oncol Lett. 16:543–551.
2018.PubMed/NCBI
|
|
71
|
Josson S, Gururajan M, Hu P, Shao C, Chu
GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al: miR-409-3p/-5p
promotes tumorigenesis, epithelial-to-mesenchymal transition, and
bone metastasis of human prostate cancer. Clin Cancer Res.
20:4636–4646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Gururajan M, Josson S, Chu GC, Lu CL, Lu
YT, Haga CL, Zhau HE, Liu C, Lichterman J, Duan P, et al: miR-154*
and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate
epithelial to mesenchymal transition and bone metastasis of
prostate cancer. Clin Cancer Res. 20:6559–6569. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Josson S, Gururajan M, Sung SY, Hu P, Shao
C, Zhau HE, Liu C, Lichterman J, Duan P, Li Q, et al: Stromal
fibroblast-derived miR-409 promotes epithelial-to-mesenchymal
transition and prostate tumorigenesis. Oncogene. 34:2690–2699.
2015. View Article : Google Scholar
|
|
74
|
Yu Q, Li P, Weng M, Wu S, Zhang Y, Chen X,
Zhang Q, Shen G, Ding X and Fu S: Nano-vesicles are a potential
tool to monitor therapeutic efficacy of carbon ion radiotherapy in
prostate cancer. J Biomed Nanotechnol. 14:168–178. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fredsøe J, Rasmussen AKI, Mouritzen P,
Bjerre MT, Østergren P, Fode M, Borre M and Sørensen KD: Profiling
of circulating microRNAs in prostate cancer reveals diagnostic
biomarker potential. Diagnostics (Basel). 10:1882020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Nguyen HCN, Xie W, Yang M, Hsieh CL,
Drouin S, Lee GS and Kantoff PW: Expression differences of
circulating microRNAs in metastatic castration resistant prostate
cancer and low-risk, localized prostate cancer. Prostate.
73:346–354. 2013. View Article : Google Scholar
|
|
77
|
Zhi F, Shao N, Li B, Xue L, Deng D, Xu Y,
Lan Q, Peng Y and Yang Y: A serum 6-miRNA panel as a novel
non-invasive biomarker for meningioma. Sci Rep. 6:320672016.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ding X, Wang X, Han L, Zhao Z, Jia S and
Tuo Y: CircRNA DOCK1 regulates miR-409-3p/MCL1 axis to modulate
proliferation and apoptosis of human brain vascular smooth muscle
cells. Front Cell Dev Biol. 9:6556282021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Cao Y, Zhang L, Wei M, Jiang X and Jia D:
MicroRNA-409-3p represses glioma cell invasion and proliferation by
targeting high-mobility group nucleosome-binding domain 5. Oncol
Res. 25:1097–1107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ma Z, Chen Z, Zhou Y, Li Y, Li S, Wang H
and Feng J: Hsa_circ_0000418 promotes the progression of glioma by
regulating microRNA-409-3p/pyruvate dehydrogenase kinase 1 axis.
Bioengineered. 13:7541–7552. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Weng C, Dong H, Chen G, Zhai Y, Bai R, Hu
H, Lu L and Xu Z: miR-409-3p inhibits HT1080 cell proliferation,
vascularization and metastasis by targeting angiogenin. Cancer
Lett. 323:171–179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Khalil S, Fabbri E, Santangelo A, Bezzerri
V, Cantù C, Di Gennaro G, Finotti A, Ghimenton C, Eccher A,
Dechecchi M, et al: miRNA array screening reveals cooperative
MGMT-regulation between miR-181d-5p and miR-409-3p in glioblastoma.
Oncotarget. 7:28195–28206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chang JH, Xu BW, Shen D, Zhao W, Wang Y,
Liu JL, Meng GX, Li GZ and Zhang ZL: BRF2 is mediated by
microRNA-409-3p and promotes invasion and metastasis of HCC through
the Wnt/β-catenin pathway. Cancer Cell Int. 23:462023. View Article : Google Scholar
|
|
84
|
Li L, Ai R, Yuan X, Dong S, Zhao D, Sun X,
Miao T, Guan W, Guo P, Yu S and Nan Y: LINC00886 facilitates
hepatocellular carcinoma tumorigenesis by sequestering
microRNA-409-3p and microRNA-214-5p. J Hepatocell Carcinoma.
10:863–881. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Pigati L, Yaddanapudi SCS, Iyengar R, Kim
DJ, Hearn SA, Danforth D, Hastings ML and Duelli DM: Selective
release of microRNA species from normal and malignant mammary
epithelial cells. PLoS One. 5:e135152010. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Witwer KW, Buzás EI, Bemis LT, Bora A,
Lässer C, Lötvall J, Nolte-'t Hoen EN, Piper MG, Sivaraman S, Skog
J, et al: Standardization of sample collection, isolation and
analysis methods in extracellular vesicle research. J Extracell
Vesicles. 2:2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gallo A, Tandon M, Alevizos I and Illei
GG: The majority of microRNAs detectable in serum and saliva is
concentrated in exosomes. PLoS One. 7:e306792012. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Mathivanan S, Ji H and Simpson RJ:
Exosomes: Extracellular organelles important in intercellular
communication. J Proteomics. 73:1907–1920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK,
Pritchard CC, Gibson DF, Mitchell PS, Bennett CF,
Pogosova-Agadjanyan EL, Stirewalt DL, et al: Argonaute2 complexes
carry a population of circulating microRNAs independent of vesicles
in human plasma. Proc Natl Acad Sci USA. 108:5003–5008. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Turchinovich A, Weiz L, Langheinz A and
Burwinkel B: Characterization of extracellular circulating
microRNA. Nucleic Acids Res. 39:7223–7233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Finak G, Bertos N, Pepin F, Sadekova S,
Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu
A, et al: Stromal gene expression predicts clinical outcome in
breast cancer. Nat Med. 14:518–527. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Goetsch L, Caussanel V and Corvaia N:
Biological significance and targeting of c-Met tyrosine kinase
receptor in cancer. Front Biosci (Landmark Ed). 18:454–473. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Cecchi F, Rabe DC and Bottaro DP:
Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer.
46:1260–1270. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Dienstmann R, Rodon J, Serra V and
Tabernero J: Picking the point of inhibition: A comparative review
of PI3K/AKT/mTOR pathway inhibitors. Mol Cancer Ther. 13:1021–1031.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Friedl P and Wolf K: Tumour-cell invasion
and migration: Diversity and escape mechanisms. Nat Rev Cancer.
3:362–374. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Singh M, Yelle N, Venugopal C and Singh
SK: EMT: Mechanisms and therapeutic implications. Pharmacol Ther.
182:80–94. 2018. View Article : Google Scholar
|
|
100
|
Liu T, Zhao X, Zheng X, Zheng Y, Dong X,
Zhao N, Liao S and Sun B: The EMT transcription factor, Twist1, as
a novel therapeutic target for pulmonary sarcomatoid carcinomas.
Int J Oncol. 56:750–760. 2020.PubMed/NCBI
|
|
101
|
Stenmark H: Rab GTPases as coordinators of
vesicle traffic. Nat Rev Mol Cell Biol. 10:513–525. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cheng HL, Trink B, Tzai TS, Liu HS, Chan
SH, Ho CL, Sidransky D and Chow NH: Overexpression of c-met as a
prognostic indicator for transitional cell carcinoma of the urinary
bladder: A comparison with p53 nuclear accumulation. J Clin Oncol.
20:1544–1550. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tan Q, Joshua AM, Wang M, Bristow RG,
Wouters BG, Allen CJ and Tannock IF: Correction to: Up-regulation
of autophagy is a mechanism of resistance to chemotherapy and can
be inhibited by pantoprazole to increase drug sensitivity. Cancer
Chemother Pharmacol. Jan 19–2024.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Helgason GV, Holyoake TL and Ryan KM: Role
of autophagy in cancer prevention, development and therapy. Essays
Biochem. 55:133–151. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhou Y, Sun K, Ma Y, Yang H, Zhang Y, Kong
X and Wei L: Autophagy inhibits chemotherapy-induced apoptosis
through downregulating Bad and Bim in hepatocellular carcinoma
cells. Sci Rep. 4:53822014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Sui X, Chen R, Wang Z, Huang Z, Kong N,
Zhang M, Han W, Lou F, Yang J, Zhang Q, et al: Autophagy and
chemotherapy resistance: A promising therapeutic target for cancer
treatment. Cell Death Dis. 4:e8382013. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Fujiwara K, Iwado E, Mills GB, Sawaya R,
Kondo S and Kondo Y: Akt inhibitor shows anticancer and
radiosensitizing effects in malignant glioma cells by inducing
autophagy. Int J Oncol. 31:753–760. 2007.PubMed/NCBI
|
|
108
|
Kim EJ, Jeong JH, Bae S, Kang S, Kim CH
and Lim YB: mTOR inhibitors radiosensitize PTEN-deficient
non-small-cell lung cancer cells harboring an EGFR activating
mutation by inducing autophagy. J Cell Biochem. 114:1248–1256.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Arico S, Petiot A, Bauvy C, Dubbelhuis PF,
Meijer AJ, Codogno P and Ogier-Denis E: The tumor suppressor PTEN
positively regulates macroautophagy by inhibiting the
phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol
Chem. 276:35243–35246. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Liberti MV and Locasale JW: The Warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Orang AV, Petersen J, McKinnon RA and
Michael MZ: Micromanaging aerobic respiration and glycolysis in
cancer cells. Mol Metab. 23:98–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhang Q, Wang L, Cao L and Wei T: Novel
circular RNA circATRNL1 accelerates the osteosarcoma aerobic
glycolysis through targeting miR-409-3p/LDHA. Bioengineered.
12:9965–9975. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Chang SH and Hla T: Gene regulation by RNA
binding proteins and microRNAs in angiogenesis. Trends Mol Med.
17:650–658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Folkman J, Watson K, Ingber D and Hanahan
D: Induction of angiogenesis during the transition from hyperplasia
to neoplasia. Nature. 339:58–61. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhang Y and Wang X: Targeting the
Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol.
13:1652020. View Article : Google Scholar
|
|
116
|
Huang Y, Yang Y, He Y and Li J: The
emerging role of Nemo-like kinase (NLK) in the regulation of
cancers. Tumour Biol. 36:9147–9152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Masoumi KC, Daams R, Sime W, Siino V, Ke
H, Levander F and Massoumi R: NLK-mediated phosphorylation of HDAC1
negatively regulates Wnt signaling. Mol Biol Cell. 28:346–355.
2017. View Article : Google Scholar :
|
|
118
|
Smit L, Baas A, Kuipers J, Korswagen H,
van de Wetering M and Clevers H: Wnt activates the Tak1/Nemo-like
kinase pathway. J Biol Chem. 279:17232–17240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Deng Y, Deng H, Liu J, Han G, Malkoski S,
Liu B, Zhao R, Wang XJ and Zhang Q: Transcriptional down-regulation
of Brca1 and E-cadherin by CtBP1 in breast cancer. Mol Carcinog.
51:500–507. 2012. View Article : Google Scholar
|
|
120
|
Ma PC, Maulik G, Christensen J and Salgia
R: c-Met: Structure, functions and potential for therapeutic
inhibition. Cancer Metastasis Rev. 22:309–325. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Revathidevi S and Munirajan AK: Akt in
cancer: Mediator and more. Semin Cancer Biol. 59:80–91. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Johnson GL and Lapadat R:
Mitogen-activated protein kinase pathways mediated by ERK, JNK, and
p38 protein kinases. Science. 298:1911–1912. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48(D1): D127–D131. 2020. View Article : Google Scholar :
|
|
124
|
McGeary SE, Lin KS, Shi CY, Pham TM,
Bisaria N, Kelley GM and Bartel DP: The biochemical basis of
microRNA targeting efficacy. Science. 366:eaav17412019. View Article : Google Scholar : PubMed/NCBI
|