|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel S: Breast cancer: Lesser-known
facets and hypotheses. Biomed Pharmacother. 98:499–506. 2018.
View Article : Google Scholar
|
|
3
|
Zhu Z, Albadawy E, Saha A, Zhang J,
Harowicz MR and Mazurowski MA: Deep learning for identifying
radiogenomic associations in breast cancer. Comput Biol Med.
109:85–90. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lord SJ, Bahlmann K, O'Connell DL, Kiely
BE, Daniels B, Pearson SA, Beith J, Bulsara MK and Houssami N: De
novo and recurrent metastatic breast cancer-A systematic review of
population-level changes in survival since 1995. EClinicalMedicine.
44:1012822022. View Article : Google Scholar
|
|
5
|
Dai D, Wang H, Zhu L, Jin H and Wang X:
N6-methyladenosine links RNA metabolism to cancer progression. Cell
Death Dis. 9:1242018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hyun K, Jeon J, Park K and Kim J: Writing,
erasing and reading histone lysine methylations. Exp Mol Med.
49:e3242017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
An Y and Duan H: The role of m6A RNA
methylation in cancer metabolism. Mol Cancer. 21:142022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin H, Wang Y, Wang P, Long F and Wang T:
Mutual regulation between N6-methyladenosine (m6A) modification and
circular RNAs in cancer: Impacts on therapeutic resistance. Mol
Cancer. 21:1482022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Z, Zou H, Dang Q, Xu H, Liu L, Zhang
Y, Lv J, Li H, Zhou Z and Han X: Biological and pharmacological
roles of m6A modifications in cancer drug resistance.
Mol Cancer. 21:2202022. View Article : Google Scholar
|
|
10
|
Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M,
Lyu WY, Qi Q, Tiwari AK, Chen JX, Zhang DM and Chen ZS: m6A
modification: Recent advances, anticancer targeted drug discovery
and beyond. Mol Cancer. 21:522022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
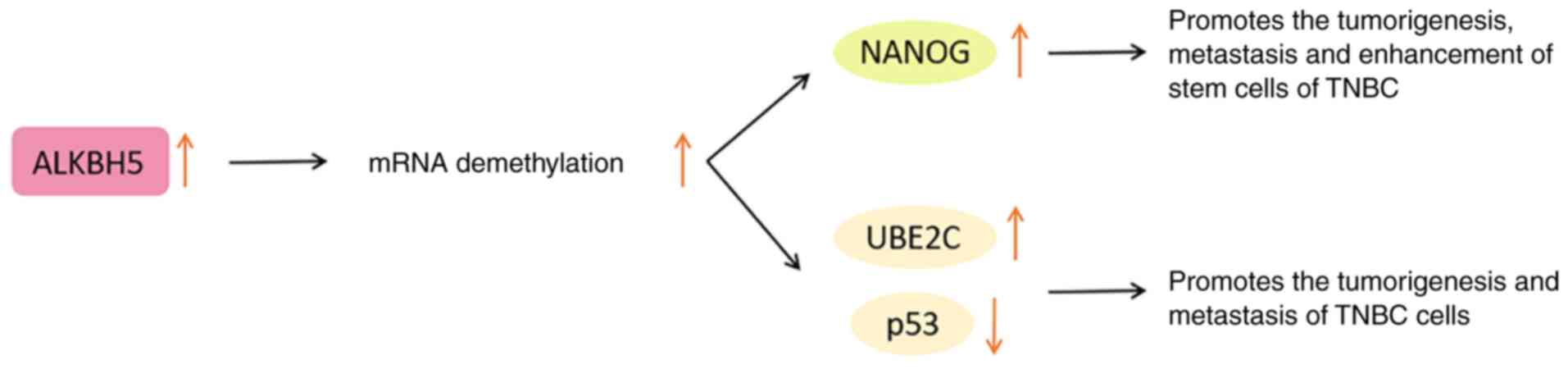
13
|
Loibl S, Poortmans P, Morrow M, Denkert C
and Curigliano G: Breast cancer. Lancet. 397:1750–1769. 2021.
View Article : Google Scholar : PubMed/NCBI
|
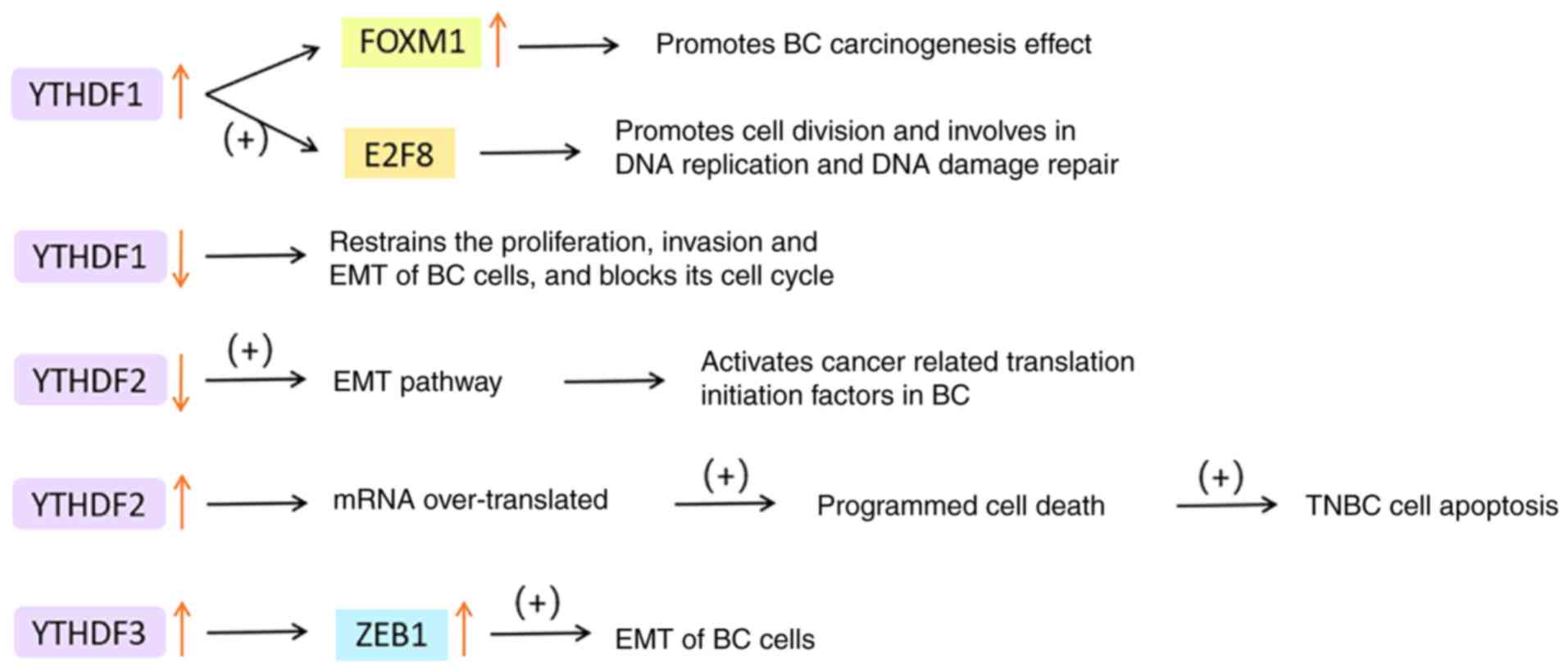
|
14
|
Hong R and Xu B: Breast cancer: an
up-to-date review and future perspectives. Cancer Commun (Lond).
42:913–936. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Golshan M, Loibl S, Wong SM, Houber JB,
O'Shaughnessy J, Rugo HS, Wolmark N, McKee MD, Maag D, Sullivan DM,
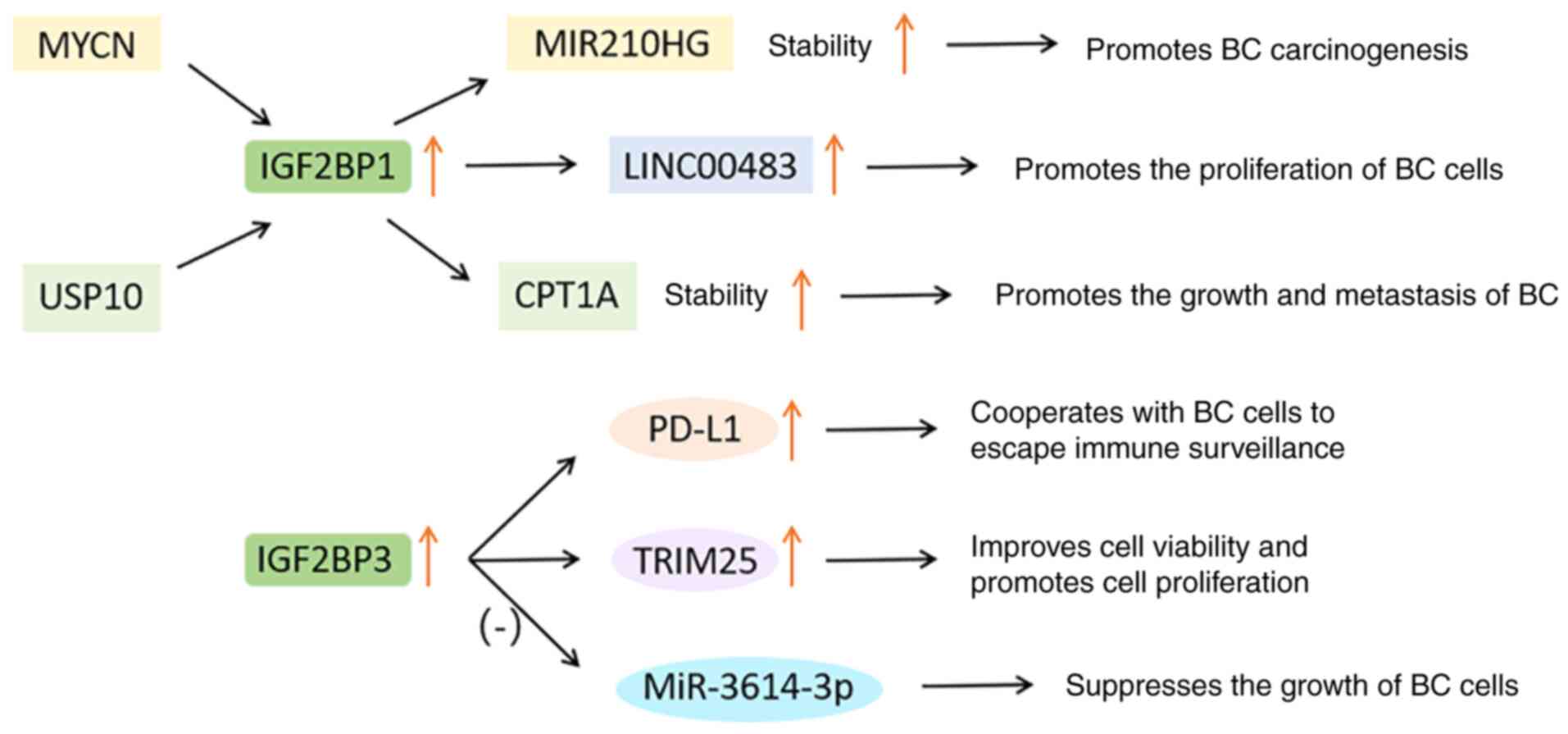
et al: Breast conservation after neoadjuvant chemotherapy for
triple-negative breast cancer: Surgical results from the brightness
randomized clinical trial. JAMA Surg. 155:e1954102020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Waks AG and Winer EP: Breast cancer
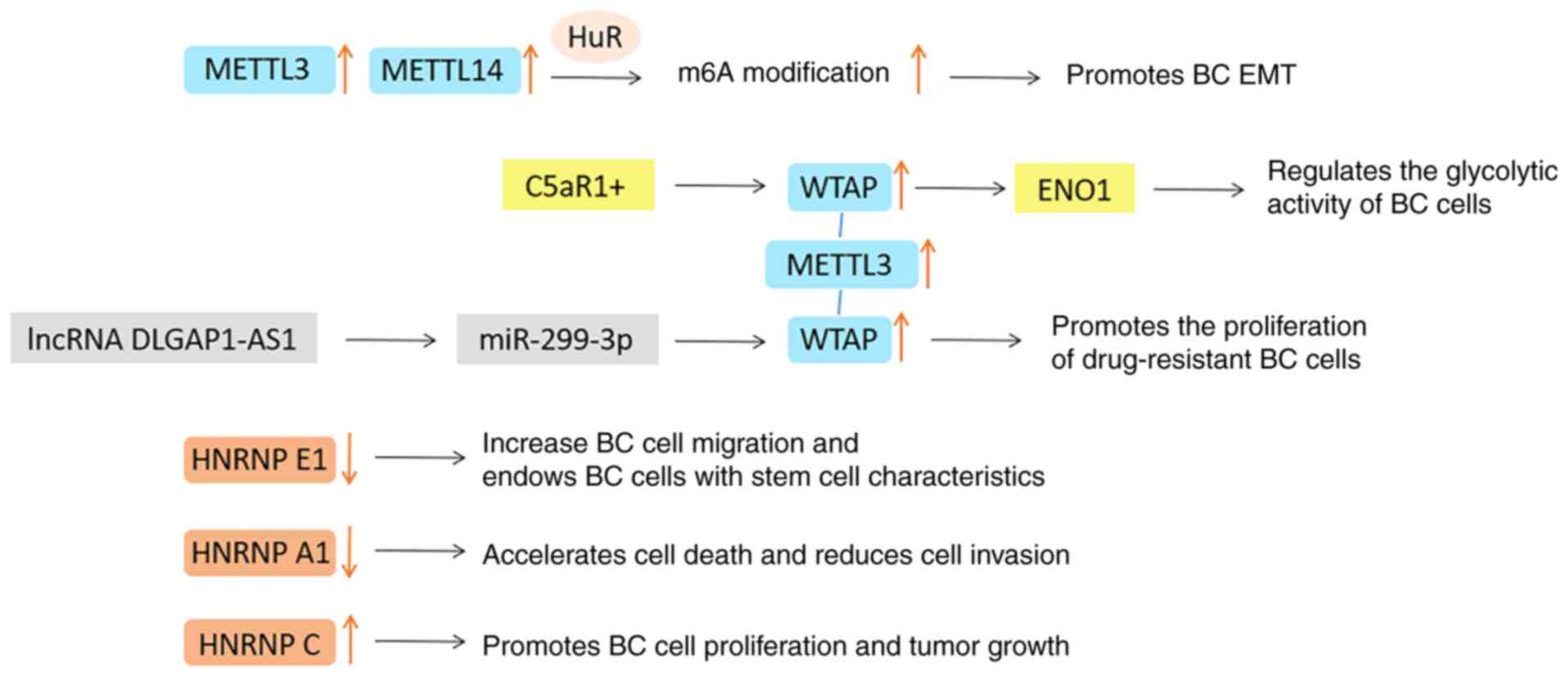
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jääskeläinen A, Roininen N, Karihtala P
and Jukkola A: High parity predicts poor outcomes in patients with
luminal B-like (HER2 negative) early breast cancer: A prospective
finnish single-center study. Front Oncol. 10:14702020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choong GM, Cullen GD and O'Sullivan CC:
Evolving standards of care and new challenges in the management of
HER2-positive breast cancer. CA Cancer J Clin. 70:355–374. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu KD, Ye FG, He M, Fan L, Ma D, Mo M, Wu
J, Liu GY, Di GH, Zeng XH, et al: Effect of adjuvant paclitaxel and
carboplatin on survival in women with triple-negative breast
cancer: A phase 3 randomized clinical trial. JAMA Oncol.
6:1390–1396. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garrido-Castro AC, Lin NU and Polyak K:
Insights into molecular classifications of triple-negative breast
cancer: Improving patient selection for treatment. Cancer Discov.
9:176–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gaudet MM, Gierach GL, Carter BD, Luo J,
Milne RL, Weiderpass E, Giles GG, Tamimi RM, Eliassen AH, Rosner B,
et al: Pooled analysis of nine cohorts reveals breast cancer risk
factors by tumor molecular subtype. Cancer Res. 78:6011–6021. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nur U, El Reda D, Hashim D and Weiderpass
E: A prospective investigation of oral contraceptive use and breast
cancer mortality: Findings from the Swedish women's lifestyle and
health cohort. BMC Cancer. 19:8072019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trabert B, Sherman ME, Kannan N and
Stanczyk FZ: Progesterone and breast cancer. Endocr Rev.
41:320–344. 2020. View Article : Google Scholar :
|
|
24
|
Reiner AS, Sisti J, John EM, Lynch CF,
Brooks JD, Mellemkjær L, Boice JD, Knight JA, Concannon P, Capanu
M, et al: Breast cancer family history and contralateral breast
cancer risk in young women: an update from the women's
environmental cancer and radiation epidemiology study. J Clin
Oncol. 36:1513–1520. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ho PJ, Ho WK, Khng AJ, Yeoh YS, Tan BK,
Tan EY, Lim GH, Tan SM, Tan VKM, Yip CH, et al: Overlap of
high-risk individuals predicted by family history, and genetic and
non-genetic breast cancer risk prediction models: Implications for
risk stratification. BMC Med. 20:1502022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu HM, Li S, Black MH, Lee S, Hoiness R,
Wu S, Mu W, Huether R, Chen J, Sridhar S, et al: Association of
breast and ovarian cancers with predisposition genes identified by
large-scale sequencing. JAMA Oncol. 5:51–57. 2019. View Article : Google Scholar :
|
|
27
|
Breast Cancer Association Consortium;
Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C,
Wahlström C, Pooley KA, Parsons MT, Fortuno C, et al: Breast cancer
risk genes-association analysis in more than 113,000 women. N Engl
J Med. 384:428–439. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ru W, Zhang X, Yue B, Qi A, Shen X, Huang
Y, Lan X, Lei C and Chen H: Insight into m6A methylation
from occurrence to functions. Open Biol. 10:2000912020. View Article : Google Scholar
|
|
29
|
Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J,
Lin S and Wang H: N6-methyladenosine regulates
glycolysis of cancer cells through PDK4. Nat Commun. 11:25782020.
View Article : Google Scholar
|
|
30
|
Huang H, Weng H and Chen J: m6A
modification in coding and non-coding RNAs: Roles and therapeutic
implications in cancer. Cancer Cell. 37:270–288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Choe J, Lin S, Zhang W, Liu Q, Wang L,
Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al: mRNA
circularization by METTL3-eIF3h enhances translation and promotes
oncogenesis. Nature. 561:556–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shi B, Liu WW, Yang K, Jiang GM and Wang
H: The role, mechanism, and application of RNA methyltransferase
METTL14 in gastrointestinal cancer. Mol Cancer. 21:1632022.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen H, Wang Y, Su H, Zhang X, Chen H and
Yu J: RNA N6-methyladenine modification, cellular
reprogramming, and cancer stemness. Front Cell Dev Biol.
10:9352242022. View Article : Google Scholar
|
|
36
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Structural basis of
N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature.
534:575–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol
Oncol. 13:1172020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Y, Peng C, Chen J, Chen D, Yang B, He
B, Hu W, Zhang Y, Liu H, Dai L, et al: WTAP facilitates progression
of hepatocellular carcinoma via m6A-HuR-dependent epigenetic
silencing of ETS1. Mol Cancer. 18:1272019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Su R, Dong L, Li Y, Gao M, He PC, Liu W,
Wei J, Zhao Z, Gao L, Han L, et al: METTL16 exerts an
m6A-independent function to facilitate translation and
tumorigenesis. Nat Cell Biol. 24:205–216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu Y, Ouyang Z, Sui X, Qi M, Li M, He Y,
Cao Y, Cao Q, Lu Q, Zhou S, et al: Oocyte competence is maintained
by m6A methyltransferase KIAA1429-mediated RNA
metabolism during mouse follicular development. Cell Death Differ.
27:2468–2483. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tan C, Xia P, Zhang H, Xu K, Liu P, Guo D
and Liu Z: YY1-Targeted RBM15B promotes hepatocellular carcinoma
cell proliferation and sorafenib resistance by promoting TRAM2
expression in an m6A-dependent manner. Front Oncol. 12:8730202022.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wen J, Lv R, Ma H, Shen H, He C, Wang J,
Jiao F, Liu H, Yang P, Tan L, et al: Zc3h13 regulates nuclear RNA
m6A methylation and mouse embryonic stem cell
self-renewal. Mol Cell. 69:1028–1038.e6. 2018. View Article : Google Scholar
|
|
43
|
Wang T, Kong S, Tao M and Ju S: The
potential role of RNA N6-methyladenosine in Cancer progression. Mol
Cancer. 19:882020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang L, Song C, Wang N, Li S, Liu Q, Sun
Z, Wang K, Yu SC and Yang Q: NADP modulates RNA m6A
methylation and adipogenesis via enhancing FTO activity. Nat Chem
Biol. 16:1394–1402. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bartosovic M, Molares HC, Gregorova P,
Hrossova D, Kudla G and Vanacova S: N6-methyladenosine demethylase
FTO targets pre-mRNAs and regulates alternative splicing and 3'-end
processing. Nucleic Acids Res. 45:11356–11370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi
Y, He S and Shimamoto F: m6A demethylase ALKBH5 inhibits
pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation
and mediating Wnt signaling. Mol Cancer. 19:32020. View Article : Google Scholar
|
|
47
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M,
Ma J and Wu L: YTHDF2 destabilizes m(6)A-containing RNA through
direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun.
7:126262016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen Z, Zhong X, Xia M and Zhong J: The
roles and mechanisms of the m6A reader protein YTHDF1 in tumor
biology and human diseases. Mol Ther Nucleic Acids. 26:1270–1279.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zaccara S and Jaffrey SR: A unified model
for the function of YTHDF proteins in regulating
m6A-modified mRNA. Cell. 181:1582–1595.e18. 2020.
View Article : Google Scholar
|
|
52
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Roundtree IA, Luo GZ, Zhang Z, Wang X,
Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al: YTHDC1
mediates nuclear export of N6-methyladenosine methylated
mRNAs. Elife. 6:e313112017. View Article : Google Scholar
|
|
54
|
Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B
and Qian SB: m6A in mRNA coding regions promotes
translation via the RNA helicase-containing YTHDC2. Nat Commun.
10:53322019. View Article : Google Scholar
|
|
55
|
Wu B, Su S, Patil DP, Liu H, Gan J,
Jaffrey SR and Ma J: Molecular basis for the specific and
multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun. 9:4202018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sun CY, Cao D, Du BB, Chen CW and Liu D:
The role of Insulin-like growth factor 2 mRNA-binding proteins
(IGF2BPs) as m6A readers in cancer. Int J Biol Sci.
18:2744–2758. 2022. View Article : Google Scholar :
|
|
57
|
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y,
Cheng C, Li L, Pi J, Si Y, et al: The m6A reader YTHDF1 promotes
ovarian cancer progression via augmenting EIF3C translation.
Nucleic Acids Res. 48:3816–3831. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Y, Zhang Y, Du Y, Zhou M, Hu Y and
Zhang S: Emerging roles of N6-methyladenosine (m6A)
modification in breast cancer. Cell Biosci. 10:1362020. View Article : Google Scholar
|
|
59
|
Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X,
Wang Q, Li X, Zhang Y and Xu J: Molecular characterization and
clinical relevance of m6A regulators across 33 cancer
types. Mol Cancer. 18:1372019. View Article : Google Scholar
|
|
60
|
Wei M, Bai JW, Niu L, Zhang YQ, Chen HY
and Zhang GJ: The complex roles and therapeutic implications of
m6A modifications in breast cancer. Front Cell Dev Biol.
8:6150712021. View Article : Google Scholar
|
|
61
|
Han H, Yang C, Zhang S, Cheng M, Guo S,
Zhu Y, Ma J, Liang Y, Wang L, Zheng S, et al: METTL3-mediated
m6A mRNA modification promotes esophageal cancer
initiation and progression via Notch signaling pathway. Mol Ther
Nucleic Acids. 26:333–346. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018. View Article : Google Scholar
|
|
63
|
Jin H, Ying X, Que B, Wang X, Chao Y,
Zhang H, Yuan Z, Qi D, Lin S, Min W, et al:
N6-methyladenosine modification of ITGA6 mRNA promotes
the development and progression of bladder cancer. EBioMedicine.
47:195–207. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C,
Li S, Tan L, Mai D, Li G, et al: Excessive miR-25-3p maturation via
N6-methyladenosine stimulated by cigarette smoke
promotes pancreatic cancer progression. Nat Commun. 10:18582019.
View Article : Google Scholar
|
|
65
|
Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G,
Yuan W, Kan Q and Sun Z: The interplay between m6A RNA methylation
and noncoding RNA in cancer. J Hematol Oncol. 12:1212019.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xie JW, Huang XB, Chen QY, Ma YB, Zhao YJ,
Liu LC, Wang JB, Lin JX, Lu J, Cao LL, et al: m6A
modification-mediated BATF2 acts as a tumor suppressor in gastric
cancer through inhibition of ERK signaling. Mol Cancer. 19:1142020.
View Article : Google Scholar
|
|
67
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation
regulates the self-renewal and tumorigenesis of glioblastoma stem
cells. Cell Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zheng W, Dong X, Zhao Y, Wang S, Jiang H,
Zhang M, Zheng X and Gu M: Multiple functions and mechanisms
underlying the role of METTL3 in human cancers. Front Oncol.
9:14032019. View Article : Google Scholar
|
|
69
|
Wang G, Dai Y, Li K, Cheng M, Xiong G,
Wang X, Chen S, Chen Z, Chen J, Xu X, et al: Deficiency of Mettl3
in bladder cancer stem cells inhibits bladder cancer progression
and angiogenesis. Front Cell Dev Biol. 9:6277062021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shi Y, Zheng C, Jin Y, Bao B, Wang D, Hou
K, Feng J, Tang S, Qu X, Liu Y, et al: Reduced expression of METTL3
promotes metastasis of triple-negative breast cancer by m6A
methylation-mediated COL3A1 up-regulation. Front Oncol.
10:11262020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W,
Guo W, Wu X, Pu C, Hu X, et al: METTL3/IGF2BP3 axis inhibits tumor
immune surveillance by upregulating N6-methyladenosine
modification of PD-L1 mRNA in breast cancer. Mol Cancer. 21:602022.
View Article : Google Scholar
|
|
72
|
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang
Z, Liu Y, Zhang X, Zhang W and Ye L: HBXIP-elevated
methyltransferase METTL3 promotes the progression of breast cancer
via inhibiting tumor suppressor let-7g. Cancer Lett. 415:11–19.
2018. View Article : Google Scholar
|
|
73
|
Ma J, Zhang J, Weng YC and Wang JC:
EZH2-mediated microRNA-139-5p regulates epithelial-mesenchymal
transition and lymph node metastasis of pancreatic cancer. Mol
Cells. 41:868–880. 2018.PubMed/NCBI
|
|
74
|
Hu S, Song Y, Zhou Y, Jiao Y and Li G:
METTL3 accelerates breast cancer progression via regulating EZH2
m6A modification. J Healthc Eng. 2022:57944222022.
|
|
75
|
Li W, Xue D, Xue M, Zhao J, Liang H, Liu Y
and Sun T: Fucoidan inhibits epithelial-to-mesenchymal transition
via regulation of the HIF-1α pathway in mammary cancer cells under
hypoxia. Oncol Lett. 18:330–338. 2019.PubMed/NCBI
|
|
76
|
Zhao C, Ling X, Xia Y, Yan B and Guan Q:
The m6A methyltransferase METTL3 controls epithelial-mesenchymal
transition, migration and invasion of breast cancer through the
MALAT1/miR-26b/HMGA2 axis. Cancer Cell Int. 21:4412021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia
TS, Zhou WB, Wang S, Ding Q and Wei JF: KIAA1429 acts as an
oncogenic factor in breast cancer by regulating CDK1 in an
N6-methyladenosine-independent manner. Oncogene. 38:6123–6141.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang X, Dai XY, Qian JY, Xu F, Wang ZW,
Xia T, Zhou XJ, Li XX, Shi L, Wei JF and Ding Q: SMC1A regulated by
KIAA1429 in m6A-independent manner promotes EMT progress in breast
cancer. Mol Ther Nucleic Acids. 27:133–146. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R,
Wang YY and Zhe H: FTO regulates the chemo-radiotherapy resistance
of cervical squamous cell carcinoma (CSCC) by targeting β-catenin
through mRNA demethylation. Mol Carcinog. 57:590–597. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu J, Ren D, Du Z, Wang H, Zhang H and
Jin Y: m6A demethylase FTO facilitates tumor progression
in lung squamous cell carcinoma by regulating MZF1 expression.
Biochem Biophys Res Commun. 502:456–464. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Shimura T, Kandimalla R, Okugawa Y, Ohi M,
Toiyama Y, He C and Goel A: Novel evidence for m6A
methylation regulators as prognostic biomarkers and FTO as a
potential therapeutic target in gastric cancer. Br J Cancer.
126:228–237. 2022. View Article : Google Scholar
|
|
82
|
Azzam SK, Alsafar H and Sajini AA: FTO m6A
demethylase in obesity and cancer: implications and underlying
molecular mechanisms. Int J Mol Sci. 23:38002022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zheng QK, Ma C, Ullah I, Hu K, Ma RJ,
Zhang N and Sun ZG: Roles of N6-methyladenosine demethylase FTO in
malignant tumors progression. Onco Targets Ther. 14:4837–4846.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun
L, Wang Y, Li X, Xiong XF, Wei B, et al: RNA N6-methyladenosine
demethylase FTO promotes breast tumor progression through
inhibiting BNIP3. Mol Cancer. 18:462019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Xu Y, Ye S, Zhang N, Zheng S, Liu H, Zhou
K, Wang L, Cao Y, Sun P and Wang T: The FTO/miR-181b-3p/ARL5B
signaling pathway regulates cell migration and invasion in breast
cancer. Cancer Commun (Lond). 40:484–500. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Basu A: The interplay between apoptosis
and cellular senescence: Bcl-2 family proteins as targets for
cancer therapy. Pharmacol Ther. 230:1079432022. View Article : Google Scholar
|
|
87
|
Gao X, Wang Y, Lu F, Chen X, Yang D, Cao
Y, Zhang W, Chen J, Zheng L, Wang G, et al: Extracellular vesicles
derived from oesophageal cancer containing P4HB promote muscle
wasting via regulating PHGDH/Bcl-2/caspase-3 pathway. J Extracell
Vesicles. 10:e120602021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu Y, Wang R, Zhang L, Li J, Lou K and
Shi B: The lipid metabolism gene FTO influences breast cancer cell
energy metabolism via the PI3K/AKT signaling pathway. Oncol Lett.
13:4685–4690. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Qu J, Yan H, Hou Y, Cao W, Liu Y, Zhang E,
He J and Cai Z: RNA demethylase ALKBH5 in cancer: From mechanisms
to therapeutic potential. J Hematol Oncol. 15:82022. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wu L, Wu D, Ning J, Liu W and Zhang D:
Changes of N6-methyladenosine modulators promote breast cancer
progression. BMC Cancer. 19:3262019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Hu Y, Liu H, Xiao X, Yu Q, Deng R, Hua L,
Wang J and Wang X: Bone marrow mesenchymal stem cell-derived
exosomes inhibit triple-negative breast cancer cell stemness and
metastasis via an ALKBH5-dependent mechanism. Cancers (Basel).
14:60592022. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Fry NJ, Law BA, Ilkayeva OR, Carraway KR
and Mansfield KD: N6-methyladenosine contributes to
cellular phenotype in a genetically-defined model of breast cancer
progression. Oncotarget. 9:31231–31243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang C, Samanta D, Lu H, Bullen JW, Zhang
H, Chen I, He X and Semenza GL: Hypoxia induces the breast cancer
stem cell phenotype by HIF-dependent and ALKBH5-mediated
m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA.
113:E2047–E2056. 2016.
|
|
94
|
Zhang C, Zhi WI, Lu H, Samanta D, Chen I,
Gabrielson E and Semenza GL: Hypoxia-inducible factors regulate
pluripotency factor expression by ZNF217- and ALKBH5-mediated
modulation of RNA methylation in breast cancer cells. Oncotarget.
7:64527–64542. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang S, You X, Zheng Y, Shen Y, Xiong X
and Sun Y: The UBE2C/CDH1/DEPTOR axis is an oncogene and tumor
suppressor cascade in lung cancer cells. J Clin Invest.
133:e1624342023. View Article : Google Scholar :
|
|
96
|
Wang Y, Xie Y, Niu Y, Song P, Liu Y,
Burnett J, Yang Z, Sun D, Ran Y, Li Y and Sun L: Carboxypeptidase
A4 negatively correlates with p53 expression and regulates the
stemness of breast cancer cells. Int J Med Sci. 18:1753–1759. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chen H, Yu Y, Yang M, Huang H, Ma S, Hu J,
Xi Z, Guo H, Yao G, Yang L, et al: YTHDF1 promotes breast cancer
progression by facilitating FOXM1 translation in an m6A-dependent
manner. Cell Biosci. 12:192022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sun Y, Dong D, Xia Y, Hao L, Wang W and
Zhao C: YTHDF1 promotes breast cancer cell growth, DNA damage
repair and chemoresistance. Cell Death Dis. 13:2302022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Anita R, Paramasivam A, Priyadharsini JV
and Chitra S: The m6A readers YTHDF1 and YTHDF3 aberrations
associated with metastasis and predict poor prognosis in breast
cancer patients. Am J Cancer Res. 10:2546–2554. 2020.PubMed/NCBI
|
|
100
|
Zhong L, Liao D, Zhang M, Zeng C, Li X,
Zhang R, Ma H and Kang T: YTHDF2 suppresses cell proliferation and
growth via destabilizing the EGFR mRNA in hepatocellular carcinoma.
Cancer Lett. 442:252–261. 2019. View Article : Google Scholar
|
|
101
|
Chen YG, Chen R, Ahmad S, Verma R, Kasturi
SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, et al:
N6-methyladenosine modification controls circular RNA immunity. Mol
Cell. 76:96–109.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Paris J, Morgan M, Campos J, Spencer GJ,
Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA,
Sepulveda C, et al: Targeting the RNA m6A reader YTHDF2
selectively compromises cancer stem cells in acute myeloid
leukemia. Cell Stem Cell. 25:137–148.e6. 2019. View Article : Google Scholar
|
|
103
|
Dixit D, Prager BC, Gimple RC, Poh HX,
Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJY, Xie Q, et al: The RNA m6A
Reader YTHDF2 maintains oncogene expression and is a targetable
dependency in glioblastoma stem cells. Cancer Discov. 11:480–499.
2021. View Article : Google Scholar :
|
|
104
|
Li J, Xie H, Ying Y, Chen H, Yan H, He L,
Xu M, Xu X, Liang Z, Liu B, et al: YTHDF2 mediates the mRNA
degradation of the tumor suppressors to induce AKT phosphorylation
in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer.
19:1522020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Einstein JM, Perelis M, Chaim IA, Meena
JK, Nussbacher JK, Tankka AT, Yee BA, Li H, Madrigal AA, Neill NJ,
et al: Inhibition of YTHDF2 triggers proteotoxic cell death in
MYC-driven breast cancer. Mol Cell. 81:3048–3064.e9. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Li A, Chen YS, Ping XL, Yang X, Xiao W,
Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, et al: Cytoplasmic
m6A reader YTHDF3 promotes mRNA translation. Cell Res.
27:444–447. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Lin Y, Jin X, Nie Q, Chen M, Guo W, Chen
L, Li Y, Chen X, Zhang W, Chen H, et al: YTHDF3 facilitates
triple-negative breast cancer progression and metastasis by
stabilizing ZEB1 mRNA in an m6A-dependent manner. Ann
Transl Med. 10:832022. View Article : Google Scholar
|
|
108
|
Chang G, Shi L, Ye Y, Shi H, Zeng L,
Tiwary S, Huse JT, Huo L, Ma L, Ma Y, et al: YTHDF3 induces the
translation of m6A-enriched gene transcripts to promote
breast cancer brain metastasis. Cancer Cell. 38:857–871.e7. 2020.
View Article : Google Scholar
|
|
109
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Müller S, Glaß M, Singh AK, Haase J, Bley
N, Fuchs T, Lederer M, Dahl A, Huang H, Chen J, et al: IGF2BP1
promotes SRF-dependent transcription in cancer in a m6A- and
miRNA-dependent manner. Nucleic Acids Res. 47:375–390. 2019.
View Article : Google Scholar :
|
|
111
|
Qiao YS, Zhou JH, Jin BH, Wu YQ and Zhao
B: LINC00483 is regulated by IGF2BP1 and participates in the
progression of breast cancer. Eur Rev Med Pharmacol Sci.
25:1379–1386. 2021.PubMed/NCBI
|
|
112
|
Shi W, Tang Y, Lu J, Zhuang Y and Wang J:
MIR210HG promotes breast cancer progression by IGF2BP1 mediated m6A
modification. Cell Biosci. 12:382022. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Shi J, Zhang Q, Yin X, Ye J, Gao S, Chen
C, Yang Y, Wu B, Fu Y, Zhang H, et al: Stabilization of IGF2BP1 by
USP10 promotes breast cancer metastasis via CPT1A in an
m6A-dependent manner. Int J Biol Sci. 19:449–464. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zeng F, Yao M, Wang Y, Zheng W, Liu S, Hou
Z, Cheng X, Sun S, Li T, Zhao H, et al: Fatty acid β-oxidation
promotes breast cancer stemness and metastasis via the
miRNA-328-3p-CPT1A pathway. Cancer Gene Ther. 29:383–395. 2022.
View Article : Google Scholar
|
|
115
|
Xiong Y, Liu Z, Li Z, Wang S, Shen N, Xin
Y and Huang T: Long non-coding RNA nuclear paraspeckle assembly
transcript 1 interacts with microRNA-107 to modulate breast cancer
growth and metastasis by targeting carnitine
palmitoyltransferase-1. Int J Oncol. 55:1125–1136. 2019.PubMed/NCBI
|
|
116
|
Wang Z, Tong D, Han C, Zhao Z, Wang X,
Jiang T, Li Q, Liu S, Chen L, Chen Y, et al: Blockade of miR-3614
maturation by IGF2BP3 increases TRIM25 expression and promotes
breast cancer cell proliferation. EBioMedicine. 41:357–369. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Wang CQ, Tang CH, Wang Y, Huang BF, Hu GN,
Wang Q and Shao JK: Upregulated WTAP expression appears to both
promote breast cancer growth and inhibit lymph node metastasis. Sci
Rep. 12:10232022. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Ou B, Liu Y, Yang X, Xu X, Yan Y and Zhang
J: C5aR1-positive neutrophils promote breast cancer glycolysis
through WTAP-dependent m6A methylation of ENO1. Cell Death Dis.
12:7372021. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Huang T, Cao L, Feng N, Xu B, Dong Y and
Wang M: N6-methyladenosine (m6A)-mediated
lncRNA DLGAP1-AS1enhances breast canceradriamycin resistance
through miR-299-3p/WTAP feedback loop. Bioengineered.
12:10935–10944. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Fan Y, Li X, Sun H, Gao Z, Zhu Z and Yuan
K: Role of WTAP in cancer: From mechanisms to the therapeutic
potential. Biomolecules. 12:12242022. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Howley BV and Howe PH: TGF-beta signaling
in cancer: Post-transcriptional regulation of EMT via hnRNP E1.
Cytokine. 118:19–26. 2019. View Article : Google Scholar
|
|
122
|
Howley BV, Mohanty B, Dalton A, Grelet S,
Karam J, Dincman T and Howe PH: The ubiquitin E3 ligase ARIH1
regulates hnRNP E1 protein stability, EMT and breast cancer
progression. Oncogene. 41:1679–1690. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Loh TJ, Moon H, Cho S, Jang H, Liu YC, Tai
H, Jung DW, Williams DR, Kim HR, Shin MG, et al: CD44 alternative
splicing and hnRNP A1 expression are associated with the metastasis
of breast cancer. Oncol Rep. 34:1231–1238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Wu Y, Zhao W, Liu Y, Tan X, Li X, Zou Q,
Xiao Z, Xu H, Wang Y and Yang X: Function of HNRNPC in breast
cancer cells by controlling the dsRNA-induced interferon response.
EMBO J. 37:e990172018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Duijf PHG, Nanayakkara D, Nones K, Srihari
S, Kalimutho M and Khanna KK: Mechanisms of genomic instability in
breast cancer. Trends Mol Med. 25:595–611. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Hong J, Xu K and Lee JH: Biological roles
of the RNA m6A modification and its implications in
cancer. Exp Mol Med. 54:1822–1832. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Xiang Y, Laurent B, Hsu CH, Nachtergaele
S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al: RNA
m6A methylation regulates the ultraviolet-induced DNA
damage response. Nature. 543:573–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhang C, Chen L, Peng D, Jiang A, He Y,
Zeng Y, Xie C, Zhou H, Luo X, Liu H, et al: METTL3 and
N6-methyladenosine promote homologous recombination-mediated repair
of DSBs by modulating DNA-RNA hybrid accumulation. Mol Cell.
79:425–442.e7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
D'Alessandro G, Whelan DR, Howard SM,
Vitelli V, Renaudin X, Adamowicz M, Iannelli F, Jones-Weinert CW,
Lee M, Matti V, et al: BRCA2 controls DNA:RNA hybrid level at DSBs
by mediating RNase H2 recruitment. Nat Commun. 9:53762018.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Abakir A, Giles TC, Cristini A, Foster JM,
Dai N, Starczak M, Rubio-Roldan A, Li M, Eleftheriou M, Crutchley
J, et al: N6-methyladenosine regulates the stability of
RNA: DNA hybrids in human cells. Nat Genet. 52:48–55. 2020.
View Article : Google Scholar
|
|
131
|
Wei J, Yin Y, Zhou J, Chen H, Peng J, Yang
J and Tang Y: METTL3 potentiates resistance to cisplatin through
m6A modification of TFAP2C in seminoma. J Cell Mol Med.
24:11366–11380. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Yang Z, Yang S, Cui YH, Wei J, Shah P,
Park G, Cui X, He C and He YY: METTL14 facilitates global genome
repair and suppresses skin tumorigenesis. Proc Natl Acad Sci USA.
118:e20259481182021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Miranda-Gonçalves V, Lobo J,
Guimarães-Teixeira C, Barros-Silva D, Guimarães R, Cantante M,
Braga I, Maurício J, Oing C, Honecker F, et al: The component of
the m6A writer complex VIRMA is implicated in aggressive
tumor phenotype, DNA damage response and cisplatin resistance in
germ cell tumors. J Exp Clin Cancer Res. 40:2682021. View Article : Google Scholar
|
|
134
|
Qu F, Tsegay PS and Liu Y:
N6-methyladenosine, DNA repair, and genome stability.
Front Mol Biosci. 8:6458232021. View Article : Google Scholar
|
|
135
|
Ji HL, Hong J, Zhang Z, de la Peña Avalos
B, Proietti CJ, Deamicis AR, Guzmán GP, Lam HM, Garcia J, Roudier
MP, et al: Regulation of telomere homeostasis and genomic stability
in cancer by N6-adenosine methylation (m6A).
Sci Adv. 7:eabg70732021. View Article : Google Scholar
|
|
136
|
Maciejowski J and de Lange T: Telomeres in
cancer: Tumour suppression and genome instability. Nat Rev Mol Cell
Biol. 18:175–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Batra RN, Lifshitz A, Vidakovic AT, Chin
SF, Sati-Batra A, Sammut SJ, Provenzano E, Ali HR, Dariush A, Bruna
A, et al: DNA methylation landscapes of 1538 breast cancers reveal
a replication-linked clock, epigenomic instability and
cis-regulation. Nat Commun. 12:54062021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Lippert TH, Ruoff HJ and Volm M: Intrinsic
and acquired drug resistance in malignant tumors. The main reason
for therapeutic failure. Arzneimittelforschung. 58:261–264.
2008.PubMed/NCBI
|
|
139
|
Taketo K, Konno M, Asai A, Koseki J,
Toratani M, Satoh T, Doki Y, Mori M, Ishii H and Ogawa K: The
epitranscriptome m6A writer METTL3 promotes chemo- and
radioresistance in pancreatic cancer cells. Int J Oncol.
52:621–629. 2018.PubMed/NCBI
|
|
140
|
Liu X, Gonzalez G, Dai X, Miao W, Yuan J,
Huang M, Bade D, Li L, Sun Y and Wang Y: Adenylate kinase 4
modulates the resistance of breast cancer cells to tamoxifen
through an m6A-based epitranscriptomic mechanism. Mol
Ther. 28:2593–2604. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Petri BJ, Piell KM, South Whitt GC, Wilt
AE and Klinge CM, Lehman NL, Clem BF, Nystoriak MA, Wysoczynski M
and Klinge CM: HNRNPA2B1 regulates tamoxifen- and
fulvestrant-sensitivity and hallmarks of endocrine resistance in
breast cancer cells. Cancer Lett. 518:152–168. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Liu X, Yuan J, Zhang X, Li L, Dai X, Chen
Q and Wang Y: ATF3 modulates the resistance of breast cancer cells
to tamoxifen through an N6-methyladenosine-based
epitranscriptomic mechanism. Chem Res Toxicol. 34:1814–1821. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Pan X, Hong X, Li S, Meng P and Xiao F:
METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells
by accelerating pri-microRNA-221-3p maturation in a m6A-dependent
manner. Exp Mol Med. 53:91–102. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Li E, Xia M, Du Y, Long F, Pan F, He L, Hu
Z and Guo Z: METTL3 promotes homologous recombination repair and
modulates chemotherapeutic response by regulating the EGF/Rad51
axis. bioRxiv. 2021.
|
|
145
|
Li S, Jiang F, Chen F, Deng Y and Pan X:
Effect of m6A methyltransferase METTL3-mediated MALAT1/E2F1/AGR2
axis on adriamycin resistance in breast cancer. J Biochem Mol
Toxicol. 36:e229222022. View Article : Google Scholar
|
|
146
|
Wu Y, Wang Z, Han L, Guo Z, Yan B, Guo L,
Zhao H, Wei M, Hou N, Ye J, et al: PRMT5 regulates RNA m6A
demethylation for doxorubicin sensitivity in breast cancer. Mol
Ther. 30:2603–2617. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Wang Y, Cheng Z, Xu J, Lai M, Liu L, Zuo M
and Dang L: Fat mass and obesity-associated protein (FTO) mediates
signal transducer and activator of transcription 3 (STAT3)-drived
resistance of breast cancer to doxorubicin. Bioengineered.
21:1874–1889. 2021. View Article : Google Scholar
|
|
148
|
Liu X, Li P, Huang Y, Li H, Liu X, Du Y,
Lin X, Chen D, Liu H and Zhou Y: M6A demethylase ALKBH5
regulates FOXO1 mRNA stability and chemoresistance in
triple-negative breast cancer. Redox Biol. 69:1029932024.
View Article : Google Scholar
|
|
149
|
Ou B, Liu Y, Gao Z, Xu J, Yan Y, Li Y and
Zhang J: Senescent neutrophils-derived exosomal piRNA-17560
promotes chemoresistance and EMT of breast cancer via FTO-mediated
m6A demethylation. Cell Death Dis. 13:9052022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zhuang H, Yu B, Tao D, Xu X, Xu Y, Wang J,
Jiao Y and Wang L: The role of m6A methylation in therapy
resistance in cancer. Mol Cancer. 22:912023. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Wang Y, Zhang L, Sun XL, Lu YC, Chen S,
Pei DS and Zhang LS: NRP1 contributes to stemness and potentiates
radioresistance via WTAP-mediated m6A methylation of Bcl-2 mRNA in
breast cancer. Apoptosis. 28:233–246. 2023. View Article : Google Scholar
|