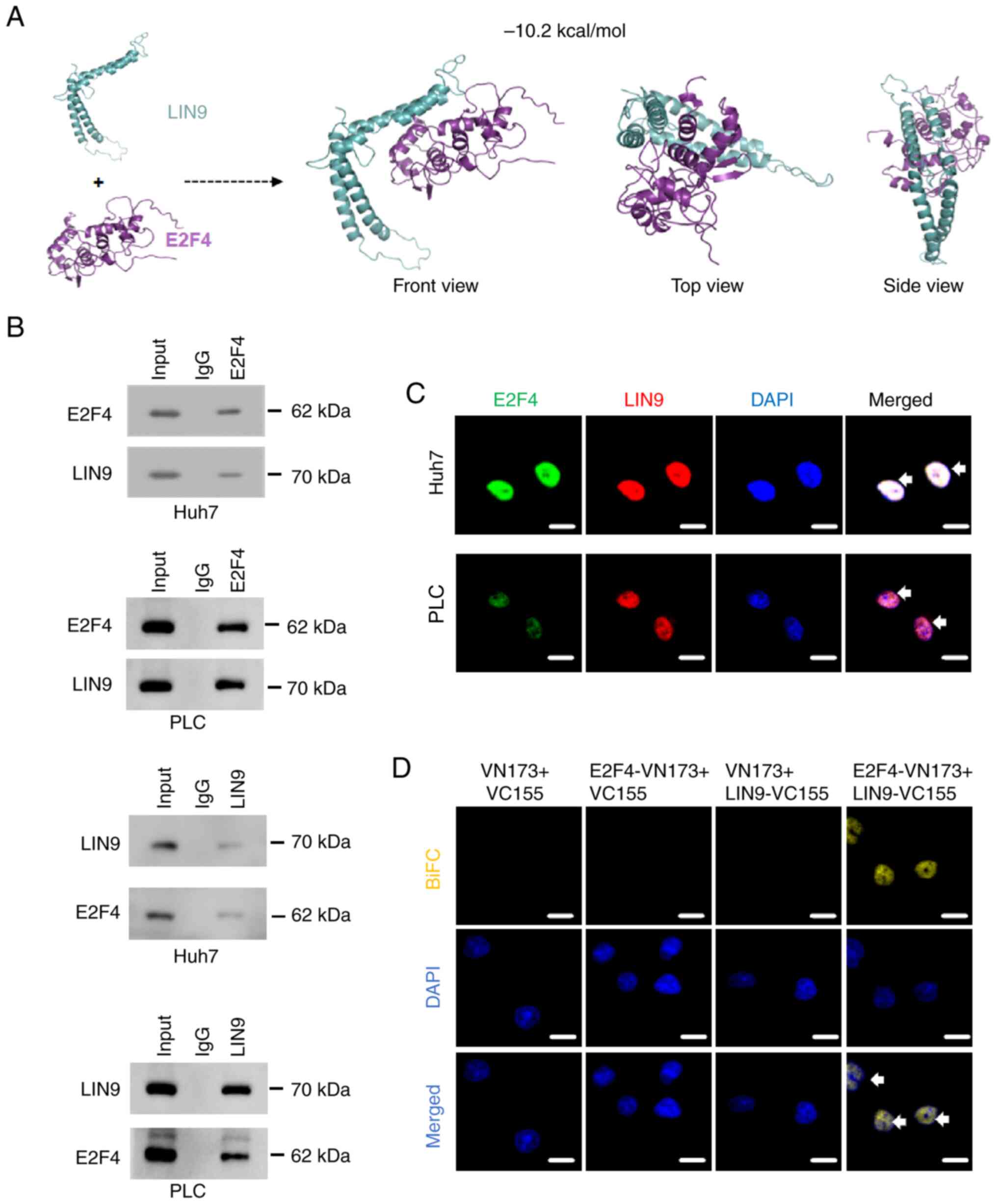
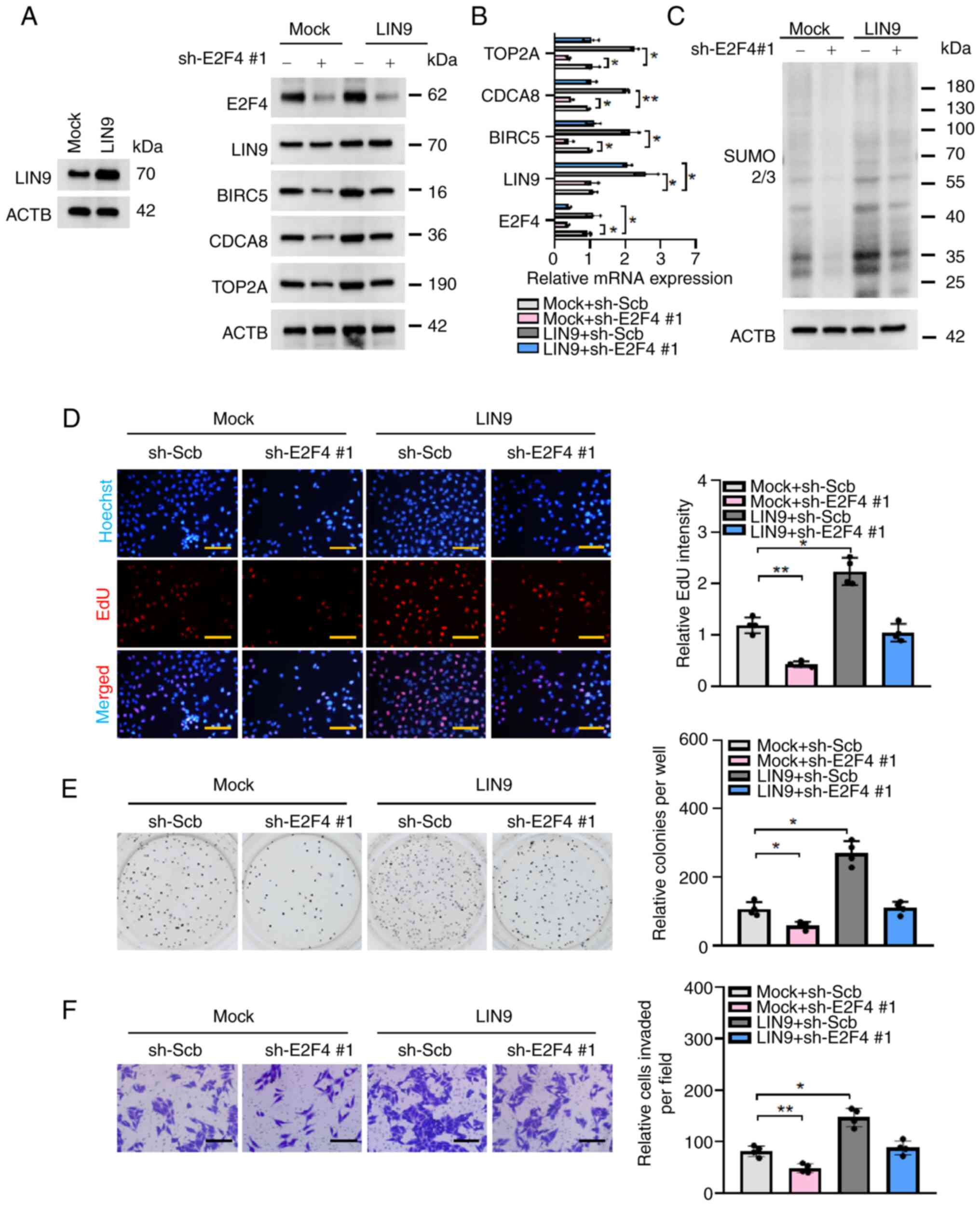
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Facciorusso A: Drug-eluting beads
transarterial chemoembolization for hepatocellular carcinoma:
Current state of the art. World J Gastroenterol. 24:161–169. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB, Marrero JA, Rudolph L and
Reddy KR: Diagnosis and treatment of hepatocellular carcinoma.
Gastroenterology. 134:1752–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shen RR, Zhou AY, Kim E, O'Connell JT,
Hagerstrand D, Beroukhim R and Hahn WC: TRAF2 is an
NF-κB-activating oncogene in epithelial cancers. Oncogene.
34:209–216. 2015. View Article : Google Scholar
|
|
5
|
Sunami Y, Ringelhan M, Kokai E, Lu M,
O'Connor T, Lorentzen A, Weber A, Rodewald AK, Müllhaupt B,
Terracciano L, et al: Canonical NF-κB signaling in hepatocytes acts
as a tumor-suppressor in hepatitis B virus surface antigen-driven
hepatocellular carcinoma by controlling the unfolded protein
response. Hepatology. 63:1592–1607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang X, Yao J, Cui D, Zheng W, Liu Y, Lou
G, Ye B, Shui L, Sun Y, Zhao Y and Zheng M: The TRAF2-p62 axis
promotes proliferation and survival of liver cancer by activating
mTORC1 pathway. Cell Death Differ. 30:1550–1562. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tian Y, Kuo CF, Sir D, Wang L,
Govindarajan S, Petrovic LM and Ou JH: Autophagy inhibits oxidative
stress and tumor suppressors to exert its dual effect on
hepatocarcinogenesis. Cell Death Differ. 22:1025–1034. 2015.
View Article : Google Scholar :
|
|
8
|
Chang HM and Yeh ETHH: SUMO: From bench to
bedside. Physiol Rev. 100:1599–1619. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seeler JS and Dejean A: SUMO and the
robustness of cancer. Nat Rev Cancer. 17:184–197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eifler K and Vertegaal ACO:
SUMOylation-mediated regulation of cell cycle progression and
cancer. Trends Biochem Sci. 40:779–793. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Souza RF, Yin J, Smolinski KN, Zou TT,
Wang S, Shi YQ, Rhyu MG, Cottrell J, Abraham JM, Biden K, et al:
Frequent mutation of the E2F-4 cell cycle gene in primary human
gastrointestinal tumors. Cancer Res. 57:2350–2353. 1997.PubMed/NCBI
|
|
12
|
Wang D, Russell JL and Johnson DG: E2F4
and E2F1 have similar proliferative properties but different
apoptotic and oncogenic properties in vivo. Mol Cell Biol.
20:3417–3424. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schwemmle S and Pfeifer GP: Genomic
structure and mutation screening of the E2F4 gene in human tumors.
Int J Cancer. 86:672–677. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zamani-Ahmadmahmudi M, Najafi A and
Nassiri SM: Reconstruction of canine diffuse large B-cell lymphoma
gene regulatory network: Detection of functional modules and hub
genes. J Comp Pathol. 152:119–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng C, Varn FS and Marsit CJ: E2F4
program is predictive of progression and intravesical immunotherapy
efficacy in bladder cancer. Mol Cancer Res. 13:1316–1324. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Molina-Privado I, Jiménez-P R,
Montes-Moreno S, Chiodo Y, Rodríguez-Martínez M, Sánchez-Verde L,
Iglesias T, Piris MA and Campanero MR: E2F4 plays a key role in
Burkitt lymphoma tumorigenesis. Leukemia. 26:2277–2285. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rakha EA, Pinder SE, Paish EC, Robertson
JF and Ellis IO: Expression of E2F-4 in invasive breast carcinomas
is associated with poor prognosis. J Pathol. 203:754–761. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiao W, Wang J, Wang X, Cai S, Guo Y, Ye
L, Li D, Hu A, Jin S, Yuan B, et al: Therapeutic targeting of the
USP2-E2F4 axis inhibits autophagic machinery essential for zinc
homeostasis in cancer progression. Autophagy. 18:2615–2635. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gong J, Fan H, Deng J and Zhang Q: LncRNA
HAND2-AS1 represses cervical cancer progression by interaction with
transcription factor E2F4 at the promoter of C16orf74. J Cell Mol
Med. 24:6015–6027. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Paquin MC, Leblanc C, Lemieux E, Bian B
and Rivard N: Functional impact of colorectal cancer-associated
mutations in the transcription factor E2F4. Int J Oncol.
43:2015–2022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feng Y, Li L, Du Y, Peng X and Chen F:
E2F4 functions as a tumour suppressor in acute myeloid leukaemia
via inhibition of the MAPK signalling pathway by binding to EZH2. J
Cell Mol Med. 24:2157–2168. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Garneau H, Paquin MC, Carrier JC and
Rivard N: E2F4 expression is required for cell cycle progression of
normal intestinal crypt cells and colorectal cancer cells. J Cell
Physiol. 221:350–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hlady RA, Sathyanarayan A, Thompson JJ,
Zhou D, Wu Q, Pham K, Lee JH, Liu C and Robertson KD: Integrating
the epigenome to identify drivers of hepatocellular carcinoma.
Hepatology. 69:639–652. 2019. View Article : Google Scholar
|
|
24
|
Cancer Genome Atlas Research Network:
Electronic address: simplewheeler@bcm.edu; Cancer
Genome Atlas Research Network: Comprehensive and integrative
genomic characterization of hepatocellular carcinoma. Cell.
169:1327–1341.e23. 2017. View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Jiang G, Zheng L, Pu J, Mei H, Zhao J,
Huang K, Zeng F and Tong Q: Small RNAs targeting transcription
start site induce heparanase silencing through interference with
transcription initiation in human cancer cells. PLoS One.
7:e313792012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fang E, Wang X, Wang J, Hu A, Song H, Yang
F, Li D, Xiao W, Chen Y, Guo Y, et al: Therapeutic targeting of
YY1/MZF1 axis by MZF1-uPEP inhibits aerobic glycolysis and
neuroblastoma progression. Theranostics. 10:1555–1571. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li H, Yang F, Hu A, Wang X, Fang E, Chen
Y, Li D, Song H, Wang J, Guo Y, et al: Therapeutic targeting of
circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma
progression. EMBO Mol Med. 11:e108352019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fang E, Wang X, Yang F, Hu A, Wang J, Li
D, Song H, Hong M, Guo Y, Liu Y, et al: Therapeutic targeting of
MZF1-AS1/PARP1/E2F1 axis inhibits proline synthesis and
neuroblastoma progression. Adv Sci (Weinh). 6:19005812019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47(W1): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Q, Li F, Zhong C, Zou Y, Li Z, Gao Y,
Zou Q, Xia Y, Wang K and Shen F: Inflammation score system using
preoperative inflammatory markers to predict prognosis for
hepatocellular carcinoma after hepatectomy: A cohort study. J
Cancer. 11:4947–4956. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang Y, Sun A, Zhao Y, Ying W, Sun H,
Yang X, Xing B, Sun W, Ren L, Hu B, et al: Proteomics identifies
new therapeutic targets of early-stage hepatocellular carcinoma.
Nature. 567:257–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Clark DJ, Dhanasekaran SM, Petralia F, Pan
J, Song X, Hu Y, da Veiga Leprevost F, Reva B, Lih TSM, Chang HY,
et al: Integrated proteogenomic characterization of clear cell
renal cell carcinoma. Cell. 180:2072020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Du Y, Hou G, Zhang H, Dou J, He J, Guo Y,
Li L, Chen R, Wang Y, Deng R, et al: SUMOylation of the m6A-RNA
methyltransferase METTL3 modulates its function. Nucleic Acids Res.
46:5195–5208. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bogachek MV, Park JM, De Andrade JP,
Lorenzen AW, Kulak MV, White JR, Gu VW, Wu VT and Weigel RJ:
Inhibiting the SUMO pathway represses the cancer stem cell
population in breast and colorectal carcinomas. Stem Cell Reports.
7:1140–1151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
He X, Riceberg J, Soucy T, Koenig E,
Minissale J, Gallery M, Bernard H, Yang X, Liao H, Rabino C, et al:
Probing the roles of SUMOylation in cancer cell biology by using a
selective SAE inhibitor. Nat Chem Biol. 13:1164–1171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hay RT: SUMO: A history of modification.
Mol Cell. 18:1–12. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gareau JR and Lima CD: The SUMO pathway:
Emerging mechanisms that shape specificity, conjugation and
recognition. Nat Rev Mol Cell Biol. 11:861–871. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Geiss-Friedlander R and Melchior F:
Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol.
8:947–956. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu B, Swatkoski S, Holly A, Lee LC, Giroux
V, Lee CS, Hsu D, Smith JL, Yuen G, Yue J, et al: Oncogenesis
driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9.
Proc Natl Acad Sci USA. 112:E1724–E1733. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang J, Tan GL, Jiang M, Wang TS, Liu GH,
Xiong SS and Qing X: Effects of SENP1-induced deSUMOylation of
STAT1 on proliferation and invasion in nasopharyngeal carcinoma.
Cell Signal. 101:1105302023. View Article : Google Scholar
|
|
44
|
Wang Z, Pan B, Su L, Yu H, Wu X, Yao Y,
Zhang X, Qiu J and Tang N: SUMOylation inhibitors activate
anti-tumor immunity by reshaping the immune microenvironment in a
preclinical model of hepatocellular carcinoma. Cell Oncol (Dordr).
47:513–532. 2024. View Article : Google Scholar
|
|
45
|
Wang B, Li X, Zhao G, Yan H, Dong P,
Watari H, Sims M, Li W, Pfeffer LM, Guo Y and Yue J: miR-203
inhibits ovarian tumor metastasis by targeting BIRC5 and
attenuating the TGFβ pathway. J Exp Clin Cancer Res. 37:2352018.
View Article : Google Scholar
|
|
46
|
Kelly RJ, Lopez-Chavez A, Citrin D, Janik
JE and Morris JC: Impacting tumor cell-fate by targeting the
inhibitor of apoptosis protein survivin. Mol Cancer. 10:352011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu R, Lin L, Zhang B, Wang J, Zhao F, Liu
X and Li Y and Li Y: Identification of prognostic markers for
hepatocellular carcinoma based on the epithelial-mesenchymal
transition-related gene BIRC5. BMC Cancer. 21:6872021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tian LL, Qian B, Jiang XH, Liu YS, Chen T,
Jia CY, Zhou YL, Liu JB, Ma YS, Fu D and Ding ST: MicroRNA-497-5p
is downregulated in hepatocellular carcinoma and associated with
tumorigenesis and poor prognosis in patients. Int J Genomics.
2021:66703902021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang M, Yan X, Wen P, Bai W and Zhang Q:
CircANKRD52 promotes the tumorigenesis of hepatocellular carcinoma
by sponging miR-497-5p and upregulating BIRC5 expression. Cell
Transplant. 30:96368972110088742021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang C, Zhao L, Leng L, Zhou Q, Zhang S,
Gong F, Xie P and Lin G: CDCA8 regulates meiotic spindle assembly
and chromosome segregation during human oocyte meiosis. Gene.
741:1444952020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yamanaka Y, Heike T, Kumada T, Shibata M,
Takaoka Y, Kitano A, Shiraishi K, Kato T, Nagato M, Okawa K, et al:
Loss of Borealin/DasraB leads to defective cell proliferation, p53
accumulation and early embryonic lethality. Mech Dev. 125:441–450.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cui Y and Jiang N: CDCA8 facilitates tumor
proliferation and predicts a poor prognosis in hepatocellular
carcinoma. Appl Biochem Biotechnol. 196:1481–1492. 2024. View Article : Google Scholar
|
|
53
|
Nielsen CF, Zhang T, Barisic M, Kalitsis P
and Hudson DF: Topoisomerase IIα is essential for maintenance of
mitotic chromosome structure. Proc Natl Acad Sci USA.
117:12131–12142. 2020. View Article : Google Scholar
|
|
54
|
Zhong W, Yang Y, Zhang A, Lin W, Liang G,
Ling Y, Zhong J, Yong J, Liu Z, Tian Z, et al: Prognostic and
predictive value of the combination of TOP2A and HER2 in
node-negative tumors 2 cm or smaller (T1N0) breast cancer. Breast
Cancer. 27:1147–1157. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shen S, Kong J, Qiu Y, Yang X, Wang W and
Yan L: Identification of core genes and outcomes in hepatocellular
carcinoma by bioinformatics analysis. J Cell Biochem.
120:10069–10081. 2019. View Article : Google Scholar
|
|
56
|
Gao X, Wang X and Zhang S: Bioinformatics
identification of crucial genes and pathways associated with
hepatocellular carcinoma. Biosci Rep. 38:BSR201814412018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Meng J, Wei Y, Deng Q, Li L and Li X:
Study on the expression of TOP2A in hepatocellular carcinoma and
its relationship with patient prognosis. Cancer Cell Int.
22:292022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang K, Jiang X, Jiang Y, Liu J, Du Y,
Zhang Z, Li Y, Zhao X, Li J and Zhang R: EZH2-H3K27me3-mediated
silencing of mir-139-5p inhibits cellular senescence in
hepatocellular carcinoma by activating TOP2A. J Exp Clin Cancer
Res. 42:3202023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhao HC, Chen CZ, Tian YZ, Song HQ, Wang
XX, Li YJ, He JF and Zhao HL: CD168+ macrophages promote
hepatocellular carcinoma tumor stemness and progression through
TOP2A/β-catenin/YAP1 axis. iScience. 26:1068622023. View Article : Google Scholar
|
|
60
|
Yang J, Song K, Krebs TL, Jackson MW and
Danielpour D: Rb/E2F4 and Smad2/3 link survivin to TGF-beta-induced
apoptosis and tumor progression. Oncogene. 27:5326–5338. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zwicker J, Lucibello FC, Wolfraim LA,
Gross C, Truss M, Engeland K and Müller R: Cell cycle regulation of
the cyclin A, cdc25C and cdc2 genes is based on a common mechanism
of transcriptional repression. EMBO J. 14:4514–4522. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ikeda MA, Jakoi L and Nevins JR: A unique
role for the Rb protein in controlling E2F accumulation during cell
growth and differentiation. Proc Natl Acad Sci USA. 93:3215–3220.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
van der Sman J, Thomas NS and Lam EW:
Modulation of E2F complexes during G0 to S phase transition in
human primary B-lymphocytes. J Biol Chem. 274:12009–12016. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Aksoy O, Chicas A, Zeng T, Zhao Z,
McCurrach M, Wang X and Lowe SW: The atypical E2F family member
E2F7 couples the p53 and RB pathways during cellular senescence.
Genes Dev. 26:1546–1557. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zheng Q, Fu Q, Xu J, Gu X, Zhou H and Zhi
C: Transcription factor E2F4 is an indicator of poor prognosis and
is related to immune infiltration in hepatocellular carcinoma. J
Cancer. 12:1792–1803. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Huang YL, Ning G, Chen LB, Lian YF, Gu YR,
Wang JL, Chen DM, Wei H and Huang YH: Promising diagnostic and
prognostic value of E2Fs in human hepatocellular carcinoma. Cancer
Manag Res. 11:1725–1740. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu J, Xia L, Wang S, Cai X, Wu X, Zou C,
Shan B, Luo M and Wang D: E2F4 promotes the proliferation of
hepatocellular carcinoma cells through upregulation of CDCA3. J
Cancer. 12:5173–5180. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wei J, Shi Y, Zou C, Zhang H, Peng H, Wang
S, Xia L, Yang Y, Zhang X, Liu J, et al: Cellular Id1 inhibits
hepatitis B virus transcription by interacting with the novel
covalently closed circular DNA-binding protein E2F4. Int J Biol
Sci. 18:65–81. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Korenjak M and Brehm A: E2F-Rb complexes
regulating transcription of genes important for differentiation and
development. Curr Opin Genet Dev. 15:520–527. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lewis PW, Beall EL, Fleischer TC,
Georlette D, Link AJ and Botchan MR: Identification of a Drosophila
Myb-E2F2/RBF transcriptional repressor complex. Genes Dev.
18:2929–2940. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Korenjak M, Taylor-Harding B, Binné UK,
Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N and
Brehm A: Native E2F/RBF complexes contain Myb-interacting proteins
and repress transcription of developmentally controlled E2F target
genes. Cell. 119:181–193. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Litovchick L, Sadasivam S, Florens L, Zhu
X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS and
DeCaprio JA: Evolutionarily conserved multisubunit RBL2/p130 and
E2F4 protein complex represses human cell cycle-dependent genes in
quiescence. Mol Cell. 26:539–551. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sadasivam S, Duan S and DeCaprio JA: The
MuvB complex sequentially recruits B-Myb and FoxM1 to promote
mitotic gene expression. Genes Dev. 26:474–489. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fischer M, Grossmann P, Padi M and
DeCaprio JA: Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F
target gene analyses identifies cell cycle gene regulatory
networks. Nucleic Acids Res. 44:6070–6086. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sadasivam S and DeCaprio JA: The DREAM
complex: Master coordinator of cell cycle-dependent gene
expression. Nat Rev Cancer. 13:585–595. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
MacDonald J, Ramos-Valdes Y, Perampalam P,
Litovchick L, DiMattia GE and Dick FA: A systematic analysis of
negative growth control implicates the DREAM complex in cancer cell
dormancy. Mol Cancer Res. 15:371–381. 2017. View Article : Google Scholar
|
|
77
|
Wang L and Liu X: Comprehensive analysis
of the expression and prognosis for the DREAM complex in human
cancers. Front Genet. 13:8147252022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tan Z, Chen M, Peng F, Yang P, Peng Z,
Zhang Z, Li X, Zhu X, Zhang L, Zhao Y and Liu Y: E2F1 as a
potential prognostic and therapeutic biomarker by affecting tumor
development and immune microenvironment in hepatocellular
carcinoma. Transl Cancer Res. 11:2713–2732. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ito Y, Miyoshi E, Takeda T, Sakon M, Noda
K, Tsujimoto M, Monden M, Taniguchi N and Matsuura N: Expression
and possible role of ets-1 in hepatocellular carcinoma. Am J Clin
Pathol. 114:719–725. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yan Y, Zheng L, Du Q, Yan B and Geller DA:
Interferon regulatory factor 1 (IRF-1) and IRF-2 regulate PD-L1
expression in hepatocellular carcinoma (HCC) cells. Cancer Immunol
Immunother. 69:1891–1903. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Qu LH, Fang Q, Yin T, Yi HM, Mei GB, Hong
ZZ, Qiu XB, Zhou R and Dong HF: Comprehensive analyses of
prognostic biomarkers and immune infiltrates among histone lysine
demethylases (KDMs) in hepatocellular carcinoma. Cancer Immunol
Immunother. 71:2449–2467. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Nakajima T, Yasui K, Zen K, Inagaki Y,
Fujii H, Minami M, Tanaka S, Taniwaki M, Itoh Y, Arii S, et al:
Activation of B-Myb by E2F1 in hepatocellular carcinoma. Hepatol
Res. 38:886–895. 2008. View Article : Google Scholar
|
|
83
|
Chen Q, Wang L, Jiang M, Huang J, Jiang Z,
Feng H and Ji Z: E2F1 interactive with BRCA1 pathway induces HCC
two different small molecule metabolism or cell cycle regulation
via mitochondrion or CD4+T to cytosol. J Cell Physiol.
233:1213–1221. 2018. View Article : Google Scholar
|
|
84
|
Arakawa Y, Kajino K, Kano S, Tobita H,
Hayashi J, Yasen M, Moriyama M, Arakawa Y and Hino O: Transcription
of dbpA, a Y box binding protein, is positively regulated by E2F1:
Implications in hepatocarcinogenesis. Biochem Biophys Res Commun.
322:297–302. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen YL, Uen YH, Li CF, Horng KC, Chen LR,
Wu WR, Tseng HY, Huang HY, Wu LC and Shiue YL: The E2F
transcription factor 1 transactives stathmin 1 in hepatocellular
carcinoma. Ann Surg Oncol. 20:4041–4054. 2013. View Article : Google Scholar
|
|
86
|
Huang Y, Tai AW, Tong S and Lok AS: HBV
core promoter mutations promote cellular proliferation through
E2F1-mediated upregulation of S-phase kinase-associated protein 2
transcription. J Hepatol. 58:1068–1073. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Farra R, Grassi G, Tonon F, Abrami M,
Grassi M, Pozzato G, Fiotti N, Forte G and Dapas B: The role of the
transcription factor E2F1 in hepatocellular carcinoma. Curr Drug
Deliv. 14:272–281. 2017.
|
|
88
|
Sun HX, Xu Y, Yang XR, Wang WM, Bai H, Shi
RY, Nayar SK, Devbhandari RP, He YZ, Zhu QF, et al: Hypoxia
inducible factor 2 alpha inhibits hepatocellular carcinoma growth
through the transcription factor dimerization partner 3/E2F
transcription factor 1-dependent apoptotic pathway. Hepatology.
57:1088–1097. 2013. View Article : Google Scholar
|
|
89
|
Choi M, Lee H and Rho HM: E2F1 activates
the human p53 promoter and overcomes the repressive effect of
hepatitis B viral X protein (Hbx) on the p53 promoter. IUBMB Life.
53:309–317. 2002. View Article : Google Scholar
|
|
90
|
Wang H, Chu F, Zhijie L, Bi Q, Lixin L,
Zhuang Y, Xiaofeng Z, Niu X, Zhang D, Xi H and Li BA: MTBP enhances
the activation of transcription factor ETS-1 and promotes the
proliferation of hepatocellular carcinoma cells. Front Oncol.
12:9850822022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Bhagyaraj E, Ahuja N, Kumar S, Tiwari D,
Gupta S, Nanduri R and Gupta P: TGF-β induced chemoresistance in
liver cancer is modulated by xenobiotic nuclear receptor PXR. Cell
Cycle. 18:3589–3602. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Shao Z, Li Y, Dai W, Jia H, Zhang Y, Jiang
Q, Chai Y, Li X, Sun H, Yang R, et al: ETS-1 induces
Sorafenib-resistance in hepatocellular carcinoma cells via
regulating transcription factor activity of PXR. Pharmacol Res.
135:188–200. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Li YH, Lv MF, Lu MS and Bi JP: Bone marrow
mesenchymal stem cell-derived exosomal MiR-338-3p represses
progression of hepatocellular carcinoma by targeting ETS1. J Biol
Regul Homeost Agents. 35:617–627. 2021.PubMed/NCBI
|
|
94
|
Jie Y, Liu G, E M, Li Y, Xu G, Guo J, Li
Y, Rong G, Li Y and Gu A: Novel small molecule inhibitors of the
transcription factor ETS-1 and their antitumor activity against
hepatocellular carcinoma. Eur J Pharmacol. 906:1742142021.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tamura T, Yanai H, Savitsky D and
Taniguchi T: The IRF family transcription factors in immunity and
oncogenesis. Annu Rev Immunol. 26:535–584. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li P, Du Q, Cao Z, Guo Z, Evankovich J,
Yan W, Chang Y, Shao L, Stolz DB, Tsung A and Geller DA:
Interferon-γ induces autophagy with growth inhibition and cell
death in human hepatocellular carcinoma (HCC) cells through
interferon-regulatory factor-1 (IRF-1). Cancer Lett. 314:213–222.
2012. View Article : Google Scholar
|
|
97
|
Guo W, Li S, Qian Y, Li L, Wang F, Tong Y,
Li Q, Zhu Z, Gao WQ and Liu Y: KDM6A promotes hepatocellular
carcinoma progression and dictates lenvatinib efficacy by
upregulating FGFR4 expression. Clin Transl Med. 13:e14522023.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Li Y, Yang J, Zhang X, Liu H and Guo J:
KDM6A suppresses hepatocellular carcinoma cell proliferation by
negatively regulating the TGF-β/SMAD signaling pathway. Exp Ther
Med. 20:2774–2782. 2020.PubMed/NCBI
|