|
1
|
Ransohoff JD, Wei Y and Khavari PA: The
functions and unique features of long intergenic non-coding RNA.
Nat Rev Mol Cell Biol. 19:143–157. 2018. View Article : Google Scholar :
|
|
2
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang D, Garcia-Bassets I, Benner C, Li W,
Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al:
Reprogramming transcription by distinct classes of enhancers
functionally defined by eRNA. Nature. 474:390–394. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou Y, Sun W, Qin Z, Guo S, Kang Y, Zeng
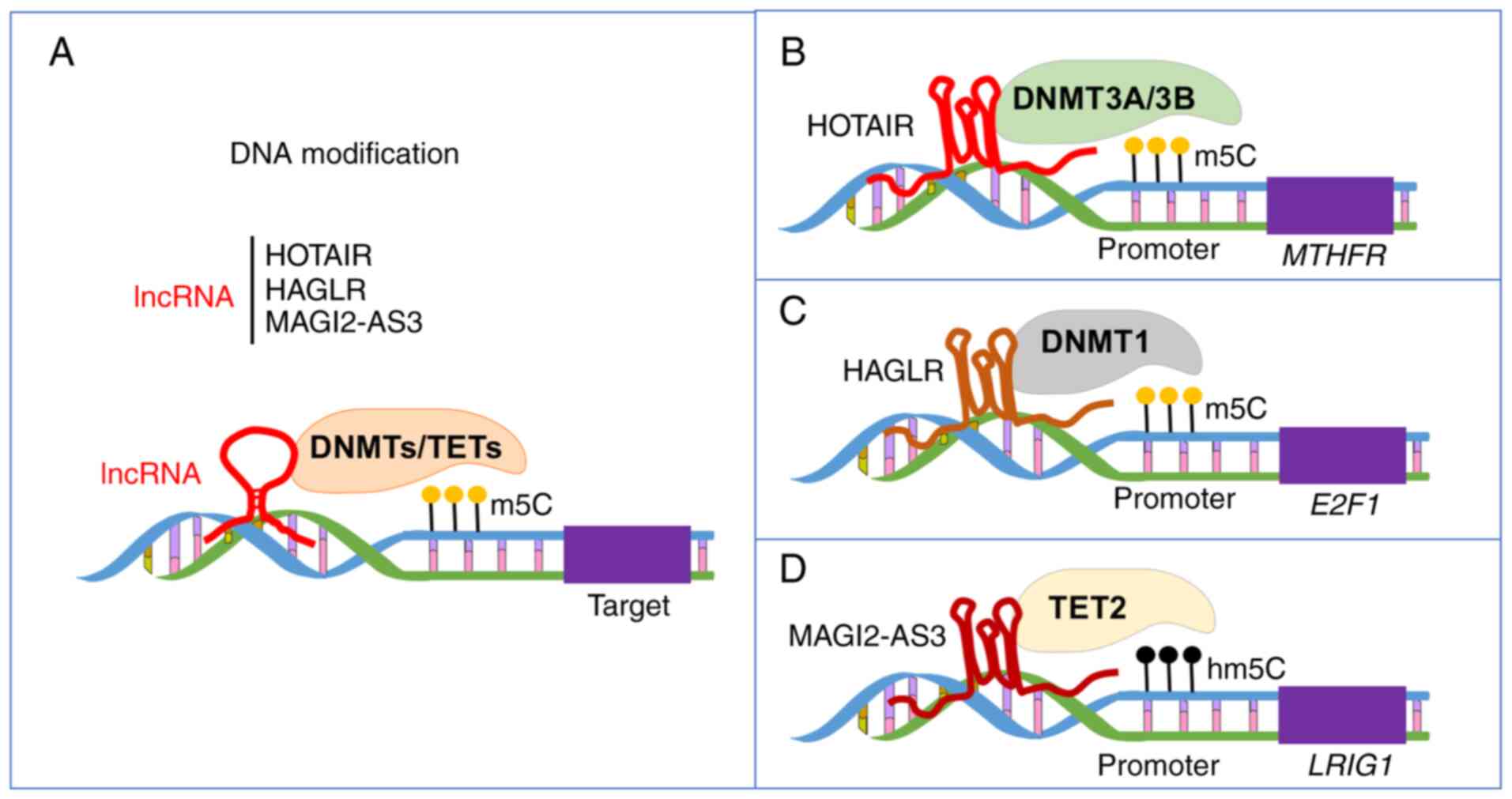
S and Yu L: LncRNA regulation: New frontiers in epigenetic
solutions to drug chemoresistance. Biochem Pharmacol.
189:1142282021. View Article : Google Scholar
|
|
5
|
Morlando M and Fatica A: Alteration of
epigenetic regulation by long noncoding RNAs in cancer. Int J Mol
Sci. 19:5702018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Khorkova O, Hsiao J and Wahlestedt C:
Basic biology and therapeutic implications of lncRNA. Adv Drug
Deliv Rev. 87:15–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Beckedorff FC, Amaral MS,
Deocesano-Pereira C and Verjovski-Almeida S: Long non-coding RNAs
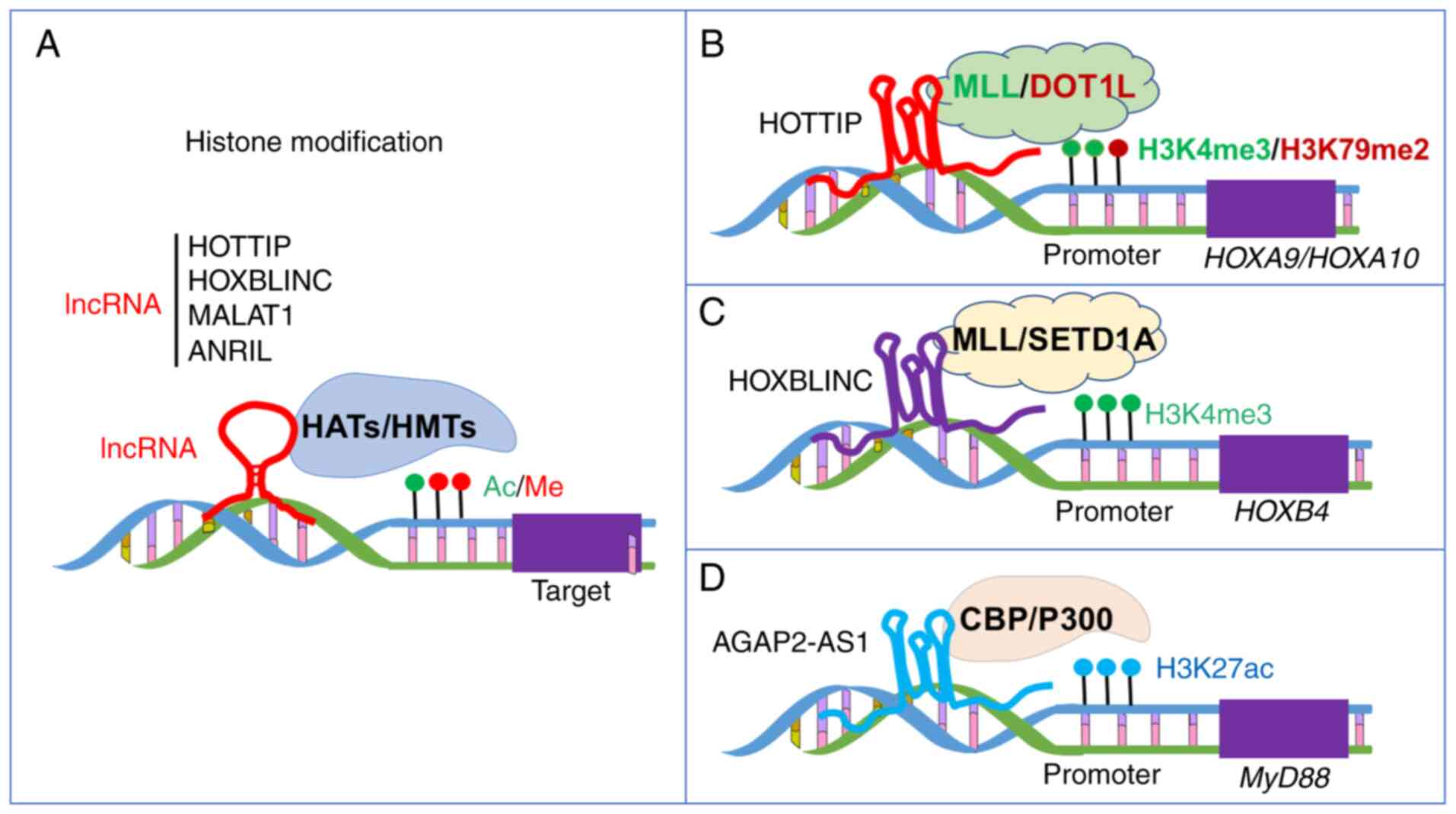
and their implications in cancer epigenetics. Biosci Rep.
33:e000612013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu Y, Chan YT, Tan HY, Li S, Wang N and
Feng Y: Epigenetic regulation in human cancer: The potential role
of epi-drug in cancer therapy. Mol Cancer. 19:792020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tachiwana H, Yamamoto T and Saitoh N: Gene
regulation by non-coding RNAs in the 3D genome architecture. Curr
Opin Genet Dev. 61:69–74. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Phillips JE and Corces VG: CTCF: Master
weaver of the genome. Cell. 137:1194–1211. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang C, Rao S, Crispino JD and
Ntziachristos P: Determinants and role of chromatin organization in
acute leukemia. Leukemia. 34:2561–2575. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hansen AS, Hsieh TS, Cattoglio C, Pustova
I, Saldaña-Meyer R, Reinberg D, Darzacq X and Tjian R: Distinct
classes of chromatin loops revealed by deletion of an RNA-binding
region in CTCF. Mol Cell. 76:395–411.e13. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
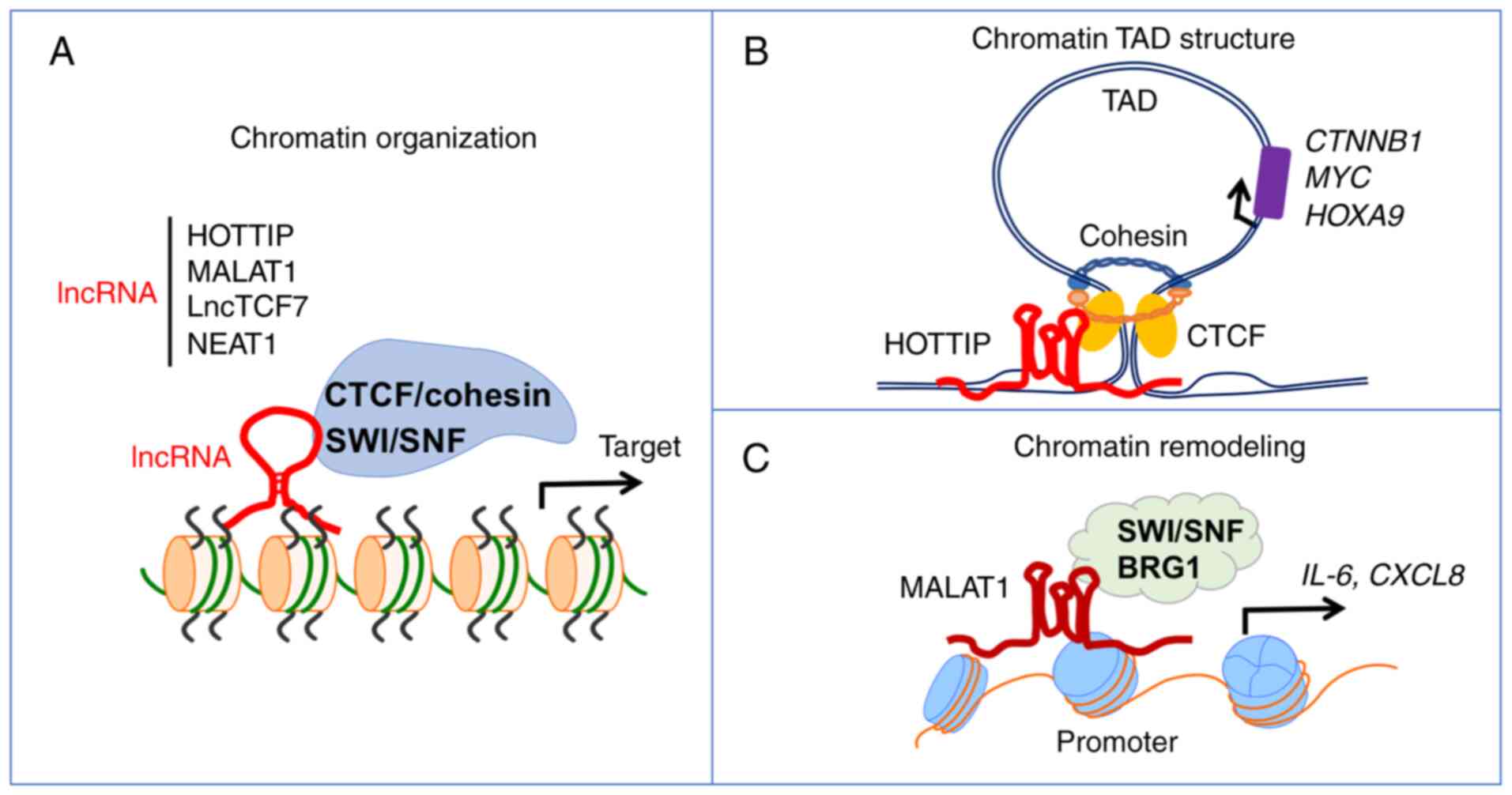
Saldana-Meyer R, Rodriguez-Hernaez J,
Escobar T, Nishana M, Jácome-López K, Nora EP, Bruneau BG, Tsirigos
A, Furlan-Magaril M, Skok J and Reinberg D: RNA interactions are
essential for CTCF-mediated genome organization. Mol Cell.
76:412–422. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui L, Ma R, Cai J, Guo C, Chen Z, Yao L,
Wang Y, Fan R, Wang X and Shi Y: RNA modifications: Importance in
immune cell biology and related diseases. Signal Transduct Target
Ther. 7:3342022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dinescu S, Ignat S, Lazar AD, Constantin
C, Neagu M and Costache M: Epitranscriptomic signatures in lncRNAs
and their possible roles in cancer. Genes (Basel). 10:522019.
View Article : Google Scholar : PubMed/NCBI
|
|
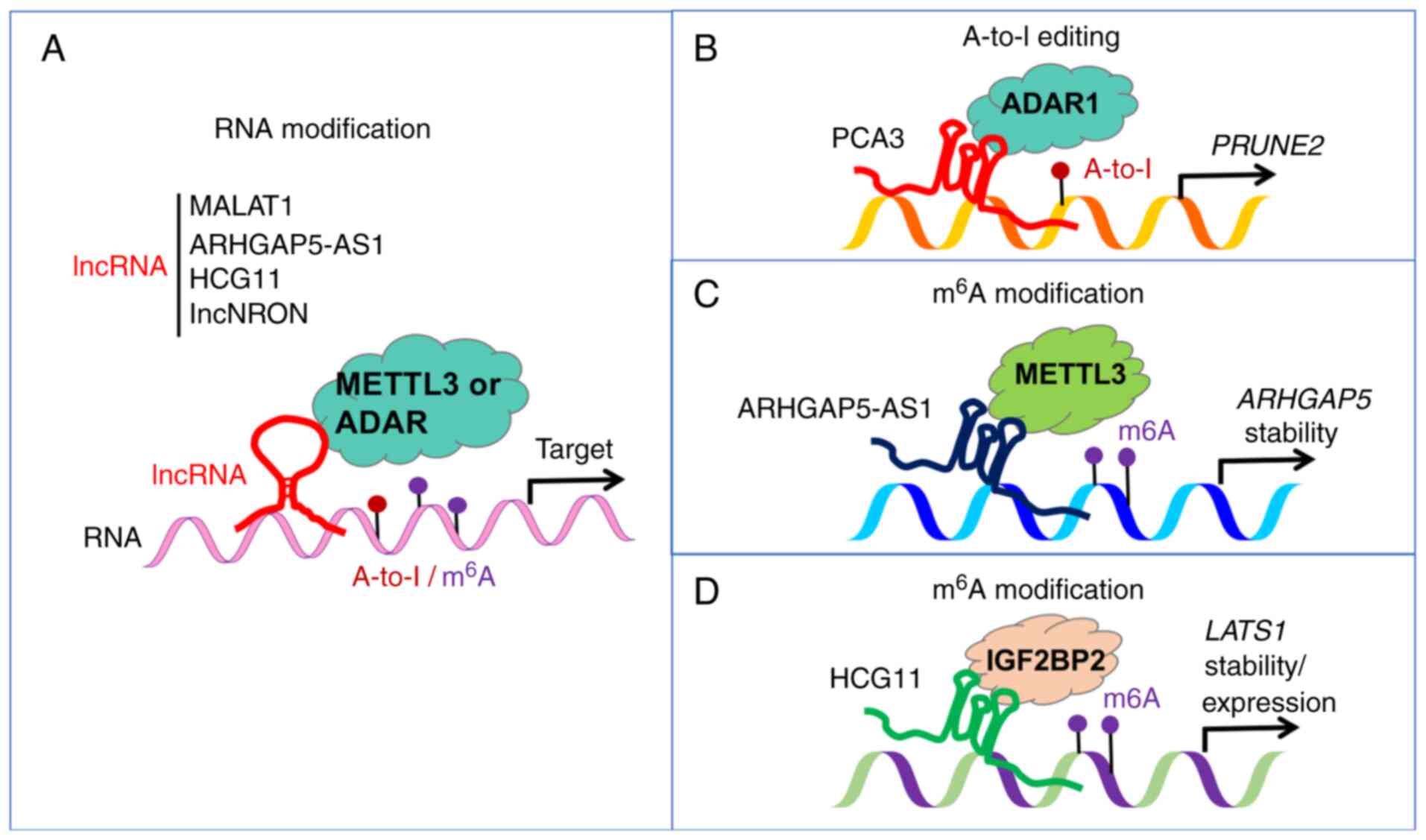
16
|
Yang Z, Xu F, Teschendorff AE, Zhao Y, Yao
L, Li J and He Y: Insights into the role of long non-coding RNAs in
DNA methylation mediated transcriptional regulation. Front Mol
Biosci. 9:10674062022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar
|
|
18
|
Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y,
Fung TK, Zeisig BB, Cui Y, Zha J, et al: HOTTIP lncRNA promotes
hematopoietic stem cell Self-renewal leading to AML-like disease in
mice. Cancer Cell. 36:645–659.e8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Greenberg MVC and Bourc'his D: The diverse
roles of DNA methylation in mammalian development and disease. Nat
Rev Mol Cell Biol. 20:590–607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao SG, Chen WS, Li H, Foye A, Zhang M,
Sjöström M, Aggarwal R, Playdle D, Liao A, Alumkal JJ, et al: The
DNA methylation landscape of advanced prostate cancer. Nat Genet.
52:778–789. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robertson KD: DNA methylation,
methyltransferases, and cancer. Oncogene. 20:3139–3155. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
van der Velden PA, Metzelaar-Blok JA,
Bergman W, Monique H, Hurks H, Frants RR, Gruis NA and Jager MJ:
Promoter hypermethylation: A common cause of reduced p16(INK4a)
expression in uveal melanoma. Cancer Res. 61:5303–5306.
2001.PubMed/NCBI
|
|
23
|
Saif I, Bouziyane A, Benhessou M, Karroumi
ME and Ennaji MM: Detection of hypermethylation BRCA1/2 gene
promoter in breast tumours among Moroccan women. Mol Biol Rep.
48:7147–7152. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang
Y, Jia L, Li S; Cancer Genome Atlas Research Network; Xie W and
Yang D: lncRNA epigenetic landscape analysis identifies EPIC1 as an
oncogenic lncRNA that interacts with MYC and promotes Cell-cycle
progression in cancer. Cancer Cell. 33:706–720.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lyko F: The DNA methyltransferase family:
A versatile toolkit for epigenetic regulation. Nat Rev Genet.
19:81–92. 2018. View Article : Google Scholar
|
|
26
|
Liu D, Wu K, Yang Y, Zhu D, Zhang C and
Zhao S: Long noncoding RNA ADAMTS9-AS2 suppresses the progression
of esophageal cancer by mediating CDH3 promoter methylation. Mol
Carcinog. 59:32–44. 2020. View Article : Google Scholar
|
|
27
|
Zhang S, Zheng F, Zhang L, Huang Z, Huang
X, Pan Z, Chen S, Xu C, Jiang Y, Gu S, et al: LncRNA
HOTAIR-mediated MTHFR methylation inhibits 5-fluorouracil
sensitivity in esophageal cancer cells. J Exp Clin Cancer Res.
39:1312020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang L, Bu P, Ai Y, Srinivasan T, Chen HJ,
Xiang K, Lipkin SM and Shen X: A long non-coding RNA targets
microRNA miR-34a to regulate colon cancer stem cell asymmetric
division. Elife. 5:e146202016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lai IL, Chang YS, Chan WL Lee YT, Yen JC,
Yang CA, Hung SY and Chang JG: Male-specific long noncoding RNA
TTTY15 inhibits Non-small cell lung cancer proliferation and
metastasis via TBX4. Int J Mol Sci. 20:34732019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Q, Su Z, Xu X, Liu G, Song X, Wang R,
Sui X, Liu T, Chang X, Huang D, et al: AS1DHRS4, a head-to-head
natural antisense transcript, silences the DHRS4 gene cluster in
cis and trans. Proc Natl Acad Sci USA. 109:14110–14115. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang
CY, Tang CH, Lee YC and Yang SF: A novel melatonin-regulated lncRNA
suppresses TPA-induced oral cancer cell motility through
replenishing PRUNE2 expression. J Pineal Res. 71:e127602021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Luo Q, Li Z, Wang Y, Zhu C, Li T and
Li X: Long Non-coding RNA IRAIN inhibits VEGFA expression via
enhancing Its DNA methylation leading to tumor suppression in renal
carcinoma. Front Oncol. 10:10822020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo X, Chen Z, Zhao L, Cheng D, Song W and
Zhang X: Long non-coding RNA-HAGLR suppressed tumor growth of lung
adenocarcinoma through epigenetically silencing E2F1. Exp Cell Res.
382:1114612019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yoon JH, You BH, Park CH, Kim YJ, Nam JW
and Lee SK: The long noncoding RNA LUCAT1 promotes tumorigenesis by
controlling ubiquitination and stability of DNA methyltransferase 1
in esophageal squamous cell carcinoma. Cancer Lett. 417:47–57.
2018. View Article : Google Scholar
|
|
35
|
Jones R, Wijesinghe S, Wilson C, Halsall
J, Liloglou T and Kanhere A: A long intergenic non-coding RNA
regulates nuclear localization of DNA methyl transferase-1.
iScience. 24:1022732021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu X, Lou Y, Tang J, Teng Y, Zhang Z, Yin
Y, Zhuo H and Tan Z: The long non-coding RNA Linc-GALH promotes
hepatocellular carcinoma metastasis via epigenetically regulating
Gankyrin. Cell Death Dis. 10:862019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vennin C, Spruyt N, Robin YM, Chassat T,
Le Bourhis X and Adriaenssens E: The long non-coding RNA 91H
increases aggressive phenotype of breast cancer cells and
up-regulates H19/IGF2 expression through epigenetic modifications.
Cancer Lett. 385:198–206. 2017. View Article : Google Scholar
|
|
38
|
Jin L, Cai Q, Wang S, Wang S, Wang J and
Quan Z: Long noncoding RNA PVT1 promoted gallbladder cancer
proliferation by epigenetically suppressing miR-18b-5p via DNA
methylation. Cell Death Dis. 11:8712020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu X and Zhang Y: TET-mediated active DNA
demethylation: Mechanism, function and beyond. Nat Rev Genet.
18:517–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rasmussen KD and Helin K: Role of TET
enzymes in DNA methylation, development, and cancer. Genes Dev.
30:733–750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Al-Imam MJ, Hussein UA, Sead FF, Faqri
AMA, Mekkey SM, Khazel AJ and Almashhadani HA: The interactions
between DNA methylation machinery and long non-coding RNAs in tumor
progression and drug resistance. DNA Repair (Amst). 128:1035262023.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou L, Ren M, Zeng T, Wang W, Wang X, Hu
M, Su S, Sun K, Wang C, Liu J, et al: TET2-interacting long
noncoding RNA promotes active DNA demethylation of the MMP-9
promoter in diabetic wound healing. Cell Death Dis. 10:8132019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Roessner A, Franke S, Schreier J, Ullmann
S, Karras F and Jechorek D: Genetics and epigenetics in
conventional chondrosarcoma with focus on non-coding RNAs. Pathol
Res Pract. 239:1541722022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu X, Du J, Yu J, Guo R, Feng Y, Qiao L,
Xu Z, Yang F, Zhong G, Liu F, et al: LncRNA NKILA regulates
endothelium inflammation by controlling a NF-κB/KLF4 positive
feedback loop. J Mol Cell Cardiol. 126:60–69. 2019. View Article : Google Scholar
|
|
45
|
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X,
Lin L, Yao H, Su F, Li D, et al: A cytoplasmic NF-kappaB
interacting long noncoding RNA blocks IkappaB phosphorylation and
suppresses breast cancer metastasis. Cancer Cell. 27:370–381. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen L, Fan X, Zhu J, Chen X, Liu Y and
Zhou H: LncRNA MAGI2-AS3 inhibits the self-renewal of leukaemic
stem cells by promoting TET2-dependent DNA demethylation of the
LRIG1 promoter in acute myeloid leukaemia. RNA Biol. 17:784–793.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nie Y, Zhou L, Wang H, Chen N, Jia L, Wang
C, Wang Y, Chen J, Wen X, Niu C, et al: Profiling the epigenetic
interplay of lncRNA RUNXOR and oncogenic RUNX1 in breast cancer
cells by gene in situ cis-activation. Am J Cancer Res. 9:1635–1649.
2019.PubMed/NCBI
|
|
48
|
Elsheikh SE, Green AR, Rakha EA, Powe DG,
Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, et
al: Global histone modifications in breast cancer correlate with
tumor phenotypes, prognostic factors, and patient outcome. Cancer
Res. 69:3802–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Deng C, Li Y, Zhou L, Cho J, Patel B,
Terada N, Li Y, Bungert J, Qiu Y, Huang S, et al: HoxBlinc RNA
recruits Set1/MLL complexes to activate hox gene expression
patterns and mesoderm lineage development. Cell Rep. 14:103–114.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nagano T, Mitchell JA, Sanz LA, Pauler FM,
Ferguson-Smith AC, Feil R and Fraser P: The Air noncoding RNA
epigenetically silences transcription by targeting G9a to
chromatin. Science. 322:1717–1720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen
J, Guo R, Si J, Li L, Xue J, et al: LINC02273 drives breast cancer
metastasis by epigenetically increasing AGR2 transcription. Mol
Cancer. 18:1872019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hu A, Hong F, Li D, Jin Y, Kon L, Xu Z, He
H and Xie Q: Long non-coding RNA ROR recruits histone
transmethylase MLL1 to up-regulate TIMP3 expression and promote
breast cancer progression. J Transl Med. 19:952021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lai IL, Yang CA, Lin PC, Chan WL, Lee YT,
Yen JC, Chang YS and Chang JG: Long noncoding RNA MIAT promotes
non-small cell lung cancer proliferation and metastasis through
MMP9 activation. Oncotarget. 8:98148–98162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang WT, Chen TQ, Zeng ZC, Pan Q, Huang W,
Han C, Fang K, Sun LY, Yang QQ, Wang D, et al: The lncRNA LAMP5-AS1
drives leukemia cell stemness by directly modulating DOT1L
methyltransferase activity in MLL leukemia. J Hematol Oncol.
13:782020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chu W, Zhang X, Qi L, Fu Y, Wang P, Zhao
W, Du J, Zhang J, Zhan J, Wang Y, et al: The
EZH2-PHACTR2-AS1-Ribosome axis induces genomic instability and
promotes growth and metastasis in breast cancer. Cancer Res.
80:2737–2750. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wanowska E, Samorowska K and Szczesniak
MW: Emerging roles of long Noncoding RNAs in breast cancer
epigenetics and epitranscriptomics. Front Cell Dev Biol.
10:9223512022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pandey RR, Mondal T, Mohammad F, Enroth S,
Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D and Kanduri C:
Antisense Noncoding RNA mediates lineage-specific transcriptional
silencing through Chromatin-Level regulation. Mol Cell. 32:232–246.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar
|
|
61
|
Niinuma T, Suzuki H, Nojima M, Nosho K,
Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki
Y, et al: Upregulation of miR-196a and HOTAIR drive malignant
character in gastrointestinal stromal tumors. Cancer Res.
72:1126–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kondo Y, Shinjo K and Katsushima K: Long
non-coding RNAs as an epigenetic regulator in human cancers. Cancer
Sci. 108:1927–1933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long Noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kumar S, Gonzalez EA, Rameshwar P and
Etchegaray JP: Non-Coding RNAs as mediators of epigenetic changes
in malignancies. Cancers (Basel). 12:36572020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W,
De W, Wang Z and Wang R: The long noncoding RNA HOTAIR contributes
to cisplatin resistance of human lung adenocarcinoma cells via
downregualtion of p21(WAF1/CIP1) expression. PLoS One.
8:e772932013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M,
De W, Wang C and Ji G: LincRNAFEZF1-AS1 represses p21 expression to
promote gastric cancer proliferation through LSD1-Mediated H3K4me2
demethylation. Mol Cancer. 16:392017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Pang B, Wang Q, Ning S, Wu J, Zhang X,
Chen Y and Xu S: Landscape of tumor suppressor long noncoding RNAs
in breast cancer. J Exp Clin Cancer Res. 38:792019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Huo Y, Li Q, Wang X, Jiao X, Zheng J, Li Z
and Pan X: MALAT1 predicts poor survival in osteosarcoma patients
and promotes cell metastasis through associating with EZH2.
Oncotarget. 8:46993–47006. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Amodio N, Raimondi L, Juli G, Stamato MA,
Caracciolo D, Tagliaferri P and Tassone P: MALAT1: A druggable long
non-coding RNA for targeted anti-cancer approaches. J Hematol
Oncol. 11:632018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chi JS, Li JZ, Jia JJ, Zhang T, Liu XM and
Yi L: Long non-coding RNA ANRIL in gene regulation and its duality
in atherosclerosis. J Huazhong Univ Sci Technolog Med Sci.
37:816–822. 2017.PubMed/NCBI
|
|
72
|
Meseure D, Vacher S, Alsibai KD, Nicolas
A, Chemlali W, Caly M, Lidereau R, Pasmant E, Callens C and Bieche
I: Expression of ANRIL-Polycomb Complexes-CDKN2A/B/ARF genes in
breast tumors: Identification of a Two-Gene (EZH2/CBX7) signature
with independent prognostic value. Mol Cancer Res. 14:623–633.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Puvvula PK, Desetty RD, Pineau P, Marchio
A, Moon A, Dejean A and Bischof O: Long noncoding RNA PANDA and
scaffold-attachment-factor SAFA control senescence entry and exit.
Nat Commun. 5:53232014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Portoso M, Ragazzini R, Brenčič Ž, Moiani
A, Michaud A, Vassilev I, Wassef M, Servant N, Sargueil B and
Margueron R: PRC2 is dispensable for HOTAIR-mediated
transcriptional repression. EMBO J. 36:981–994. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang XH and Li J: CircAGFG1 aggravates the
progression of cervical cancer by downregulating p53. Eur Rev Med
Pharmacol Sci. 24:1704–1711. 2020.PubMed/NCBI
|
|
77
|
Zhang G, Chen X, Ma L, Ding R, Zhao L, Ma
F and Deng X: LINC01419 facilitates hepatocellular carcinoma growth
and metastasis through targeting EZH2-regulated RECK. Aging (Albany
NY). 12:11071–11084. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Chen XJ and An N: Long noncoding RNA ATB
promotes ovarian cancer tumorigenesis by mediating histone H3
lysine 27 trimethylation through binding to EZH2. J Cell Mol Med.
25:37–46. 2021. View Article : Google Scholar :
|
|
79
|
Wu L, Gong Y, Yan T and Zhang H: LINP1
promotes the progression of cervical cancer by scaffolding EZH2,
LSD1, and DNMT1 to inhibit the expression of KLF2 and PRSS8.
Biochem Cell Biol. 98:591–599. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhang J, Li WY, Yang Y, Yan LZ, Zhang SY,
He J and Wang JX: LncRNA XIST facilitates cell growth, migration
and invasion via modulating H3 histone methylation of DKK1 in
neuroblastoma. Cell Cycle. 18:1882–1892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li Z, Yu D, Li H, Lv Y and Li S: Long
non-coding RNA UCA1 confers tamoxifen resistance in breast cancer
endocrinotherapy through regulation of the EZH2/p21 axis and the
PI3K/AKT signaling pathway. Int J Oncol. 54:1033–1042.
2019.PubMed/NCBI
|
|
82
|
Dong H, Wang W, Mo S, Chen R, Zou K, Han
J, Zhang F and Hu J: SP1-induced lncRNA AGAP2-AS1 expression
promotes chemoresistance of breast cancer by epigenetic regulation
of MyD88. J Exp Clin Cancer Res. 37:2022018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Loe AKH, Zhu L and Kim TH: Chromatin and
noncoding RNA-mediated mechanisms of gastric tumorigenesis. Exp Mol
Med. 55:22–31. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Qiu Y, Xu M and Huang S: Long noncoding
RNAs: Emerging regulators of normal and malignant hematopoiesis.
Blood. 138:2327–2336. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Qiu Y and Huang S: CTCF-mediated genome
organization and leukemogenesis. Leukemia. 34:2295–2304. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ribeiro-Silva C, Vermeulen W and Lans H:
SWI/SNF: Complex complexes in genome stability and cancer. DNA
Repair (Amst). 77:87–95. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Bammidi LS and Gayen S: Multifaceted role
of CTCF in X-chromosome inactivation. Chromosoma. 133:217–231.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Martitz A and Schulz EG: Spatial
orchestration of the genome: Topological reorganisation during
X-chromosome inactivation. Curr Opin Genet Dev. 86:1021982024.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Engreitz JM, Pandya-Jones A, McDonel P,
Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander
ES, et al: The Xist lncRNA exploits three-dimensional genome
architecture to spread across the X chromosome. Science.
341:12379732013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kung JT, Kesner B, An JY, Ahn JY,
Cifuentes-Rojas C, Colognori D, Jeon Y, Szanto A, del Rosario BC,
Pinter SF, et al: Locus-specific targeting to the X chromosome
revealed by the RNA interactome of CTCF. Mol Cell. 57:361–375.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Luo H, Zhu G, Eshelman MA, Fung TK, Lai Q,
Wang F, Zeisig BB, Lesperance J, Ma X, Chen S, et al:
HOTTIP-dependent R-loop formation regulates CTCF boundary activity
and TAD integrity in leukemia. Mol Cell. 82:833–851 e811. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhu G, Luo H, Feng Y, Guryanova OA, Xu J,
Chen S, Lai Q, Sharma A, Xu B, Zhao Z, et al: HOXBLINC long
non-coding RNA activation promotes leukemogenesis in NPM1-mutant
acute myeloid leukemia. Nat Commun. 12:19562021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lai Q, Hamamoto K, Luo H, Zaroogian Z,
Zhou C, Lesperance J, Zha J, Qiu Y, Guryanova OA, Huang S and Xu B:
NPM1 mutation reprograms leukemic transcription network via
reshaping TAD topology. Leukemia. 37:1732–1736. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Venkatraman A, He XC, Thorvaldsen JL,
Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu
JY, et al: Maternal imprinting at the H19-Igf2 locus maintains
adult haematopoietic stem cell quiescence. Nature. 500:345–349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Hacisuleyman E, Goff LA, Trapnell C,
Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG,
Sauvageau M, Kelley DR, et al: Topological organization of
multichromosomal regions by the long intergenic noncoding RNA
Firre. Nat Struct Mol Biol. 21:198–206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hanly DJ, Esteller M and Berdasco M:
Interplay between long non-coding RNAs and epigenetic machinery:
Emerging targets in cancer? Philos Trans R Soc Lond B Biol Sci.
373:2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Reddy D, Bhattacharya S, Levy M, Zhang Y,
Gogol M, Li H, Florens L and Workman JL: Paraspeckles interact with
SWI/SNF subunit ARID1B to regulate transcription and splicing. EMBO
Rep. 24:e553452023. View Article : Google Scholar :
|
|
98
|
Bhattacharya A, Wang K, Penailillo J, Chan
CN, Fushimi A, Yamashita N, Daimon T, Haratake N, Ozawa H,
Nakashoji A, et al: MUC1-C regulates NEAT1 lncRNA expression and
paraspeckle formation in cancer progression. Oncogene.
43:2199–2214. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lee VH, Tsang RK, Lo AWI, Chan SY, Chung
JC, Tong CC, Leung TW and Kwong DL: SMARCB1 (INI-1)-Deficient
sinonasal carcinoma: A systematic review and pooled analysis of
treatment outcomes. Cancers (Basel). 14:32852022. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang X, Gong Y, Jin B, Wu C, Yang J, Wang
L, Zhang Z and Mao Z: Long non-coding RNA urothelial carcinoma
associated 1 induces cell replication by inhibiting BRG1 in 5637
cells. Oncol Rep. 32:1281–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Huang M, Wang H, Hu X and Cao X: lncRNA
MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically
promote inflammation-related hepatocellular carcinoma progression.
Oncoimmunology. 8:e15186282019. View Article : Google Scholar
|
|
102
|
Lino Cardenas CL, Kessinger CW, Cheng Y,
MacDonald C, MacGillivray T, Ghoshhajra B, Huleihel L, Nuri S, Yeri
AS, Jaffer FA, et al: An HDAC9-MALAT1-BRG1 complex mediates smooth
muscle dysfunction in thoracic aortic aneurysm. Nat Commun.
9:10092018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Neve B, Jonckheere N, Vincent A and Van
Seuningen I: Epigenetic regulation by lncRNAs: An overview focused
on UCA1 in colorectal cancer. Cancers (Basel). 10:10092018.
View Article : Google Scholar
|
|
104
|
Chiba H, Muramatsu M, Nomoto A and Kato H:
Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and
Drosophila brahma are transcriptional coactivators cooperating with
the estrogen receptor and the retinoic acid receptor. Nucleic Acids
Res. 22:1815–1820. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J,
Yan X, Ye B, Li C, Xia P, et al: The long noncoding RNA lncTCF7
promotes self-renewal of human liver cancer stem cells through
activation of Wnt signaling. Cell Stem Cell. 16:413–425. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Li Y, Li W, Hoffman AR, Cui J and Hu JF:
The Nucleus/Mitochondria-Shuttling LncRNAs function as new
epigenetic regulators of mitophagy in cancer. Front Cell Dev Biol.
9:6996212021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Prensner JR, Iyer MK, Sahu A, Asangani IA,
Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al:
The long noncoding RNA SChLAP1 promotes aggressive prostate cancer
and antagonizes the SWI/SNF complex. Nat Genet. 45:1392–1398. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Cimadamore A, Gasparrini S, Mazzucchelli
R, Doria A, Cheng L, Lopez-Beltran A, Santoni M, Scarpelli M and
Montironi R: Long Non-coding RNAs in prostate cancer with emphasis
on second chromosome locus associated with Prostate-1 expression.
Front Oncol. 7:3052017. View Article : Google Scholar
|
|
109
|
Boo SH and Kim YK: The emerging role of
RNA modifications in the regulation of mRNA stability. Exp Mol Med.
52:400–408. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
de Santiago PR, Blanco A, Morales F,
Marcelain K, Harismendy O, Sjöberg Herrera M and Armisén R:
Immune-related IncRNA LINC00944 responds to variations in ADAR1
levels and it is associated with breast cancer prognosis. Life Sci.
268:1189562021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Sun Y, Ge J, Shao F, Ren Z, Huang Z, Ding
Z, Dong L, Chen J, Zhang J and Zang Y: Long noncoding RNA AI662270
promotes kidney fibrosis through enhancing METTL3-mediated
m6A modification of CTGF mRNA. FASEB J. 37:e230712023.
View Article : Google Scholar
|
|
112
|
Cao Y, Di X, Cong S, Tian C, Wang Y, Jin
X, Zhao M, Zhou X, Li R and Wang K: RBM10 recruits METTL3 to induce
N6-methyladenosine-MALAT1-dependent modification, inhibiting the
invasion and migration of NSCLC. Life Sci. 315:1213592023.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Fang Y, Wu X, Gu Y, Shi R, Yu T, Pan Y,
Zhang J, Jing X, Ma P and Shu Y: LINC00659 cooperated with ALKBH5
to accelerate gastric cancer progression by stabilising JAK1 mRNA
in an m6 A-YTHDF2-dependent manner. Clin Transl Med.
13:e12052023. View Article : Google Scholar
|
|
114
|
Salameh A, Lee AK, Cardo-Vila M, Nunes DN,
Efstathiou E, Staquicini FI, Dobroff AS, Marchiò S, Navone NM,
Hosoya H, et al: PRUNE2 is a human prostate cancer suppressor
regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad
Sci USA. 112:8403–8408. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhu L, Zhu Y, Han S, Chen M, Song P, Dai
D, Xu W, Jiang T, Feng L, Shin VY, et al: Impaired autophagic
degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in
gastric cancer. Cell Death Dis. 10:3832019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Mao J, Qiu H and Guo L: LncRNA HCG11
mediated by METTL14 inhibits the growth of lung adenocarcinoma via
IGF2BP2/LATS1. Biochem Biophys Res Commun. 580:74–80. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Picardi E, D'Erchia AM, Gallo A, Montalvo
A and Pesole G: Uncovering RNA editing sites in long Non-Coding
RNAs. Front Bioeng Biotechnol. 2:642014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Ma CP, Liu H, Yi-Feng Chang I, Wang WC,
Chen YT, Wu SM, Chen HW, Kuo YP, Shih CT, Li CY and Tan BC: ADAR1
promotes robust hypoxia signaling via distinct regulation of
multiple HIF-1alpha-inhibiting factors. EMBO Rep. 20:e471072019.
View Article : Google Scholar
|
|
119
|
Salameh A, Lee AK, Cardó-Vila M, Nunes DN,
Efstathiou E, Staquicini FI, Dobroff AS, Marchiò S, Navone NM,
Hosoya H, et al: PRUNE2 is a human prostate cancer suppressor
regulated by the intronic long noncoding RNA. Proc Natl Acad Sci
USA. 112:8403–8408. 2015. View Article : Google Scholar
|
|
120
|
Arun G, Diermeier S, Akerman M, Chang KC,
Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR and Norton L:
Differentiation of mammary tumors and reduction in metastasis upon
Malat1 lncRNA loss. Genes Dev. 30:34–51. 2016. View Article : Google Scholar :
|
|
121
|
Zhao C, Ling X, Xia Y, Yan B and Guan Q:
The m6A methyltransferase METTL3 controls epithelial-mesenchymal
transition, migration and invasion of breast cancer through the
MALAT1/miR-26b/HMGA2 axis. Cancer Cell Int. 21:4412021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W,
Lu S, Xu D, Wu Y, Chen Q, et al: LNC942 promoting METTL14-mediated
m6A methylation in breast cancer cell proliferation and
progression. Oncogene. 39:5358–5372. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Pu J, Xu Z, Huang Y, Nian J, Yang M, Fang
Q, Wei Q, Huang Z, Liu G, Wang J, et al:
N6-methyladenosine-modified FAM111A-DT promotes
hepatocellular carcinoma growth via epigenetically activating
FAM111A. Cancer Sci. 114:3649–3665. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zhou L, Jiang J, Huang Z, Jin P, Peng L,
Luo M, Zhang Z, Chen Y, Xie N, Gao W, et al: Hypoxia-induced lncRNA
STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal
cancer progression by preventing m6A-mediated degradation of STEAP3
mRNA. Mol Cancer. 21:1682022. View Article : Google Scholar
|
|
125
|
Wang X, Liu C, Zhang S, Yan H, Zhang L,
Jiang A, Liu Y, Feng Y, Li D, Guo Y, et al:
N6-methyladenosine modification of MALAT1 promotes
metastasis via reshaping nuclear speckles. Dev Cell. 56:702–715.e8.
2021. View Article : Google Scholar
|
|
126
|
Yang H, Hu Y, Weng M, Liu X, Wan P, Hu Y,
Ma M, Zhang Y, Xia H and Lv K: Hypoxia inducible lncRNA-CBSLR
modulates ferroptosis through m6A-YTHDF2-dependent modulation of
CBS in gastric cancer. J Adv Res. 37:91–106. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Wang S, Wang Y, Zhang Z, Zhu C, Wang C, Yu
F and Zhao E: Long Non-Coding RNA NRON promotes tumor proliferation
by regulating ALKBH5 and nanog in gastric cancer. J Cancer.
12:6861–6872. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A
Demethylase ALKBH5 maintains tumorigenicity of glioblastoma
Stem-like cells by sustaining FOXM1 expression and cell
proliferation program. Cancer Cell. 31:591–606.e6. 2017. View Article : Google Scholar
|
|
129
|
Zou Z, Zhou S, Liang G, Tang Z, Li K, Tan
S, Zhang X and Zhu X: The pan-cancer analysis of the two types of
uterine cancer uncovered clinical and prognostic associations with
m6A RNA methylation regulators. Mol Omics. 17:438–453. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zhu P, He F, Hou Y, Tu G, Li Q, Jin T,
Zeng H, Qin Y, Wan X, Qiao Y, et al: A novel hypoxic long noncoding
RNA KB-1980E6.3 maintains breast cancer stem cell stemness via
interacting with IGF2BP1 to facilitate c-Myc mRNA stability.
Oncogene. 40:1609–1627. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Nadhan R, Isidoro C, Song YS and
Dhanasekaran DN: LncRNAs and the cancer epigenome: Mechanisms and
therapeutic potential. Cancer Lett. 605:2172972024. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Yang L, Tang L, Min Q, Tian H, Li L, Zhao
Y, Wu X, Li M, Du F, Chen Y, et al: Emerging role of RNA
modification and long noncoding RNA interaction in cancer. Cancer
Gene Ther. 31:816–830. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Kim SY, Na MJ, Yoon S, Shin E, Ha JW, Jeon
S and Nam SW: The roles and mechanisms of coding and noncoding RNA
variations in cancer. Exp Mol Med. 56:1909–1920. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar
|
|
135
|
Liu S, Jiao B, Zhao H, Liang X, Jin F, Liu
X and Hu JF: LncRNAs-circRNAs as rising epigenetic binary
superstars in regulating lipid metabolic reprogramming of cancers.
Adv Sci (Weinh). 11:e23035702024. View Article : Google Scholar
|
|
136
|
Mattick JS, Amaral PP, Carninci P,
Carpenter S, Chang HY, Chen LL, Chen R, Dean C, Dinger ME,
Fitzgerald KA, et al: Long non-coding RNAs: Definitions, functions,
challenges and recommendations. Nat Rev Mol Cell Biol. 24:430–447.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Liu SJ, Dang HX, Lim DA, Feng FY and Maher
CA: Long noncoding RNAs in cancer metastasis. Nat Rev Cancer.
21:446–460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Qian Y, Shi L and Luo Z: Long Non-coding
RNAs in Cancer: Implications for diagnosis, prognosis, and therapy.
Front Med (Lausanne). 7:6123932020. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Gong C, Li Z, Ramanujan K, Clay I, Zhang
Y, Lemire-Brachat S and Glass DJ: A long non-coding RNA, LncMyoD,
regulates skeletal muscle differentiation by blocking IMP2-mediated
mRNA translation. Dev Cell. 34:181–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Davodabadi F, Farasati Far B, Sargazi S,
Fatemeh Sajjadi S, Fathi-Karkan S, Mirinejad S, Ghotekar S, Sargazi
S and Rahman MM: Nanomaterials-based targeting of long Non-Coding
RNAs in cancer: A Cutting-edge review of current trends.
ChemMedChem. 19:e2023005282024. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Han S, Cao Y, Guo T, Lin Q and Luo F:
Targeting lncRNA/Wnt axis by flavonoids: A promising therapeutic
approach for colorectal cancer. Phytother Res. 36:4024–4040. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Parashar D, Singh A, Gupta S, Sharma A,
Sharma MK, Roy KK, Chauhan SC and Kashyap VK: Emerging roles and
potential applications of Non-Coding RNAs in Cervical cancer. Genes
(Basel). 13:12542022. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Goyal A, Myacheva K, Gross M, Klingenberg
M, Duran Arque B and Diederichs S: Challenges of CRISPR/Cas9
applications for long non-coding RNA genes. Nucleic Acids Res.
45:e122017.PubMed/NCBI
|
|
144
|
Cetinkaya M and Baran Y: MicroRNAs and
long non-coding RNAs as novel targets in Anti-cancer drug
development. Curr Pharm Biotechnol. 24:913–925. 2023. View Article : Google Scholar
|
|
145
|
Ozcan G, Ozpolat B, Coleman RL, Sood AK
and Lopez-Berestein G: Preclinical and clinical development of
siRNA-based therapeutics. Adv Drug Deliv Rev. 87:108–119. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Wahlestedt C: Targeting long non-coding
RNA to therapeutically upregulate gene expression. Nat Rev Drug
Discov. 12:433–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Alegra-Torres JA, Baccarelli A and Bollati
V: Epigenetics and lifestyle. Epigenomics. 3:267–277. 2011.
View Article : Google Scholar
|
|
148
|
Basu AK: DNA Damage, mutagenesis and
cancer. Int J Mol Sci. 19:9702018. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Cheng D, Deng J, Zhang B, He X, Meng Z, Li
G, Ye H, Zheng S, Wei L, Deng X, et al: LncRNA HOTAIR
epigenetically suppresses miR-122 expression in hepatocellular
carcinoma via DNA methylation. EBioMedicine. 36:159–170. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Vaasjo LO: LncRNAs and chromatin
modifications pattern m6A methylation at the untranslated regions
of mRNAs. Front Genet. 13:8667722022. View Article : Google Scholar :
|
|
151
|
Vaid R, Thombare K, Mendez A,
Burgos-Panadero R, Djos A, Jachimowicz D, Lundberg KI, Bartenhagen
C, Kumar N, Tümmler C, et al: METTL3 drives telomere targeting of
TERRA lncRNA through m6A-dependent R-loop formation: A therapeutic
target for ALT-positive neuroblastoma. Nucleic Acids Res.
52:2648–2671. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar
|
|
153
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Cho SW, Xu J, Sun R, Mumbach MR, Carter
AC, Chen YG, Yost KE, Kim J, He J, Nevins SA, et al: Promoter of
lncRNA Gene PVT1 is a Tumor-suppressor DNA boundary element. Cell.
173:1398–1412.e22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Arun G, Aggarwal D and Spector DL: MALAT1
Long Non-coding RNA: Functional implications. Noncoding RNA.
6:222020.PubMed/NCBI
|
|
156
|
Chen PB, Chen HV, Acharya D, Rando OJ and
Fazzio TG: R loops regulate promoter-proximal chromatin
architecture and cellular differentiation. Nat Struct Mol Biol.
22:999–1007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Chu C, Qu K, Zhong FL, Artandi SE and
Chang HY: Genomic maps of long Noncoding RNA occupancy reveal
principles of RNA-Chromatin interactions. Mol Cell. 44:667–678.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Chu C and Chang HY: ChIRP-MS: RNA-directed
proteomic discovery. Methods Mol Biol. 1861:37–45. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
de Lera AR and Ganesan A: Epigenetic
polypharmacology: From combination therapy to multitargeted drugs.
Clin Epigenetics. 8:1052016. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Delgado-Morales R, Agís-Balboa RC,
Esteller M and Berdasco M: Epigenetic mechanisms during ageing and
neurogenesis as novel therapeutic avenues in human brain disorders.
Clin Epigenetics. 9:2017. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Wu SC, Kallin EM and Zhang Y: Role of
H3K27 methylation in the regulation of lncRNA expression. Cell Res.
20:1109–1116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Kornienko AE, Dotter CP, Guenzl PM,
Gisslinger H, Gisslinger B, Cleary C, Kralovics R, Pauler FM and
Barlow DP: Long non-coding RNAs display higher natural expression
variation than protein-coding genes in healthy humans. Genome Biol.
17:142016. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Pang KC, Frith MC and Mattick JS: Rapid
evolution of noncoding RNAs: Lack of conservation does not mean
lack of function. Trends Genet. 22:1–5. 2006. View Article : Google Scholar
|
|
164
|
Bohmdorfer G and Wierzbicki AT: Control of
chromatin structure by long noncoding RNA. Trends Cell Biol.
25:623–632. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Xing Z, Lin A, Li C, Liang K, Wang S, Liu
Y, Park PK, Qin L, Wei Y, Hawke DH, et al: lncRNA directs
cooperative epigenetic regulation downstream of chemokine signals.
Cell. 159:1110–1125. 2014. View Article : Google Scholar : PubMed/NCBI
|