Introduction
Cancer stem cells (CSCs) play a pivotal role in
tumorigenesis and have been extensively investigated as a
prospective target for anticancer therapy (1,2).
An association exists between the presence of CSCs and tendencies
towards recurrence, treatment resistance and immunological
tolerance in bladder cancer (BC); nevertheless, the mechanisms by
which bladder CSCs (BCSCs) sustain their stemness are not fully
elucidated (3).
Pyrroline-5-carboxylate reductase 1 (PYCR1) has been identified as
a carcinogenic factor of BC and has been shown to promote BC
proliferation, metastasis and epithelial-mesenchymal transition
(EMT) (4-6). PYCR1 can promote proliferation,
metastasis and EMT of BC, and its role is similar to that of tumor
stemness; therefore (7), it is
plausible to hypothesize the potential role of PYCR1 in the
regulation of BCSC stemness.
Histone methylation, a well-known epigenetic
modification mediated by histone demethylases, methyltransferases
and methylation reader proteins, is instrumental in cancer
pathogenesis (8). SET and MYND
domain-containing protein 2 (SMYD2) is a protein methyltransferase,
including a catalytic SET domain, known to facilitate
monomethylation of lysine residues on both histone and non-histone
proteins (8,9). Expression of SMYD2 is significantly
enhanced in breast cancer tissues, and patients with BC with
elevated SMYD2 expression show a poor prognosis (10). Elevated levels of SMYD2 may
promote the proliferation of papillary thyroid carcinoma cells
(11), as well as regulate gene
expression by modulating histone H3 lysine 4 trimethylation
(H3K4me3) modification in the promoter region of its target gene,
thereby affecting gastric cancer cell glycolysis and proliferation
(12). It was proposed that SMYD2
may stimulate the transcriptional expression of PYCR1 by regulating
the levels of H3K4me3 in the promoter region of PYCR1 to regulate
BC stemness.
Mitophagy facilitates maintenance of stemness in
CSCs (13,14). Mitophagy enhances the plasticity
of CSCs via metabolic recombination, thereby improving their
adaptation to the tumor microenvironment (15). The putative kinase 1
(PINK1)/Parkin signaling pathway is one of the key mechanisms
regulating mitophagy (16,17).
When the PINK1/Parkin mitophagy pathway is stimulated, SRY-box
transcription factor 2 (Sox2) promotes Nanog expression,
potentiates CSC self-renewal and preserves CSC stemness (18). PYCR1/2 serves as a crucial enzyme
in the proline biosynthetic pathway, and proline is capable of
inducing mitophagy via activation of AMPKα and the upregulation of
the Parkin expression (19). Our
previous study revealed that overexpression of PYCR1 has the
potential to boost the protein expression of PINK1 and Parkin,
leading to mitophagy in BC (4).
Also, PYCR1-augmented proline synthesis serves a crucial role in
sustaining the stemness of CSCs (20). Therefore, PYCR1 might regulate
BCSC stemness by controlling mitophagy via the PINK1/Parkin
pathway. It was hypothesized that SMYD2 may increase PYCR1
expression by upregulating the H3K4me3 levels in the PYCR1 promoter
region and promote BCSC stemness through PINK1/Parkin-regulated
mitophagy.
Materials and methods
Bioinformatics analysis
SMYD2 expression in BC and normal bladder tissue and
the association between SMYD2 and PYCR1 in BC were analyzed using
the GEPIA database (gepia.cancer-pku.cn/).
Cell culture and grouping
Human BC cell lines T24 (SCSP-536) and EJ (YS1803C)
were procured from Institute of Chinese Academy of Sciences and
Yaji Biotechnology Co., Ltd. (both Shanghai, China), respectively.
T24 and EJ cells were cultured in DMEM/Ham's F-12 with 10% fetal
bovine serum (both Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin solution (Shanghai Zeye Biotechnology Co.,
Ltd.) at 37°C with 5% CO2. Cells from the P3 generation
with 80% confluence were collected for subsequent experiments.
Human BC cell lines were grouped as follows: Blank
(untreated); small interfering (si)-PYCR1 (cells were transfected
with PYCR1 siRNA for 6 h and cultured for 18 h].); si-SMYD2 (BC
cells were transfected with SMYD2 siRNA for 24 h); si-negative
control (NC) (transfected with scramble siRNA for 24 h);
overexpression (oe)-PYCR1 (pcDNA3.1-PYCR1-transfected for 24 h);
oe-SMYD2 (transfected with pcDNA3.1-SMYD2 for 24 h); oe-NC
(transfected with pcDNA3.1-NC for 24 h); oe-SMYD2 + si-PYCR1
(transfection of pcDNA3.1-SMYD2 while being treated with PYCR1
siRNA for 24 h); oe-SMYD2 + si-NC (treated with scramble siRNA for
24 h while being transfected with pcDNA3.1-SMYD2); oe-PYCR1 +
si-PINK1 (24-h simultaneous treatment with pcDNA3.1-PYCR1 and PINK1
siRNA) and oe-PYCR1 + si-NC group (co-treatment with pcDNA3.1-PYCR1
and scramble siRNA for 24 h). si-PYCR1 forward 5′-UGC UAU CAA CGC
UGU GG-3′ and reverse 5′-CCA CAG CGU UGA UAG CA-3′; si-SMYD2:
forward: 5′-CAC CAG UUC UAC UCC AAG UTT-3′, reverse: 5′-ACU UGG AGU
AGA ACU GGU GTT-3′; si-NC: forward 5′-UUC UCC GAA CGU GUC ACG
UTT-3′ and reverse 5′-ACG UGA CAC GUU CGG AGA ATT-3′.
Cell transfection was performed using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 24 h
with 10 nmol/l pcDNA3.1-PYCR1, pcDNA3.1-SMYD2, pcDNA3.1-NC, PYCR1
siRNA, SMYD2 siRNA, PINK1 siRNA, and scramble siRNA, with a final
concentration of 3.75 µl/ml, as previously described
(21).
Flow cytometry
Cells were prepared into a single-cell suspension
for the screening of BCSCs. After being washed twice with PBS,
cells were resuspended in PBS to adjust the cell concentration to
1×109/l. Cells were cultured in the presence of
antibodies labeled with CSC markers, such as CD44 (IM7) Rat mAb
(Alexa Fluor® 555 Conjugate; cat. no. #95235) and CD133
(A8N6N) Mouse mAb (Alexa Fluor® 647 Conjugate; cat. no.
#53276; both 1:50; both Cell Signaling Technology, Inc.) on ice
without light for 30 min, followed by PBS rinsing three times.
Fluorescence-activated cell sorting Aria III (BD Biosciences) was
employed to sort CD44+CD133+ cells.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA extraction from cells was performed using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA
underwent RT to complementary DNA using a PrimeScript RT reagent
kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's instructions. qPCR was performed using TB
Green® Premix Ex Taq II (Takara Biotechnology Co., Ltd.)
and primers (Table I) on the
ABI7900 fluorescence PCR instrument. Thermocycling conditions were
as follows: Initial denaturation at 95°C for 10 min, followed by 40
cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 20
sec and extension at 72°C for 34 sec. β-actin was used as the
internal reference, and data analysis was conducted using the
2−ΔΔCq method (22).
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene | Forward, 5′→3′ | Reverse, 5′→3′ |
|---|
| PYCR1 |
GGCTGCCCACAAGATAATGGC |
GGCTGCCCACAAGATAATGGC |
| SMYD2 |
ATCTCCTGTACCCAACGGAAG |
CACCTTGGCCTTATCCTTGTCC |
| β-actin |
TGGCACCCAGCACAATGAA |
TGGCACCCAGCACAATGAA |
| Nanog |
CCCCAGCCTTTACTCTTCCT |
CCAGGTTGAATTGTTCCAGGTC |
| Sox2 |
ATCAGGAGTTGTCAAGGCAGAG |
AGAGGCAAACTGGAATCAGGA |
Colony formation assay
BC cells were seeded in 6 cm culture dishes at a
density of 1×103 cells/ml and cultured at 37°C for 14
days to colony formation. After the culture dish was washed with
distilled water, cells were immobilized at room temperature with 4%
paraformaldehyde (Sigma-Aldrich; Merck KGaA) for 15 min and then
stained with crystal violet (Beyotime Institute of Biotechnology)
for 30 min at room temperature. Images were captured and the
quantification of cell colonies was performed manually. The colony
was defined as a cell population large enough to be observed with
the naked eye.
CSC sphere-forming assay
Serum-free DMEM/F-12 (Gibco) was supplemented with
20 ng/ml each epidermal growth factor (Invitrogen) and basic
fibroblast growth factor (Gibco; all Thermo Fisher Scientific,
Inc.). Then, 5 µg/ml insulin (Procell Life Science &
Technology Co., Ltd.) and 100 µg/ml penicillin/streptomycin
were added to a low adhesion 6-well plate. Cells were re-suspended
with 1 ml medium added to the plate every 3 days. Afterward, cells
were seeded at 1,000 cells/well into DMEM/F-12 (serum-free).
Following 7 days of culture at 37°C, tumor spheres were collected
once the diameter reached 50-100 µm. Spheres were detached
and dispersed into single cells and re-seeded to form new tumor
spheres once again. After 14 days at 37°C, the culture was
terminated and the number of tumor sphere (diameter >50
µm) was calculated manually under the Leica DMi8 inverted
light microscope (200X magnification).
Cell mitophagy analysis
Cells were stained at 37°C with 400 nM Mito-Tracker
Green (cat. no. #C1046, Beyotime Institute of Biotechnology) for 40
min, and then blocked at 37°C for 1 h with 1% bovine serum albumin
(BSA, cat. no. #735094; Amresco, LLC) in PBS. Following 1 h
incubation at 37°C with the primary antibody light chain-3B (LC3B;
cat. no. ab192890, 1:100), the secondary antibody 6478-Conjugated
Goal Anti-Rabbit IgG (cat. no. ab150083, 1:200; both Abcam) was
added to cells for 30-min at 37°C and nuclei were stained with 5
µg/ml DAPI (cat. no. #C1002; Beyotime Institute of
Biotechnology) at room temperature for 3 min. Finally, the
immunofluorescence was visualized using a laser confocal microscope
(200×).
Chromatin immunoprecipitation (ChIP)
assay
Analysis was conducted utilizing a
simpleChIP®Plus ultrasonic ChIP kit (cat. no. #56383,
Cell Signaling Technology, Inc.). Cells at 80% confluence were
fixed using 4% paraformaldehyde at room temperature for 15 min and
treated with glycine at room temperature for 15 sec at a final
concentration of 0.125 M (both Beyotime Institute of Biotechnology)
to halt the fixation process. Cells were lysed and centrifuged at
300 × g at 4°C for 5 min. Chromatin was resuspended in ChIP lysis
buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8, 1%
Triton X-100, 0.1% deoxycholate, 0.1% SDS), 1% protease inhibitor)
and sonicated on ice for 2 min (sonication: 3 sec on, 2 s off) to
shear DNA-protein fragments to approximately 500 bp. 20 µl
of cell lysate was incubated overnight at 4°C with 5 µg of
rabbit Anti-H3K4me3 antibody (#9751S, 1:50, Cell Signaling
Technology) or rabbit IgG (2729S, 1:100, Cell Signaling
Technology). Protein-A beads (20 µl) were then used to
capture the antibody complex. The beads were washed 7 times with
wash buffer (50 mM HEPES, pH 7.5; 500 mM LiCl; 1 mM EDTA; 1% NP-40
and 0.7% deoxycholate), followed by one wash with Tris-EDTA buffer
(10 mM Tris pH 8.0, 1 mM EDTA). The precipitated DNA was purified
using a purification column and the expression of PYCR1 was
detected by RT-qPCR as aforementioned. Primers were as follows:
PYCR1-forward, 5′-CCT GTC ATC ATC TAA GAT AC-3′; PYCR1-reverse,
5′-CCC TGT CGG TGG CCA AGA TT-3′.
In vivo xenograft experiment
BALB/C nude mice (male, n=30; age, 5 weeks; weight
20-22 g) were purchased from Experimental Animal Resource Platform
of the Chinese Academy of Sciences [animal license no. SYXK
(Beijing) 2021-0063, Beijing, China]. Nude mice were housed in a
standard animal house (25±2°C, 50±10% relative humidity, 12/12-h
light-dark cycle and ad libitum access to food and
water).
The BC xenograft model was established by
subcutaneously injecting T24 cells (2×106 cells in 100
µl PBS) into nude mice. Mice were randomly divided (six
mice/group) into groups as follows: BC; BC + lentivirus
(Lv)-oe-SMYD2 (T24 cells delivered with pcDNA3.1-SMYD2 Lv were
subcutaneously injected); BC + Lv-oe-NC (injection of T24 cells
transfected with pcDNA3.1-NC Lv plasmid); BC + Lv-oe-SMYD2 +
Lv-si-PYCR1 (T24 cells transfected with pcDNA3.1-SMYD2 + si-PYCR1
Lv plasmids were hypodermically injected); BC + Lv-oe-SMYD2 +
Lv-si-NC (T24 cells transfected with pcDNA3.1-SMYD2 + si-NC Lv
plasmids were subcutaneously injected). After 4 weeks, mice were
euthanized via intraperitoneal injection of 100 mg/kg sodium
pentobarbital and tumor tissue was obtained to measure the longest
longitudinal (a) and transverse diameter (b). The tumor volume was
calculated and weighed using the formula (a x b2)/2
(23), followed by protein
extraction, hematoxylin-eosin (HE) staining and
immunohistochemistry (IHC). The humane endpoints were body weight
of the mice decreased by 40% or the tumor diameter more than 3 cm).
No mice reached the humane endpoints.
Western blotting
The homogenates of cells or tumor tissues were lysed
using RIPA lysis solution (Beyotime Institute of Biotechnology)
containing protease inhibitors (Sigma-Aldrich; Merck KGaA) and
supernatants were obtained upon 20 min centrifugation at 12,000 × g
at 4°C. Protein concentration was determined using a BCA kit. The
protein loading amount of each well was 10 µg. Following
separation via 10% SDS-PAGE, protein bands were moved to a PVDF
membrane and incubated overnight at 4°C with the following primary
antibodies: Anti-PYCR1 (cat. no. ab226340, 1:2,000), anti-SMYD2
(cat. no. ab108217, 1:2,000), anti-Nanog (cat. no. ab21624, 1:200),
anti-Sox2 (cat. no. ab92494, 1:1,000), anti-H3K4me3 (cat. no.
ab272143, 1:500), anti-PINK1 (cat. no. ab216144, 1:1,000, all
Abcam), anti-Parkin (cat. no. PA5-13399, 1:2,000, Invitrogen;
Thermo Fisher Scientific, Inc.), anti-LC3B (cat. no. ab192890,
1:2,000), anti-p62 (cat. no. ab109012, 1:10,000) and β-actin (cat.
no. ab8227, 1:1,000; all Abcam). Subsequently, the membrane was
washed with TBS-tween (0.1% Tween 20; cat. no. ST673; Beyotime) and
incubated with horseradish peroxidase-labeled goat anti-rabbit IgG
(cat. no. ab205718, 1:2,000, Abcam) for 1 h at room temperature.
Protein bands were visualized using the ultrasensitive enhanced
chemiluminescence kit (cat. no. P0018S, Beyotime) and the gray
values were quantified using Image J 1.8 software (National
Institutes of Health), with β-actin serving as the internal
reference. Cell experiments were repeated three times.
HE staining
Rat tumor tissues were subjected to paraffin
embedding after fixation with 4% paraformaldehyde at room
temperature for 24 hand cut into 5-µm sections. The sections
were stained at room temperature for 20 min with HE Stain kit (cat.
no. G1120; Beijing Solarbio Science & Technology Co., Ltd.) and
photographed under a light microscope (200×) (Olympus
Corporation).
IHC
The tumor tissue subjected to 24-h fixation with 4%
paraformaldehyde at room temperature was embedded in paraffin,
serially sectioned (4 µm), dewaxed with xylene, and hydrated
with gradient alcohol. Sections were treated with 3%
H2O2 for 10 min to neutralize endogenous
peroxidase activity. The samples were subjected to antigen
retrieval in 0.01 mol/l sodium citrate buffer (pH=6.0, 15 min)
using microwave heating at 95°C. The sections were blocked at room
temperature for 20 min using 5% BSA (cat. no. A602440-0050, Sangon
Biotechnology Co., Ltd.) and incubated overnight at 4°C with
primary antibodies against H3K4me3 (cat. no. ab8580, 1:100), SMYD2
(cat. no. ab234862, 1:200), CD44 (cat no. ab316123, 1:5,000) and
CD133 (cat. no. ab19898, 1:500; all Abcam). HRP labeled goat anti
rabbit IgG (cat. no. ab6721, 1:1,000; Abcam) was incubated at room
temperature for 20 min. After sections were rinsed with PBS, the
color was developed using 3,3′-diaminobenzidine (cat. no.
B-IMWRS51, AmyJet Scientific, Inc.). The staining was observed
under a light microscope (200X magnification; Olympus Corporation).
The percentages of positive cells were counted and averaged using
Image J 1.8 (National Institutes of Health).
Statistical analysis
All data analysis was performed using SPSS 21.0 (IBM
Corp.) and GraphPad Prism 9.5 (GraphPad Software, Inc.; Dotmatics).
The data are presented as the mean ± SD of three independent
experiments. Comparisons between two groups were performed using
unpaired t-test, while multi-group comparisons were analyzed using
one-way ANOVA followed by Tukey's multiple comparison test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
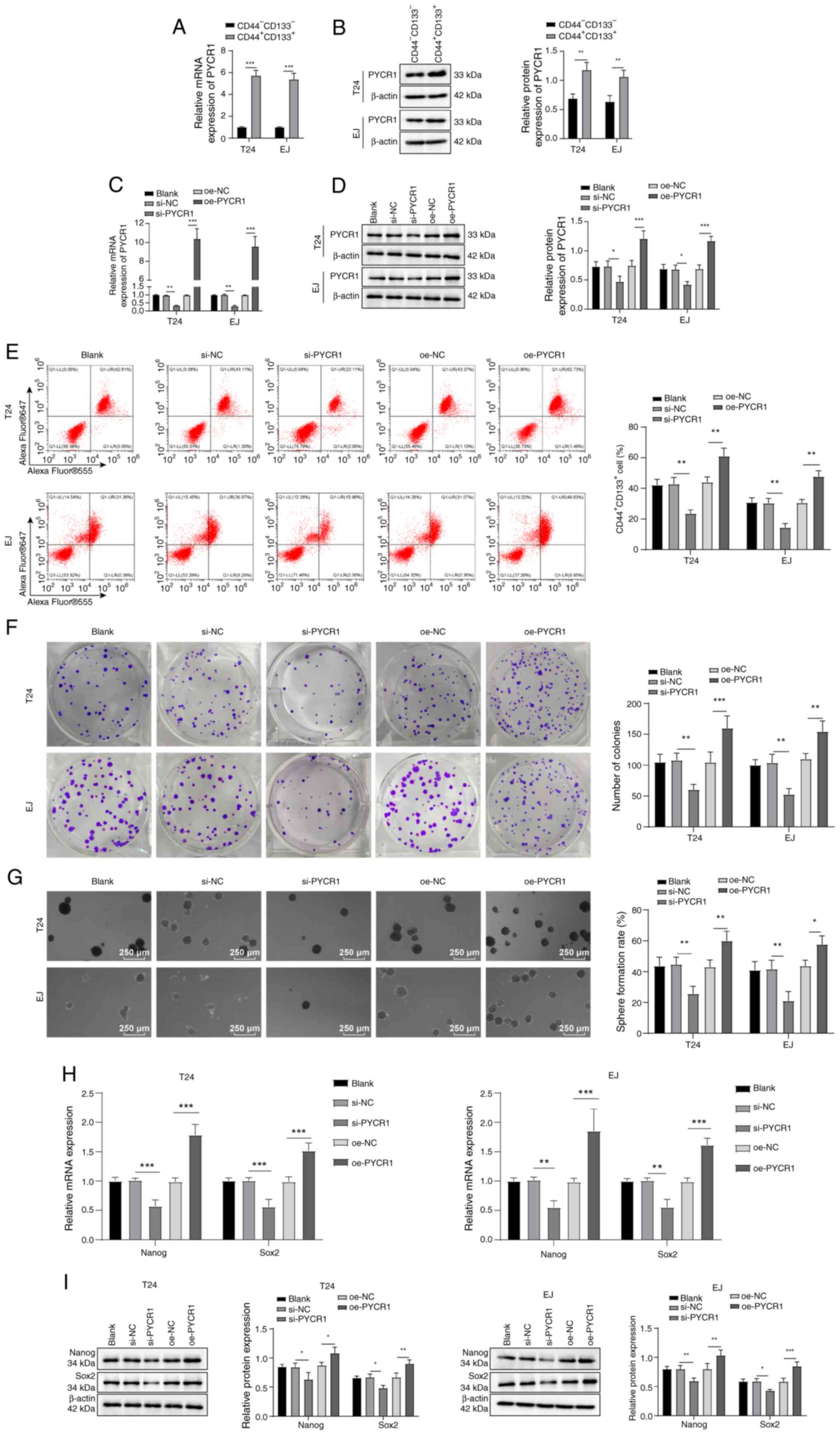
PYCR1 promotes stemness maintenance of
BCSCs
To investigate whether PYCR1 is involved in
regulating BCSC stemness, BC cell lines (EJ and T24) were cultured
in vitro. Subpopulations exhibiting SC characteristics
(CD44+CD133+) were sorted from BC cells by
flow cytometry and expression of PYCR1 mRNA and proteins in
CD44+CD133+ and
CD44−CD133− cells was assessed via RT-qPCR
and western blot. The results showed that PYCR1 mRNA and its
protein expression were higher in the
CD44+CD133+ BC cell subpopulation than in
CD44−CD133− cells (Fig. 1A and B). Following PYCR1 siRNA
treatment, PYCR1 mRNA and protein expression in BC cells (Fig. 1C and D),
CD44+CD133+ cell levels (Fig. 1E) and colony tumor sphere
formation (Fig. 1F and G) and
Nanog and Sox2 expression was reduced (Fig. 1H and I). Following transfection of
pcDNA3.1-PYCR1, BC cells had increased expression of PYCR1 mRNA and
protein (Fig. 1C and D) levels of
CD44+CD133+ cells (Fig. 1E), increased formation of cancer
cell colonies and tumor spheres (Fig.
1F and G) and elevated expression of Nanog and Sox2 (Fig. 1H and I). These results indicated
that PYCR1 enhanced BCSC stemness maintenance.
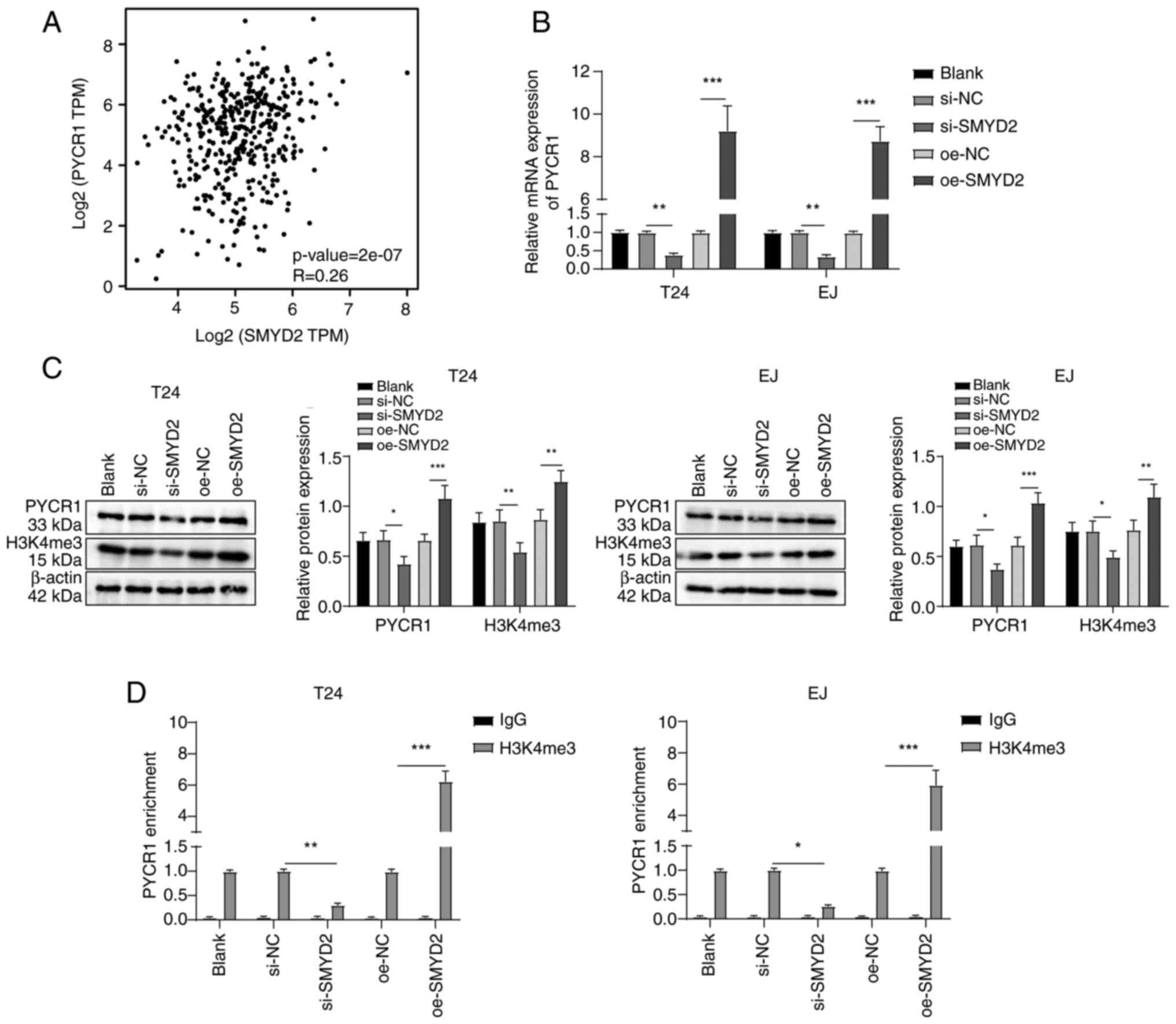
SMYD2 promotes stemness maintenance of
BCSCs
Multiple post-translational modifications of
proteins serve crucial roles in tumor progression, with methylation
modification being common (24).
SMYD2 is a protein lysine methyltransferase involved in the
advancement of BC (10). SMYD2
was found to be significantly highly expressed in BC through the
analysis of the GEPIA database (Fig.
2A). When BC cells were treated with SMYD2 siRNA, SMYD2 mRNA
and protein expression (Fig. 2B and
C), CD44+CD133+ cell levels (Fig. 2D), colony and tumor sphere
formation (Fig. 2E and F) and
Sox2 and Nanog expression decreased (Fig. 2G and H). However, following
transfection of pcDNA3.1-SMYD2 into BC cells, SMYD2 mRNA and
protein expression (Fig. 2B and
C), levels of CD44+CD133+ cells (Fig. 2D), colony and tumor sphere
formation (Fig. 2E and F) and the
protein expression of stemness markers Nanog and Sox2 was increased
(Fig. 2G and H). Altogether,
these findings illustrated that the stemness retention of BCSCs was
promoted by SMYD2.
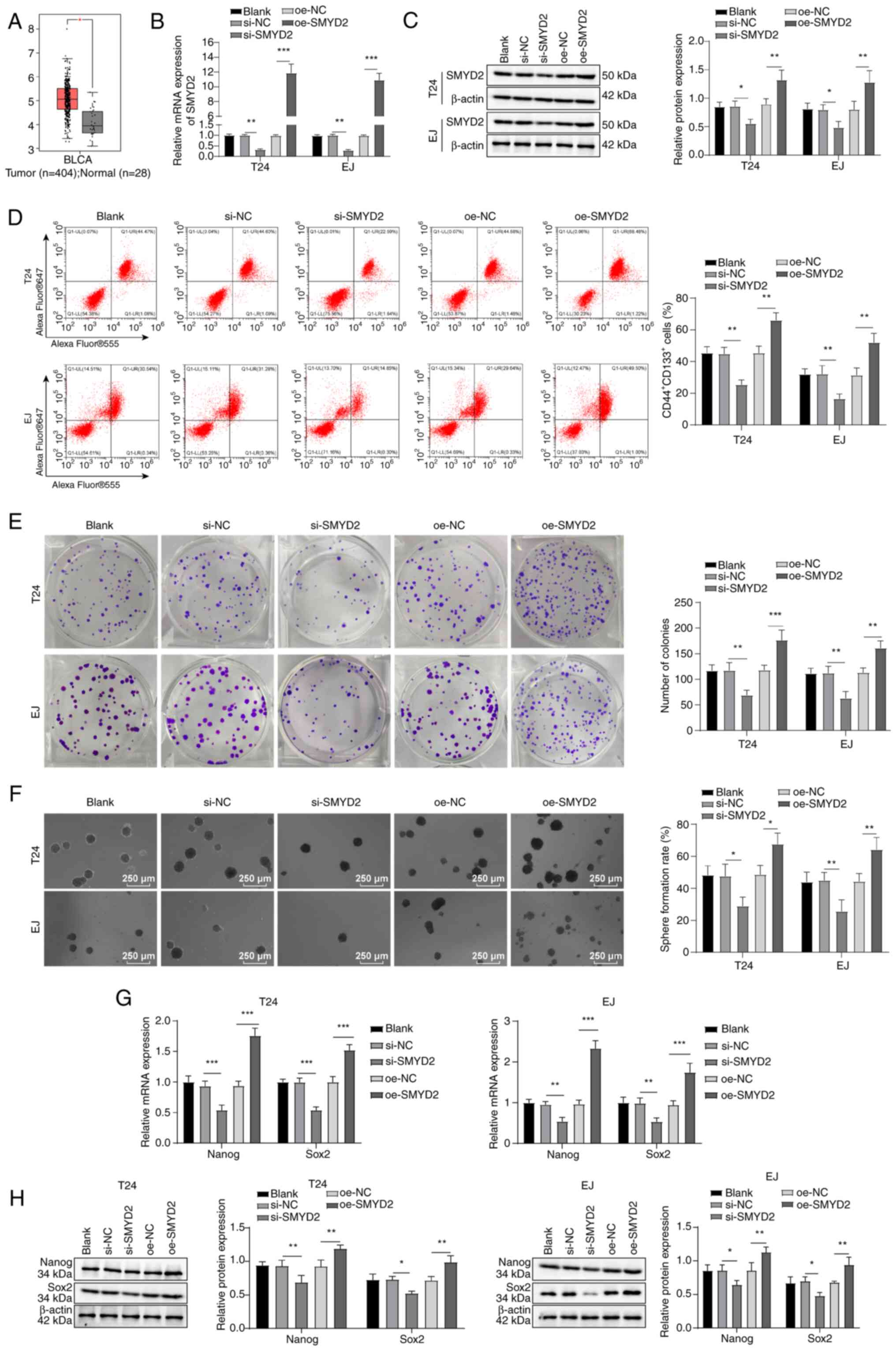 | Figure 2SMYD2 promotes stemness retention in
bladder CSCs. (A) Analysis of SMYD2 expression in normal bladder
tissue and bladder cancer using Gene Expression Profiling
Interactive Analysis database. SMYD2 (B) mRNA and (C) protein
expression using RT-qPCR and western blotting. (D) Flow cytometry
to determine CD44+CD133+ cell levels. (E)
In vitro colony formation capability. (F) CSC sphere-forming
assay to examine tumor stemness. (G) RT-qPCR and (H) western
blotting were used to evaluate the expression of Nanog and Sox2.
*P<0.05, **P<0.01,
***P<0.001. SMYD2, SET and MYND domain-containing
protein 2; CSC, cancer stem cell; RT-qPCR, reverse transcription
quantitative polymerase chain reaction; si, small-interfering; NC,
negative control; oe, overexpression; ns, not significant; BLCA,
bladder urothelial carcinoma. |
SMYD2 modulates H3K4me3 to upregulate
PYRC1 expression in BC cells
SMYD2 was positively associated with PYCR1
expression in BC according to the GEPIA database analysis (Fig. 3A). Following transfection of SMYD2
siRNA in BC cells, PYCR1 mRNA and its protein expression and
H3K4me3 levels decreased, while transfection of pcDNA3.1-SMYD2 into
BC cells increased PYCR1 mRNA and protein expression and H3K4me3
levels (Fig. 3B and C). ChIP
assay demonstrated enrichment of H3K4me3 in the PYCR1 promoter
region was decreased following SMYD2 siRNA but increased following
pcDNA3.1-SMYD2 transfection (Fig.
3D). Overall, SMYD2 modulated the expression of PYRC1 in BC
cells by controlling the H3K4me3 levels in the PYRC1 promoter
region.
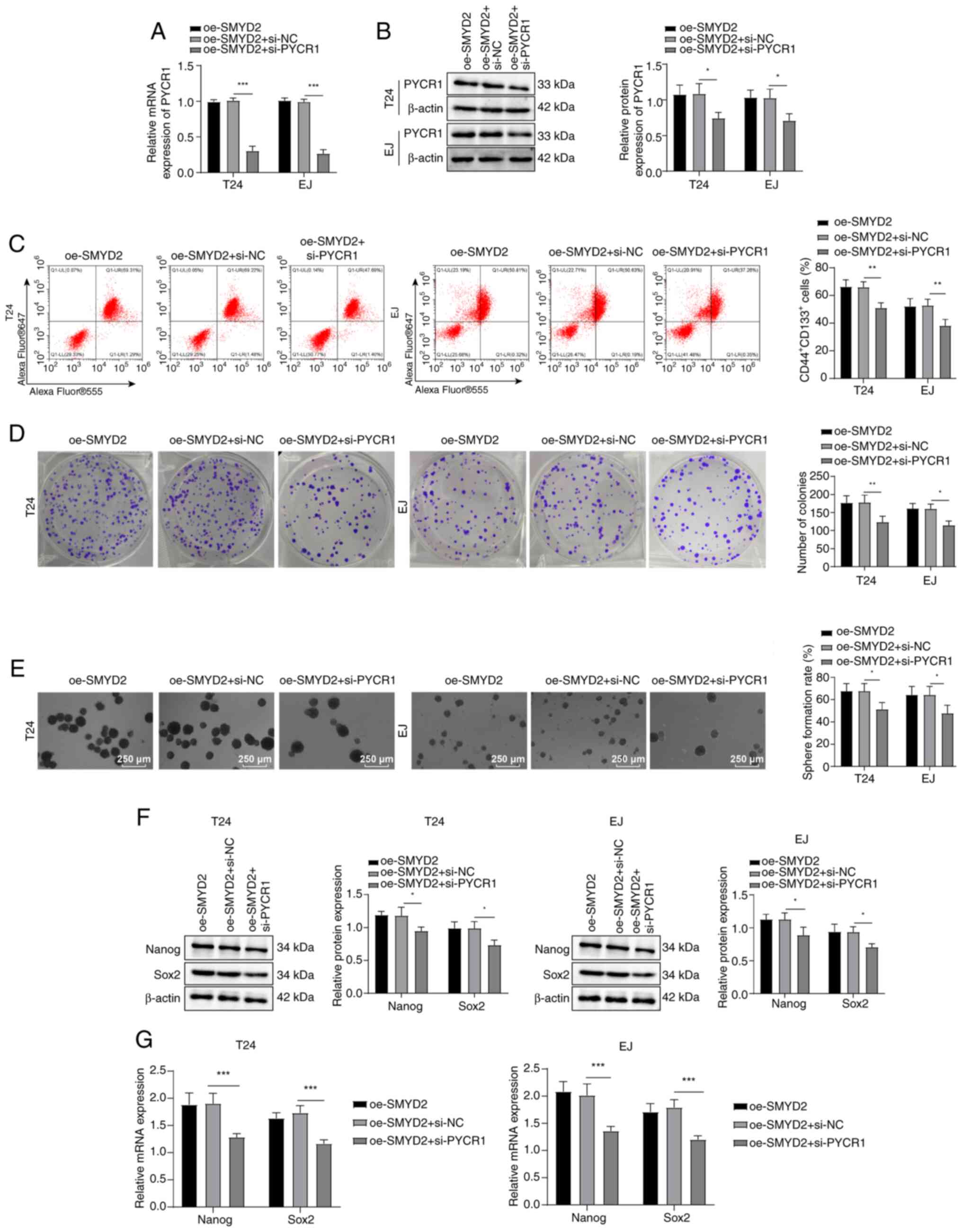
SMYD2 facilitates BCSC stemness
maintenance by regulating PYCR1
To explore whether SMYD2 can promote stemness
maintenance of BCSCs by modulating PYCR1, BC cells were transfected
with pcDNA3.1-SMYD2 and PYCR1 siRNA. Compared with oe-SMYD2 + si-NC
group, cells in the oe-SMYD2 + si-PYCR1 group exhibited decreased
expression of PYCR1 mRNA and protein (Fig. 4A and B),
CD44+CD133+ cell levels (Fig. 4C), colony and tumor sphere
formation (Fig. 4D and E) and
Nanog/Sox2 expression Fig. 4F and
G). These results suggested that SMYD2 contributed to BCSC
stemness maintenance by modulating expression of PYCR1.
PYCR1 potentiates BCSC stemness
maintenance via the PINK1/Parkin mitophagy pathway
Following PYCR1 siRNA treatment, mitochondrial LC3B
expression (Fig. 5A) and LC3B
II/I ratio decreased and p62 protein expression was augmented
(Fig. 5B); these alterations were
reversed following pcDNA3.1-PYCR1 transfection. Additionally,
following treatment with PYCR1 siRNA, expression of PINK1 and
Parkin proteins was decreased; after transfection of
pcDNA3.1-PYCR1, expression of PINK1 and Parkin proteins increased
(Fig. 5B). BC cells were
transfected with PINK1 siRNA and pcDNA3.1-PYCR1. In comparison with
the oe-PYCR1 + si-NC group, oe-PYCR1 + si-PINK1 group exhibited
reduced PINK1/Parkin protein (Fig.
5B) and LC3B expression (Fig.
5A) and LC3B II/I ratio, raised p62 protein expression
(Fig. 5B) and inhibited
CD44+CD133+ cell levels (Fig. 5C), formation of tumor spheres and
colonies (Fig. 5D and E) and
expression of Sox2 and Nanog (Fig. 5F
and G). Collectively, these findings demonstrated that PYCR1
enhanced BCSC stemness maintenance via regulation of the
PINK1/Parkin mitophagy pathway.
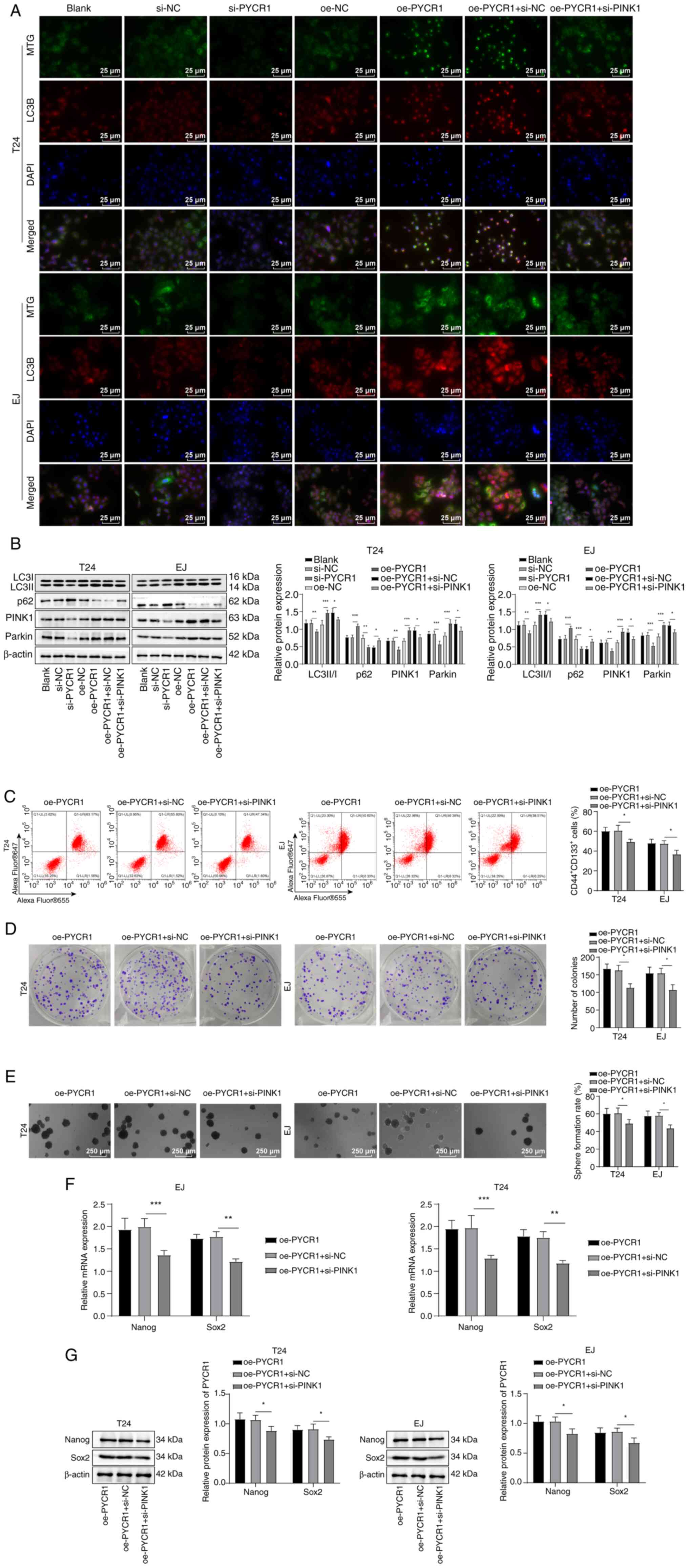 | Figure 5PYCR1 promotes bladder CSC stemness
sustenance via the PINK1/Parkin pathway. (A) Immunofluorescence
detection of mitochondrial (MTG; green) and autophagy marker (LC3B;
red) expression. (B) Assessment of the protein levels of LC3B II/I,
p62, PINK1 and Parkin by western blot. (C) Measurement of
CD44+CD133+ cell level by flow cytometry. (D)
Colony formation assay. (E) CSC sphere-forming assay to estimate
tumor stemness. Expression of stemness marker proteins Nanog and
Sox2 (F) mRNA and (G) protein was assessed by reverse
transcription-quantitative PCR and western blot.
*P<0.05, **P<0.01,
***P<0.001. SMYD2, SET and MYND domain-containing
protein 2; PYRC1, pyrroline-5-carboxylate reductase 1; si,
small-interfering; NC, negative control; oe, overexpression; PINK1,
PTEN-induced putative kinase 1; MTG, Mito-Tracker Green; CSC,
cancer stem cell. |
SMYD2 strengthens BCSC stemness
sustenance by activating the PINK1/Parkin mitophagy pathway via
upregulation of PYCR1
A BC xenograft model was established for in
vivo validation experiments by subcutaneously injecting T24
cells transfected with pcDNA3.1-SMYD2 or si-PYCR1 Lv plasmid into
nude mice. Tumor size and weight in the BC + Lv-oe-SMYD2 group were
higher than those in the BC + Lv-oe-NC group; tumor weight and size
in the BC + Lv-oe-SMYD2 + Lv-si-PYCR1 group were lower than those
in the BC + Lv-oe-SMYD2 + si-NC group (Fig. 6A-C). HE staining showed that
compared with the BC + Lv-oe-NC group, the tumor cell density of
the BC + Lv-oe-SMYD2 group was elevated and the proportion of
necrotic cells (unstructured eosinophilic substances) were
decreased; compared with the BC + Lv-oe-SMYD2 + si-NC group, the
tumor cell density of nude mice in the BC + Lv-oe-SMYD2 +
Lv-si-PYCR1 group was decreased and the proportion of necrotic
cells increased (Fig. 6D). By
contrast with the BC + Lv-oe-NC group, protein levels of SMYD2,
PYCR1, PINK1 and Parkin in tissue homogenate of nude mice in the BC
+ Lv-oe-SMYD2 group were elevated (Fig. 6E). SMYD2, Parkin, PYCR1 and PINK1
expression in the BC + Lv-oe-SMYD2 + Lv-si-PYCR1 group was lower
than in the BC + Lv-oe-SMYD2 + si-NC group (Fig. 6E). As demonstrated by IHC results,
SMYD2-, H3K4me3-, CD44- and CD133-positive cell numbers in the BC +
Lv-oe-SMYD2 group were significantly augmented relative to the BC +
Lv-oe-NC group; nude mice in the BC + Lv-oe-SMYD2 + Lv-si-PYCR1
group displayed lower CD133- and CD44-positive cell numbers than
those in the BC + Lv-oe-SMYD2 + si-NC group (Fig. 6F). Western blot results revealed
that nude mice in the BC + Lv-oe-SMYD2 group exhibited a higher
LC3B II/I ratio and lower p62 protein levels than the BC + Lv-oe-NC
group; BC + Lv-oe-SMYD2 + Lv-si-PYCR1 group exhibited a higher LC3B
II/I ratio and lower p62 protein expression compared with BC +
Lv-oe-SMYD2 + si-NC group (Fig.
6G). Moreover, Nanog and Sox2 protein levels were raised in the
BC + Lv-oe-SMYD2 group vs. BC + Lv-oe-NC group but these protein
levels dropped in the BC + Lv-oe-SMYD2 + Lv-si-PYCR1 compared with
BC + Lv-oe-SMYD2 + si-NC group (Fig.
6H and I). In summary, these results indicated that SMYD2
sustained BCSC stemness by increasing PYCR1 expression to stimulate
the PINK1/Parkin mitophagy pathway.
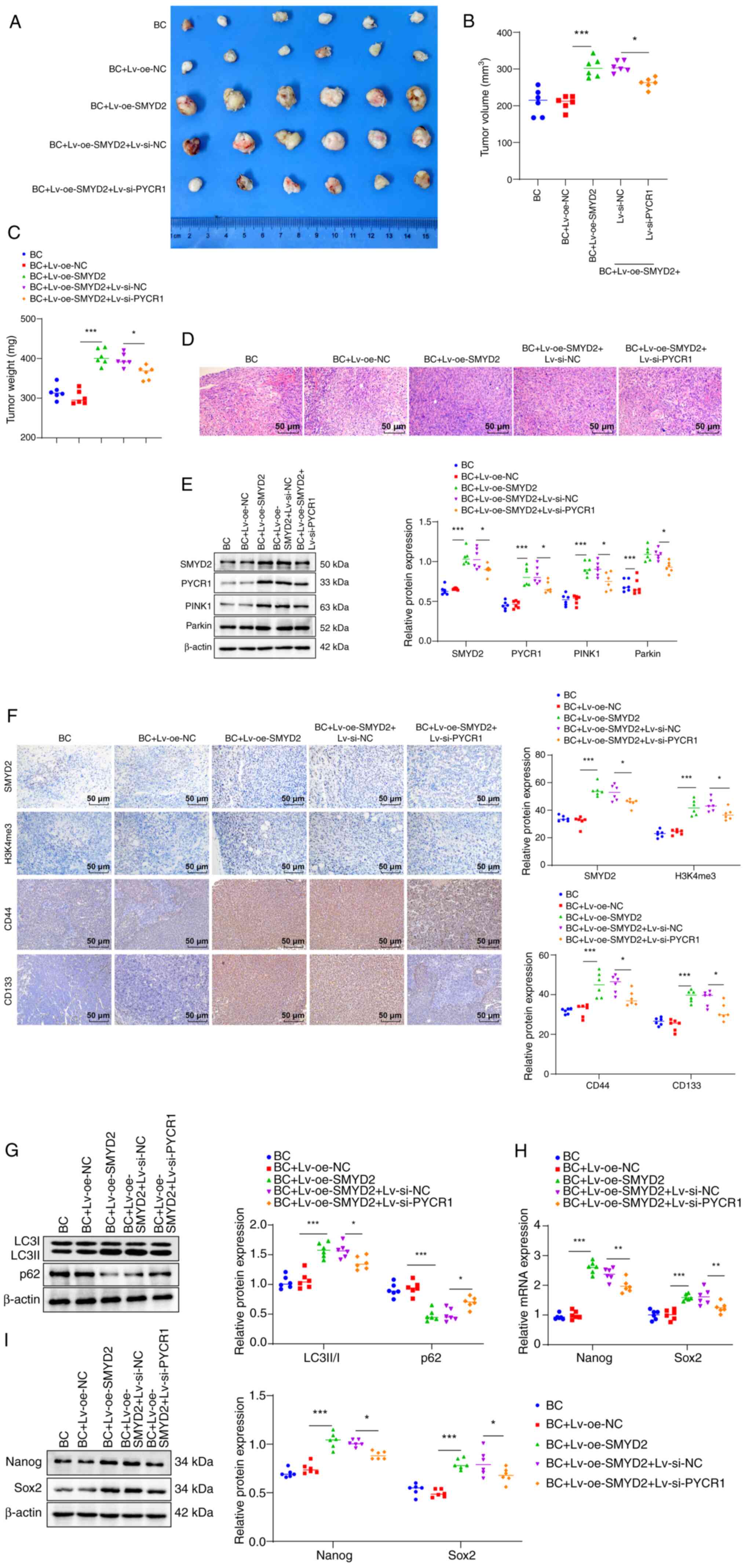 | Figure 6SMYD2 increases the maintenance of
bladder cancer stem cell stemness by upregulating PYCR1 to
stimulate the PINK1/Parkin pathway. Tumor (A) size, (B) volume and
(C) weight. (D) Hematoxylin-eosin staining to detect pathological
changes in tumor tissue. (E) Western blot to measure SMYD2, PYCR1,
PINK1 and Parkin levels in nude mouse tissue homogenate. (F)
Immunohistochemistry was performed to detect the number of SMYD2-,
H3K4me3-, CD44- and CD133-positive cells in tumor tissues. (G)
Protein expression of autophagy markers LC3B II/I and p62 was
determined by western blotting. (H) Expression of stemness marker
proteins Nanog and Sox2 mRNA was measured by reverse
transcription-quantitative PCR. (I) Western blot assay to detect
protein levels of Nanog and Sox2 in tissue homogenates of nude
mice. *P<0.05, **P<0.01,
***P<0.001. SMYD2, SET and MYND domain-containing
protein 2; PYCR1, pyrroline-5-carboxylate reductase 1; PINK1,
PTEN-induced putative kinase 1; H3K4me3, histone H3 lysine 4
trimethylation; BC, bladder cancer; Lv, lentiviral; oe,
overexpression; NC, negative control; si, small-interfering. |
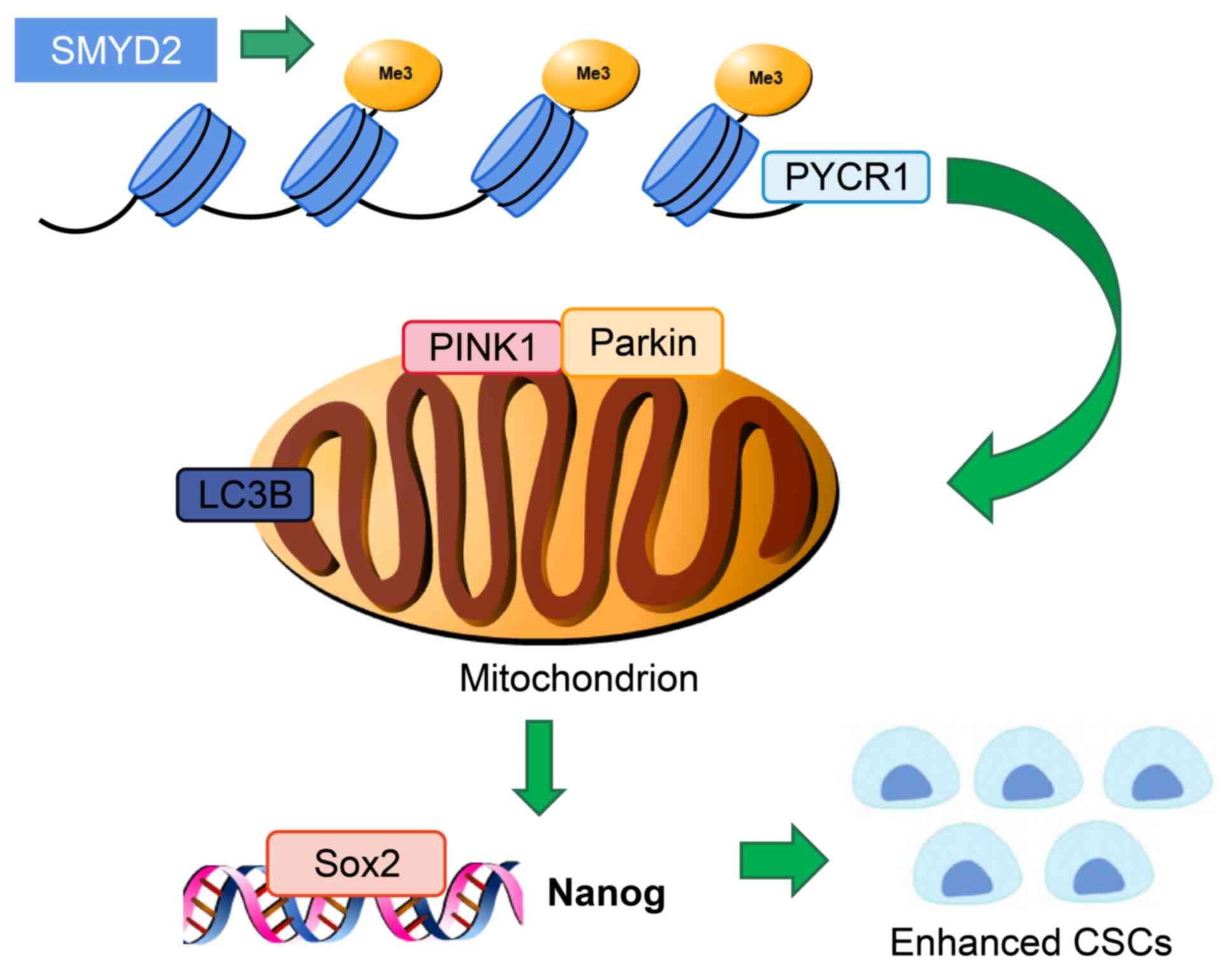
Discussion
Lysine methyltransferases, which affect numerous
oncoproteins and tumor suppressor proteins, play a key role in the
development of various types of malignancies, such as colorectal
cancer and lung cancer (25-27). SMYD2, an extensively studied
lysine methyltransferase, is associated with regulation of
transcription, epigenetics and tumorigenesis (28,29). However, the potential impact of
SMYD2 on the stemness maintenance of BCSCs remains unclear. The
present study demonstrated that histone methyltransferase SMYD2 is
involved in the stemness of BCSCs by upregulating PYCR1 expression
and activating the PINK1/Parkin pathway (Fig. 7).
PYCR1 strengthens BC proliferation and EMT, which is
associated with expression of stemness markers Nanog and Sox2
(4,30), suggesting that PYCR1 plays a role
in BCSC stemness. GEPIA database analysis demonstrated that SMYD2
was significantly highly expressed in BC. Correspondingly, SMYD2
expression is significantly increased in human BC compared with
normal bladder tissue, underscoring that inhibiting SMYD2 may have
therapeutic promise for BC (31).
The present study assessed the mechanism of SMYD2/PYCR1 in
maintaining BCSC stemness. CD44, a hallmark of BCSCs, indicates
stemness and activates many signaling pathways to maintain
self-renewal capacity (32,33). The potential role of CD44 and
CD133 as markers for BCSCs has been proposed (34). PYCR1 and SMYD2 knockdown in
CD44+CD133+ BCSC subpopulation with stem
cell-like characteristics decreased
CD44+CD133+ cell levels, Nanog and Sox2
expression and colony and sphere formation, whereas overexpressing
PYCR1 or SMYD2 resulted the opposite effects. Similarly,
suppression of PYCR1 in vitro decreases colony formation and
proliferation and induces cell cycle arrest (35). Cui et al (20) confirmed that ablation of PYCR1 can
lead to reduced expression of Sox2 and Nanog, CSC-associated
aldehyde dehydrogenase+ population and ability of breast
CSCs to form spheres, suggesting that PYCR1 is involved in
preserving the stemness of breast cancer cells. In addition, Shang
and Wei (36) demonstrated that
inhibition of SMYD2 impedes tumor sphere formation and cell
migration in non-small-cell lung cancer cells. To the best of our
knowledge, the impact of SMYD2 on the stemness of BCSCs has not
been reported. It was hypothesized that both SMYD2 and PYCR1
amplified maintenance of stemness in BCSCs.
H3K4me3 is abnormally expressed in lung, liver and
colon cancer (37-39). SMYD2 serves as a histone
methyltransferase of H3K4me3, facilitating transcriptional
activation of its downstream target genes by catalyzing
trimethylation of histone H3K4, which is associated with modulation
of target gene transcription (12,40). A notable positive association
between SMYD2 and PYRC1 was observed in the present study. To the
best of our knowledge, the present study is the first to
demonstrate that SMYD2 may promote PYCR1 expression via H3K4me3;
in vitro demonstrated increased PYCR1 and H3K4me3 levels,
along with heightened enrichment levels of H3K4me3 in the PYCR1
promoter region in SMYD2-overexpressing BC cells. Furthermore,
previous research has confirmed that proline produced by PYCR1 can
activate the cGMP/cGMP-dependent protein kinase signaling pathway,
thereby enhancing breast cancer stemness (20). Here, SMYD2 heightened stemness in
BCSCs by regulating PYCR1.
The PINK1/Parkin signaling pathway is one of the
primary regulatory mechanisms of mitophagy (17). The accumulation of PINK1 augments
PINK1/Parkin-mediated mitophagy, which efficiently removes impaired
mitochondria (41). The
feedforward signaling pathway, including PINK1 and Parkin, is
responsible for facilitating mitophagy (42). Here, PYCR1 silencing resulted in
elevated p62 and Parkin/PINK1 expression, as well as lessened LC3B
expression and LC3B II/I ratios, which were negated by PYCR1
overexpression. This indicated that PYCR1 could mediate the
Parkin/PINK1 mitophagy axis. Moreover, it is well-established that
mitophagy encourages the obtainment of tumor cell stemness
(14,43). The present experiments verified
the promoting effect of the PINK1/Parkin mitophagy pathway in BCSC
stemness. Other investigations have established a connection
between mitophagy and SC stemness (44,45). Augmentation of mitophagy can
contribute to the stemness of mesenchymal SCs (46). Conversely, the reduction of
mitophagy-related PINK1/Parkin leads to diminished stemness of bone
marrow-derived mesenchymal SCs, as indicated by decreased
expression of stemness markers Sox2 and Octamer-binding
transcription factor 4 (44).
Here, PYCR1 stimulated stemness maintenance in BCSCs via the
PINK1/Parkin mitophagy pathway. Furthermore, in vivo
verification demonstrated that SMYD2 preserved BCSC stemness by
activating the PINK1/Parkin mitophagy pathway via enhancement of
PYCR1 expression.
In conclusion, the present study demonstrated the
regulatory effect and mechanism by which SMYD2 affects PYCR1.
Additionally, in vitro and in vivo experiments
demonstrated the role of PYCR1 in modulating the PINK1/Parkin
signaling pathway, which affects mitophagy and regulates stemness
of BCSCs. However, the present study only verified that PYCR1
regulates mitophagy via regulation of PINK1/Parkin to regulate BCSC
stemness; other target genes and signaling pathways regulated by
PYCR1 require further investigation. Upstream influencing factors
of PYCR1 also need to be further explored. Future investigations
investigate the role of mitophagy in preserving the stemness of
CSCs, as well as the mechanisms by which PYCR1 modulates other
target genes and signaling pathways.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
All authors have read and approved the final
manuscript. JC contributed to the study concepts, study design; SX
and XY performed the literature review. JC and YW performed
experiments. JC and WS contributed to the data analysis. JC and SX
wrote and revised the manuscript. JC and SX confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was reviewed and approved by the
Animal Ethics Committee of Hunan Provincial People's Hospital, The
First Affiliated Hospital of Hunan Normal University (approval no.
2024-84).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by Natural Science Foundation of
Hunan Province (grant no. 2025JJ50640), Hunan Provincial Health
High-Level Talent Scientific Research Project (grant no. R2023154)
and Hunan Provincial People's Hospital Doctoral Fund Project (grant
no. BSJJ202106).
References
|
1
|
Chen S, Zhu J, Wang F, Guan Z, Ge Y, Yang
X and Cai J: LncRNAs and their role in cancer stem cells.
Oncotarget. 8:110685–110692. 2017. View Article : Google Scholar
|
|
2
|
Eid RA, Alaa Edeen M, Shedid EM, Kamal AS,
Warda MM, Mamdouh F, Khedr SA, Soltan MA, Jeon HW, Zaki MS and Kim
B: Targeting Cancer Stem Cells as the Key Driver of Carcinogenesis
and Therapeutic Resistance. Int J Mol Sci. 24:2023. View Article : Google Scholar
|
|
3
|
Aghaalikhani N, Rashtchizadeh N, Shadpour
P, Allameh A and Mahmoodi M: Cancer stem cells as a therapeutic
target in bladder cancer. J Cell Physiol. 234:3197–3206. 2019.
View Article : Google Scholar
|
|
4
|
Li Z, Liu J, Fu H, Li Y, Liu Q, Song W and
Zeng M: SENP3 affects the expression of PYCR1 to promote bladder
cancer proliferation and EMT transformation by deSUMOylation of
STAT3. Aging (Albany NY). 14:8032–8045. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song W, Li Z, Yang K, Gao Z, Zhou Q and Li
P: Antisense lncRNA-RP11-498C9.13 promotes bladder cancer
progression by enhancing reactive oxygen species-induced mitophagy.
J Gene Med. 25:e35272023. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song W, Yang K, Luo J, Gao Z and Gao Y:
Dysregulation of USP18/FTO/PYCR1 signaling network promotes bladder
cancer development and progression. Aging (Albany NY).
13:3909–3925. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Zhang X, Huang X, Tang X, Zhang
M, Li Z, Hu X, Zhang M, Wang X and Yan Y: Tumor stemness score to
estimate epithelial-to-mesenchymal transition (EMT) and cancer stem
cells (CSCs) characterization and to predict the prognosis and
immunotherapy response in bladder urothelial carcinoma. Stem Cell
Res Ther. 14:152023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye Y, Li L, Dai Q, Liu Y and Shen L:
Comprehensive analysis of histone methylation modification
regulators for predicting prognosis and drug sensitivity in lung
adenocarcinoma. Front Cell Dev Biol. 10:9919802022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McCabe MT, Mohammad HP, Barbash O and
Kruger RG: Targeting Histone Methylation in Cancer. Cancer J.
23:292–301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan Z, Fu S, Feng R, Huang Y, Li N, Wang H
and Wang J: Identification of potential biomarkers for progression
and prognosis of bladder cancer by comprehensive bioinformatics
analysis. J Oncol. 2022:18027062022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu W, Chen F, Fei X, Yang X and Lu X:
Overexpression of SET and MYND domain-containing protein 2 (SMYD2)
is associated with tumor progression and poor prognosis in patients
with papillary thyroid carcinoma. Med Sci Monit. 24:7357–7365.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu H, Ba Z, Liu C and Yu X: Long noncoding
RNA DLEU1 promotes proliferation and glycolysis of gastric cancer
cells via APOC1 upregulation by recruiting SMYD2 to induce
trimethylation of H3K4 modification. Transl Oncol. 36:1017312023.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Whelan KA, Chandramouleeswaran PM, Tanaka
K, Natsuizaka M, Guha M, Srinivasan S, Darling DS, Kita Y, Natsugoe
S, Winkler JD, et al: Autophagy supports generation of cells with
high CD44 expression via modulation of oxidative stress and
Parkin-mediated mitochondrial clearance. Oncogene. 36:4843–4858.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu K, Lee J, Kim JY, Wang L, Tian Y, Chan
ST, Cho C, Machida K, Chen D and Ou JJ: Mitophagy controls the
activities of tumor suppressor p53 to regulate hepatic cancer stem
cells. Mol Cell. 68:281–292. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Panigrahi DP, Praharaj PP, Bhol CS,
Mahapatra KK, Patra S, Behera BP, Mishra SR and Bhutia SK: The
emerging, multifaceted role of mitophagy in cancer and cancer
therapeutics. Semin Cancer Biol. 66:45–58. 2020. View Article : Google Scholar
|
|
16
|
Lou Y, Ma C, Liu Z, Shi J, Zheng G, Zhang
C and Zhang Z: Antimony exposure promotes bladder tumor cell growth
by inhibiting PINK1-Parkin-mediated mitophagy. Ecotoxicol Environ
Saf. 221:1124202021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nguyen TN, Padman BS and Lazarou M:
Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends
Cell Biol. 26:733–744. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee J, Liu K, Stiles B and Ou JJ:
Mitophagy and hepatic cancer stem cells. Autophagy. 14:715–716.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Choudhury D, Rong N, Senthil Kumar HV,
Swedick S, Samuel RZ, Mehrotra P, Toftegaard J, Rajabian N,
Thiyagarajan R, Podder AK, et al: Proline restores mitochondrial
function and reverses aging hallmarks in senescent cells. Cell Rep.
43:1137382024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui B, He B, Huang Y, Wang C, Luo H, Lu J,
Su K, Zhang X, Luo Y, Zhao Z, et al: Pyrroline-5-carboxylate
reductase 1 reprograms proline metabolism to drive breast cancer
stemness under psychological stress. Cell Death Dis. 14:6822023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zietzer A, Hosen MR, Wang H, Goody PR,
Sylvester M, Latz E, Nickenig G, Werner N and Jansen F: The
RNA-binding protein hnRNPU regulates the sorting of microRNA-30c-5p
into large extracellular vesicles. J Extracell Vesicles.
9:17869672020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Han Y, Liu C, Zhang D, Men H, Huo L, Geng
Q, Wang S, Gao Y, Zhang W, Zhang Y and Jia Z: Mechanosensitive ion
channel Piezo1 promotes prostate cancer development through the
activation of the Akt/mTOR pathway and acceleration of cell cycle.
Int J Oncol. 55:629–644. 2019.PubMed/NCBI
|
|
24
|
Dai X, Ren T, Zhang Y and Nan N:
Methylation multiplicity and its clinical values in cancer. Expert
Rev Mol Med. 23:e22021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Komatsu S, Ichikawa D, Hirajima S, Nagata
H, Nishimura Y, Kawaguchi T, Miyamae M, Okajima W, Ohashi T,
Konishi H, et al: Overexpression of SMYD2 contributes to malignant
outcome in gastric cancer. Br J Cancer. 112:357–364. 2015.
View Article : Google Scholar :
|
|
26
|
Zhang Y, Zhou L, Xu Y, Zhou J, Jiang T,
Wang J, Li C, Sun X, Song H and Song J: Targeting SMYD2 inhibits
angiogenesis and increases the efficiency of apatinib by
suppressing EGFL7 in colorectal cancer. Angiogenesis. 26:1–18.
2023. View Article : Google Scholar
|
|
27
|
Kim K, Ryu TY, Jung E, Han TS, Lee J, Kim
SK, Roh YN, Lee MS, Jung CR, Lim JH, et al: Epigenetic regulation
of SMAD3 by histone methyltransferase SMYD2 promotes lung cancer
metastasis. Exp Mol Med. 55:952–964. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sakamoto LH, Andrade RV, Felipe MS,
Motoyama AB and Pittella Silva F: SMYD2 is highly expressed in
pediatric acute lymphoblastic leukemia and constitutes a bad
prognostic factor. Leuk Res. 38:496–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yi X, Jiang XJ and Fang ZM: Histone
methyltransferase SMYD2: Ubiquitous regulator of disease. Clin
Epigenetics. 11:1122019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Migita T, Ueda A, Ohishi T, Hatano M,
Seimiya H, Horiguchi SI, Koga F and Shibasaki F:
Epithelial-mesenchymal transition promotes SOX2 and NANOG
expression in bladder cancer. Lab Invest. 97:567–576. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cho HS, Hayami S, Toyokawa G, Maejima K,
Yamane Y, Suzuki T, Dohmae N, Kogure M and Kang D: RB1 methylation
by SMYD2 enhances cell cycle progression through an increase of RB1
phosphorylation. Neoplasia. 14:476–486. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao RL, Chen XR, Li YN, Yan XY, Sun JG, He
QL and Cai FZ: Upregulation of miR-543-3p promotes growth and stem
cell-like phenotype in bladder cancer by activating the
Wnt/β-catenin signaling pathway. Int J Clin Exp Pathol.
10:9418–9426. 2017.
|
|
33
|
Hu Y, Zhang Y, Gao J, Lian X and Wang Y:
The clinicopathological and prognostic value of CD44 expression in
bladder cancer: A study based on meta-analysis and TCGA data.
Bioengineered. 11:572–581. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xia P, Liu DH, Xu ZJ and Ren F: Cancer
stem cell markers for urinary carcinoma. Stem Cells Int.
2022:36116772022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cai F, Miao Y, Liu C, Wu T, Shen S, Su X
and Shi Y: Pyrroline-5-carboxylate reductase 1 promotes
proliferation and inhibits apoptosis in non-small cell lung cancer.
Oncol Lett. 15:731–740. 2018.PubMed/NCBI
|
|
36
|
Shang L and Wei M: Inhibition of SMYD2
sensitized cisplatin to resistant cells in NSCLC through activating
p53 pathway. Front Oncol. 9:3062019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin X, Chen JD, Wang CY, Cai Z, Zhan R,
Yang C, Zhang LY, Li LY, Xiao Y, Chen MK and Wu M: Cooperation of
MLL1 and Jun in controlling H3K4me3 on enhancers in colorectal
cancer. Genome Biol. 24:2682023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Phoyen S, Sanpavat A, Ma-On C, Stein U,
Hirankarn N, Tangkijvanich P, Jindatip D, Whongsiri P and Boonla C:
H4K20me3 upregulated by reactive oxygen species is associated with
tumor progression and poor prognosis in patients with
hepatocellular carcinoma. Heliyon. 9:e225892023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou Z, Zhang B, Deng Y, Deng S, Li J, Wei
W, Wang Y, Wang J, Feng Z, Che M, et al: FBW7/GSK3 β mediated
degradation of IGF2BP2 inhibits IGF2BP2-SLC7A5 positive feedback
loop and radioresistance in lung cancer. J Exp Clin Cancer Res.
43:342024. View Article : Google Scholar
|
|
40
|
Abu-Farha M, Lambert JP, Al-Madhoun AS,
Elisma F, Skerjanc IS and Figeys D: The tale of two domains:
proteomics and genomics analysis of SMYD2, a new histone
methyltransferase. Mol Cell Proteomics. 7:560–572. 2008. View Article : Google Scholar
|
|
41
|
Wang S, Long H, Hou L, Feng B, Ma Z, Wu Y,
Zeng Y, Cai J, Zhang DW and Zhao G: The mitophagy pathway and its
implications in human diseases. Signal Transduct Target Ther.
8:3042023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Silvian LF: PINK1/parkin pathway
activation for mitochondrial quality control-which is the best
molecular target for therapy? Front Aging Neurosci. 14:8908232022.
View Article : Google Scholar
|
|
43
|
Yuan X, Chen K, Zheng F, Xu S, Li Y, Wang
Y, Ni H, Wang F, Cui Z, Qin Y, et al: Low-dose BPA and its
substitute BPS promote ovarian cancer cell stemness via a
non-canonical PINK1/p53 mitophagic signaling. J Hazard Mater.
452:1312882023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Feng X, Yin W, Wang J, Feng L and Kang YJ:
Mitophagy promotes the stemness of bone marrow-derived mesenchymal
stem cells. Exp Biol Med (Maywood). 246:97–105. 2021. View Article : Google Scholar
|
|
45
|
Liu D, Sun Z, Ye T, Li J, Zeng B, Zhao Q,
Wang J and Xing HR: The mitochondrial fission factor FIS1 promotes
stemness of human lung cancer stem cells via mitophagy. FEBS Open
Bio. 11:1997–2007. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Vazquez-Martin A, Van den Haute C, Cufi S,
Corominas-Faja B, Cuyàs E, Lopez-Bonet E, Rodr iguez-Gallego E,
Fernández-Arroyo S, Joven J, Baekelandt V and Menendez JA:
Mitophagy-driven mitochondrial rejuvenation regulates stem cell
fate. Aging (Albany NY). 8:1330–1352. 2016. View Article : Google Scholar : PubMed/NCBI
|