Introduction
Ferroptosis differs from apoptosis, autophagy and
necrotic apoptosis (1). It is a
type of programmed cell death consisting of lipid peroxidation
induced by iron-dependent ions and reactive oxygen species (ROS)
(2,3). Ferroptosis can be triggered by
blocking the Xc-system [glutamate cystine reverse transporter, a
dimer consisting of solute carrier (SLC)7A11 and SLC3A2, which
mainly relies on SLC7A11 for its function] or by inhibiting
glutathione (GSH)-dependent antioxidant reductase 4 (GPX4)
(4,5). It has been reported that ferroptosis
serves a key role in neurodegenerative diseases,
ischemia-reperfusion injury, acute kidney injury and several types
of cancer (6). Moreover, it has
been reported that accumulation of Fe2+ notably
contributes to the development of several cancers such as lung,
liver and breast cancers (7,8).
Cancer chemotherapeutic resistance is currently a major challenge
in cancer treatment, and ferroptosis serves a pivotal role in
cancer drug resistance, which in recent years has provided new
opportunities for chemotherapy of insensitive cancers (9). Therefore, modulation of key
ferroptosis genes to induce ferroptosis has emerged as a
therapeutic strategy to combat cancer progression and chemotherapy
resistance (10,11).
Glioma (neuroglioma) is currently the most common
intracranial primary cancer in adults worldwide (12,13). Glioblastoma (GBM) is a highly
aggressive cerebral malignant cancer characterized by massive
neovasculogenesis, necrosis and a strong resistance to treatment
(14). However, ferroptosis is
understudied in gliomas. It has been reported that after analyzing
five major types of programmed cell death in 1,750 patients with
glioma from four independent databases, ferroptosis was identified
as the most common type of programmed cell death (encompassing
apoptosis, autophagy, ferroptosis, necrotic apoptosis and
pyroptosis) in gliomas, and ferroptosis was associated with
malignant cancer progression, a poor outcome and immunosuppression
(15). Moreover, it has been
reported that chemotherapeutics drugs (16) and cryptotanshinone (17) can have an effect on glioma cell
ferroptosis (18). In addition,
previous studies demonstrated that polypyrimidine tract binding
protein 1 and NADPH regulate glioma ferroptosis by modulating
oxidative stress pathways (19,20). Several recent studies have also
reported that nanoparticle drugs (21) can target GBM across the
blood-brain barrier and markedly optimize antitumor efficacy by
activating ferroptosis (22,23). Furthermore, certain researchers
have used nanosensitizers (24)
to disrupt drug resistance mechanisms in GBM, providing new
therapeutic avenues for glioma treatment (25). A study reported that circLRFN5
promotes ferroptosis by inhibiting the paired related homeobox
2/GTP cyclohydrolase 1 pathway, which in turn inhibits GBM
progression (26). Nevertheless,
although the role of ferroptosis in the pathogenesis of glioma and
in targeted therapies has been studied in recent years, research
has made limited progress. Moreover, exploring the role and
mechanism of ferroptosis in the development of gliomas can help to
improve glioma treatment.
The zinc finger family of proteins is one of the
most important families regulating gene transcription in eukaryotes
(27). Zinc finger and BTB
domain-containing protein 20 (ZBTB20) is a member of this
family whose main function is transcriptional repression (28). It may also serve an important role
in hematopoiesis, oncogenesis and development, immune response and
neurodevelopment (29). Previous
studies have reported that mutations in the ZBTB20 gene can
lead to primrose syndrome (30,31). Furthermore, it has been reported
that ZBTB20 gene deletion can lead to anterior pituitary
hypoplasia, developmental dwarfism and complete loss of mature
prolactin (32). In addition,
ZBTB20 serves a crucial role in the development of several
cancers (33). It has also been
reported that ZBTB20 can promote the growth of human
hepatocellular carcinoma through the inhibition of forkhead box O1
(34,35), and ZBTB20 promotes cell
migration and invasion in gastric cancer through the inhibition of
IKBα-induced NF-κB activation (30). In neural precursor cells, the
overexpression of ZBTB20 has been reported to promote cell
differentiation to astrocytes, and knockdown of ZBTB20
inhibits cell differentiation to astrocytes (36). However, no association between
ZBTB20 and ferroptosis in glioma cells has been reported, to the
best of our knowledge. However, it has been demonstrated that, in
glioma cells, ZBTB20 regulates the expression of oxidative
stress-related proteins, which are important genes for ROS
regulation (37). As ROS has been
reported to be an important causative agent and signature feature
in the process of ferroptosis (38,39), it was hypothesized that
ZBTB20 may be associated with the process of ferroptosis in
glioma cells.
Transmembrane protein 109 (TMEM109), also
known as Mitsugumin23, is a transmembrane protein found in the
sarcoplasmic reticulum, endoplasmic reticulum (ER) and nuclear
membrane of cells (40-43). It is involved in regulating the
activity of voltage-gated ion channels and it has been reported
that knockdown of TMEM109 increases ultraviolet C-induced
cellular DNA damage, leading to cell death (44). Previous research has also reported
that TMEM109 can promote Ca2+ release in skeletal
muscle to cause muscle contraction (41). There are few studies on
TMEM109, with one reporting that the ferroptosis pathway was
altered in human cervical cancer cells with TMEM109
knockdown (40). Therefore, it
was hypothesized that TMEM109 may be associated with
ferroptosis.
To date, the association between ZBTB20 and
TEMEM109 is unclear. Therefore, the present study aimed to
assess the role of ZBTB20 and TMEM109 in GBM cells,
and the association between ZBTB20, ferroptosis and TMEM109 in
GBM.
Materials and methods
Cell cultures and reagents
The U251 glioma cells (cat no. TCHu58) and U87MG
cells [cat. no. TCHu138], the cell line as a GBM of unknown origin)
were purchased from the National Collection of Authenticated Cell
Cultures. The cell line was authenticated by short tandem repeat
analysis and subjected to mycoplasma detection at the National
Collection of Authenticated Cell Cultures. The normal human glial
cells HA1800 (cat no. 1800; ScienCell; https://sciencellonline.com/en/human-astrocytes/)
and glioma cells were cultured in high-sugar DMEM medium
(Biological Industries; Sartorius AG) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin
(Corning, Inc.) and 1% streptomycin (Corning, Inc.) at 37°C in a 5%
CO2 incubator (Thermo Fisher Scientific, Inc.).
ZBTB20 overexpression and knockdown
transfection
The lentiviral supernatants used for ZBTB20
overexpression and knockdown were purchased from Shanghai Jikai
Genetics Corporation. The biotech company has not disclosed the
sequence of the negative control (NC) short hairpin RNA (shRNA).
The target sequences of ZBTB20 knockdown were as follows:
shZBTB20-1, 5'-GCATGTGTCTGACGGATAAGT-3'; shZBTB20-2,
5'-GCACTGGACTTCAGGATAAGT-3'; and shZBTB20-3,
5'-GCCAAACACTTCTAGAGAAAT-3'. The control group (Vector group) of
the overexpression group (ZBTB20 group) was transfected with cells
containing empty pReceiver-Lv105 lentivirus, and the control group
(shZBTB20 group) of the knockdown group (shZBTB20 group) was
transfected with cells containing psi-LVRU6GP with a scrambled
sequence (5'-GCTTCGCGCCGTAGTCTTA-3'; GeneCopoeia, Inc.). When the
cell confluence reached 50%, the viral transfection operation
(MOI=10) was carried out. A total of 48 h after transfection, the
fluorescence intensity was observed under microscope to evaluate
the transfection effect. After successful transfection, the cells
can be passaged and cultured for one week before proceeding with
subsequent experimental operations.
TMEM109 vectors and lentiviral
transfection
The lentiviral supernatants used for TMEM109
overexpression and knockdown were purchased from Shanghai Jikai
Genetics Corporation. The biotech company has not disclosed the
sequence of the NC shRNA. The target sequences of TMEM109
knockdown were as follows: shTMEM109-1,
5'-GCATGTGTTCAAAGCCATTCT-3'; shTMEM109-2,
5'-GCCATCTCATCAGCCATTTCT-3'; and shTMEM109-3,
5'-ACCCAGATAGGTCGATCTGTG-3'. Uniformly planted glioma cells with
favorable growth status were cultured in 6-well plates (Wuxi NEST
Biotechnology Co., Ltd.), with ~2×105 cells per well.
Once the cells adhered to the plate and reached ~60% confluence,
the old medium was discarded. Newly prepared lentiviral mixture was
then added to the cells, which consisted of DMEM, lentiviral
solution and transfection reagent. The mixture was placed in an
incubator and incubation was continued. After 48 h, the culture
plate was removed and the fluorescence intensity was observed under
the microscope to determine the infection efficiency of the cells.
The old medium containing the virus solution was aspirated and
replaced with serum-free DMEM to continue culturing and passaging.
Half of the infected cells were used to determine infection
efficiency, whilst the remaining cells were screened for stable
infection using neomycin or puromycin (2 μg/ml) screening,
depending on the virus design. Blank control wells were set up, and
screening was stopped when the cells in the blank group were
completely eliminated.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells that had been
administered TRIzol® reagent (Corning, Inc.) after being
washed three times with PBS. The RNA was then used to create cDNA
using reverse transcription, following the instructions of the
Reverse Transcription Kit (Takara Bio, Inc.). The conditions were
set as follows: 37°C for 15 min; 85°C for 5 sec; and cooling down
to 4°C. The cDNA was amplified using an Amplification Kit for PCR
experiments (Takara Bio, Inc.). The procedure was set to tick SYBR
Green method, CT method. The conditions were set as follows: 95°C
for 5 sec; 95°C for 5 sec; 55°C for 30 sec; and 72°C for 30 sec.
The number of cycles was set to 40. The primer sequences used were
as follows: ZBTB20 forward, 5'-GACAGGATCTACTCGGCACTC-3' and
reverse, 5'-ACTGCGCCGCTGTAAAAAGA-3'; TMEM109 forward,
5'-TGGGGAAAGCATGTGTTCAAA-3' and reverse, 5'-TGGTGCAAAGTCTCGACGG-3';
and GAPDH forward, 5'-GGAAGCTTGTCATCAATGGAAATC-3' and reverse,
5'-TGATGACCCTTTTGGCTCCC-3'.
JASPAR
The promoter sequence of the target gene is obtained
from NCBI database, and the upstream 2 kb region is adjusted
according to the direction of the gene chain. The target
transcription factor was searched on the JASPAR website (https://jaspar.elixir.no/), and the appropriate motif
version was selected. Then the promoter sequence of the target gene
in FASTA format was analyzed by the JASPAR-Scan tool to find the
possible binding sites of transcription factors.
Western blotting
The cultured cells were collected using a cell
scraper and lysed using RIPA lysate (cat. no. P0013B; Beyotime
Institute of Biotechnology). The protein concentration was
determined using bicinchoninic acid (BCA) method. Proteins (20
μg/20 μl) were then extracted and solubilized using
SDS-PAGE (10% acrylamide), and then transferred onto a PVDF
membrane (MiliporeSigma; Merck KGaA). The membrane was blocked with
5% bovine serum albumin at room temperature for 2 h in TBST (0.05%
Tween; Beijing Solarbio Science & Technology Co., Ltd.) and
subsequently incubated at 4°C for 16 h in primary antibody diluent
(1:1,000-10,000) after the blocking process was completed. Western
blot analysis employed several primary antibodies, including
β-actin (cat. no. AC026; ABclonal Biotech Co., Ltd.), ZBTB20 (cat.
no. A7970; ABclonal Biotech Co., Ltd.), GPX4 (cat. no. A1933;
ABclonal Biotech Co., Ltd.), SLC7A11 (cat. no. 26864-1-AP,
Proteintech Group, Inc.) and TMEM109 (cat. no. bs-19950R; BIOSS).
Peroxidase-coupled secondary antibodies (1:1,000-2,000, cat. no.
ab6721; Abcam) were then used for 1 h incubation at room
temperature, and the signals were detected using ECL Ultra
Sensitive Luminescent Liquid (Strong; Applygen Technologies, Inc.).
Exposure imaging was performed using the fully automatic
chemiluminescence imaging system (Tanon 4600, Tanon Science and
Technology Co., Ltd.) to analyze relative intensities with the
assistance of ImageJ Lab software (ImageJ 2.0; National Institutes
of Health).
ROS fluorescence imaging
Cells were pre-cultured on 96-well plates at a
density of 3,000 cells per well for 24 h. After washing the plates
with PBS, they were incubated with DMEM containing 10 μM
dihydroethidium (cat. no. 50102ES02; Shanghai Yeasen Biotechnology
Co., Ltd.) for 2 h at room temperature or 37°C. Subsequently,
images of the plates were captured using a confocal microscope (GE
Healthcare).
Fe2+ detection experiment
Intracellular Fe2+ content was determined
using an Fe2+ content detection kit (cat. no. BC5415;
Beijing Solarbio Science & Technology Co., Ltd.). Cells were
treated with Fe2+ detection reagent 1, broken up using
ultrasonic crushing and centrifuged at 4°C (1,000 × g, 20 min).
Reagent 2 was then added and mixed and left to stand for 10 min.
The treated reagent was spread onto 96-well plates, incubated at
37°C for 30 min and then the absorbance at 593 nm was detected
using an Automatic Elisa Analyzer (PERLONG, http://www.prolong.com.cn/productinfor.php?anclss=5&nclass=5&id=5).
Finally, the Fe2+ content of the cells was calculated
according to the formula of the kit.
Malondialdehyde (MDA) detection
experiment
MDA content was determined using an MDA content
detection kit (cat. no. S0131S; Beyotime Institute of
Biotechnology). The thiobarbituric acid storage solution and MDA
detection working solution were prepared in advance and then the
standard sample was used to make a standard curve. The sample was
subsequently processed according to the instructions in the reagent
kit and an Automatic Elisa Analyzer was used to detect the
absorbance of the sample at 532 nm. Finally, the MDA content of the
cells was calculated according to the formula of the kit.
Dual luciferase reporter gene assay
Plasmids purchased from Shanghai GeneChem Co., Ltd.
were constructed using the TMEM109 gene sequence (GenBank
accession no. NC_000011.10) and the ZBTB20 gene sequence
(GenBank accession no. NC_000003.12) obtained from GenBank. These
plasmids were used to create cell lines with overexpression and
knockdown of TMEM109 for experimental purposes. 293T cells
(Stem Cell Center of Zhengzhou University) in logarithmic growth
phase were suspended and plated in 12-well plates at a density of
~2.5×105 cells per well, depending on cell morphology.
The cells were cultured overnight in a 37°C, 5% CO2
incubator. The cells were then treated with a dual luciferase
reporter gene kit (cat. no. RG027; Beyotime Institute of
Biotechnology) and divided into four groups: Promoter-NC + NC,
promoter-NC + ZBTB20, promoter-NC and promoter-ZBTB20. The prepared
DNA was added into 100 μl PBS and mixed evenly.
Subsequently, 4.8 μl HiGene transfection reagent (cat. no.
C1506; Applygen Technologies, Inc.) was applied and the mixture was
added to the well plates of the divided groups after 15 min of
mixing and resting. After 48 h, the medium was replaced with DMEM
medium and incubated further. Finally, the plate was removed and
washed with PBS three times. The appropriate amount of lysis
solution was then added to fully lyse the cells. The solution was
collected in a centrifuge tube and centrifuged at low temperature
at 12,000 × g for 5 min. The supernatant was then collected for
subsequent experiments. Subsequently, the firefly and sea kidney
luciferase reagents were configured. Then, 50 μl supernatant
were taken and added to the special 96-well plate. A total of 5
duplicate wells were set up for each sample. Finally, detection was
performed using a multifunctional Automatic Elisa Analyzer with a
2-sec interval and 10-sec assay time. Then calculated following the
provided instructions.
Chromatin immunoprecipitation (ChIP)
assay
U251 cells with a healthy growth status were
selected and fixed at room temperature for 10 min in a 1%
formaldehyde solution once they reached ~90% confluence. After
cross-linking, the nucleus and chromatin were digested, followed by
ChIP, elution and de-cross-linking. The DNA was then purified and
analyzed using RT-qPCR (cat. no. RR037A; Takara Bio, Inc.). The
transcription conditions were as follows: The reverse transcription
reaction was carried out at 37°C for 15 min for 3 cycles, followed
by the period of reverse transcriptase inactivation at 85°C for 5
sec. Additionally, 10 μl PCR products were extracted from
each tube for PCR experiments; the cycling conditions included an
initial holding period at 95°C for 5 min, followed by a two-step
PCR program consisting of 40 cycles of 95°C for 5 sec and 60°C for
10 sec.
Statistical analysis
Statistical analyses were performed using Prism
8.0.2 software (Dotmatics). Statistical significance was evaluated
using one-way analysis of variance, followed by Tukey's post hoc
test for multiple comparisons. The results are presented as the
mean ± standard deviation and were repeated in ≥3 independent
experiments. P<0.05 was considered to indicate a statistically
significant difference.
Results
ZBTB20 expression and ferroptosis level
is decreased in GBM cells
To assess the role of ZBTB20 in ferroptosis, the
expression of ZBTB20 and ferroptosis level between GBM cells (U251
and U87MG) and normal glial cells (HA1800) was first compared. The
results revealed that the expression of ZBTB20 in U251 and U87MG
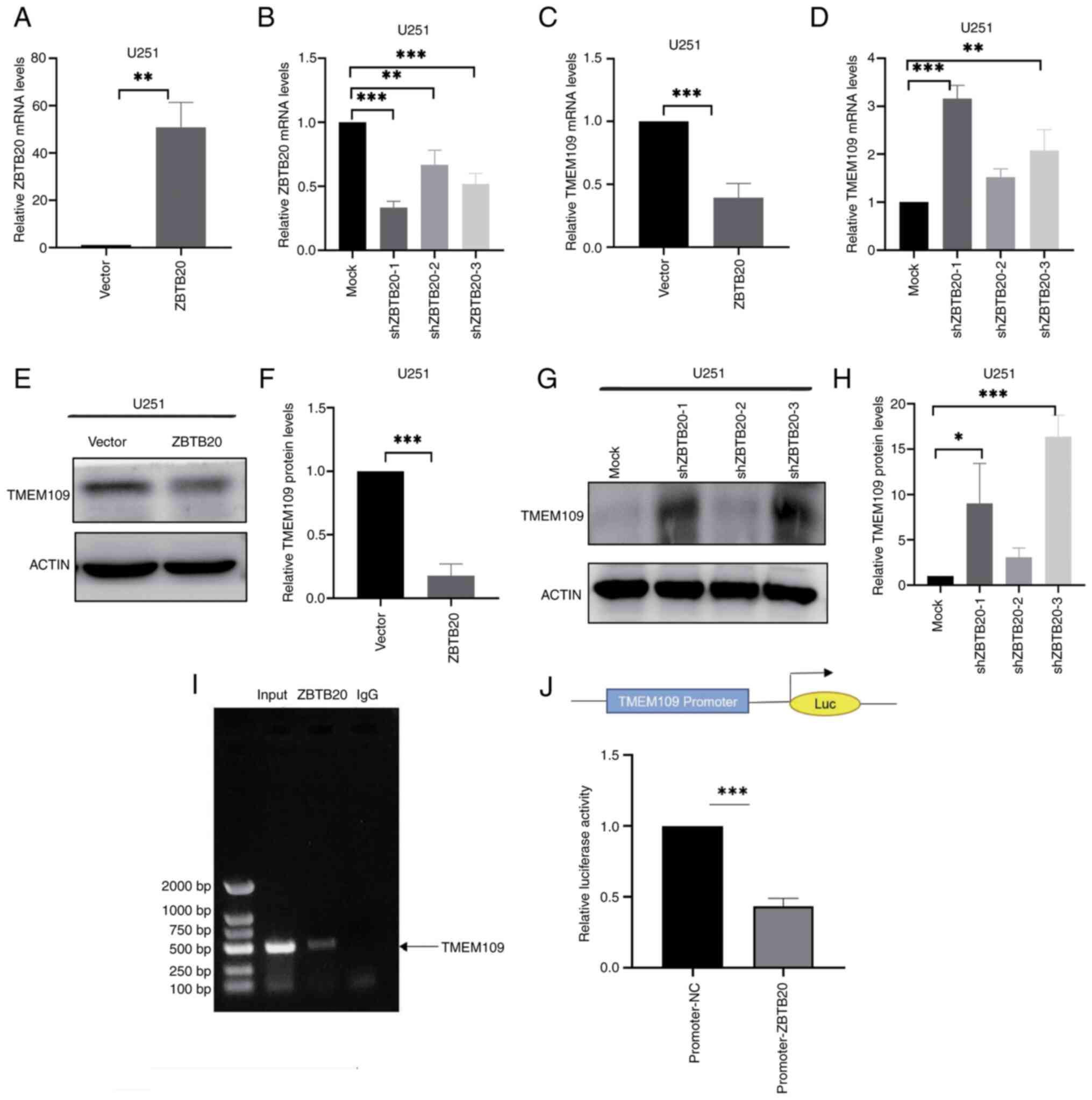
cells was lower than that in HA1800 cells (Fig. 1A, B, F and G), whilst the
expression of TMEM109, SLC7A11 and GPX4 was higher (Fig. 1A, C-F and H-J). GPX4 is an
intracellular antioxidant enzyme and its reduced level leads to the
accumulation of lipid peroxides, which promotes ferroptosis
(45). SLC7A11, an ferroptosis
regulator, is a regulator upstream of GPX4 (46).
 | Figure 1ZBTB20 expression and ferroptosis
level in GBM cells. (A-E) Expression of ZBTB20, TMEM109, SLC7A11
and GPX4 in U251 and HA1800 cells detected by (A) western blotting
and (B-E) the corresponding histograms. (F-J) Expression of ZBTB20,
TMEM109, SLC7A11 and GPX4 in U87MG and HA1800 cells detected by (F)
western blotting and (G-J) the corresponding histograms. (K and L)
ROS level detected by fluorescence microscopy and the corresponding
histogram. (M) Histogram of Fe2+ content. (N) Overall
survival of ZBTB20. (O) Overall survival of TMEM109. Data are shown
as the mean ± standard deviation, n=3. *P<0.05,
**P<0.01 and ***P<0.001. ZBTB20, zinc
finger and BTB domain containing 20, GBM, glioblastoma; TMEM109,
transmembrane protein 109; SLC7A11, solute carrier family 7 member
11, GPX4, glutathione peroxidase 4; ROS, reactive oxygen
species. |
The results also demonstrated that both ROS
(Fig. 1K and L) and
Fe2+ (Fig. 1M) levels
were lower in U251 and U87MG cells compared with HA1800 cells.
Moreover, patients with high ZBTB20 expression had longer overall
survival (Fig. 1N), but those
with high TMEM109 expression had shorter overall survival (Fig. 1O). These results indicated that a
significant reduction in the level of ferroptosis in GBM cells was
accompanied by an increase in ZBTB20 expression, suggesting that
ZBTB20 may be involved in the regulation of ferroptosis.
ZBTB20 promotes ferroptosis in U251 GBM
cells
Previous research reported that ZBTB20 inhibits cell
proliferation and promotes apoptosis in GBM cells (47). The present study evaluated the
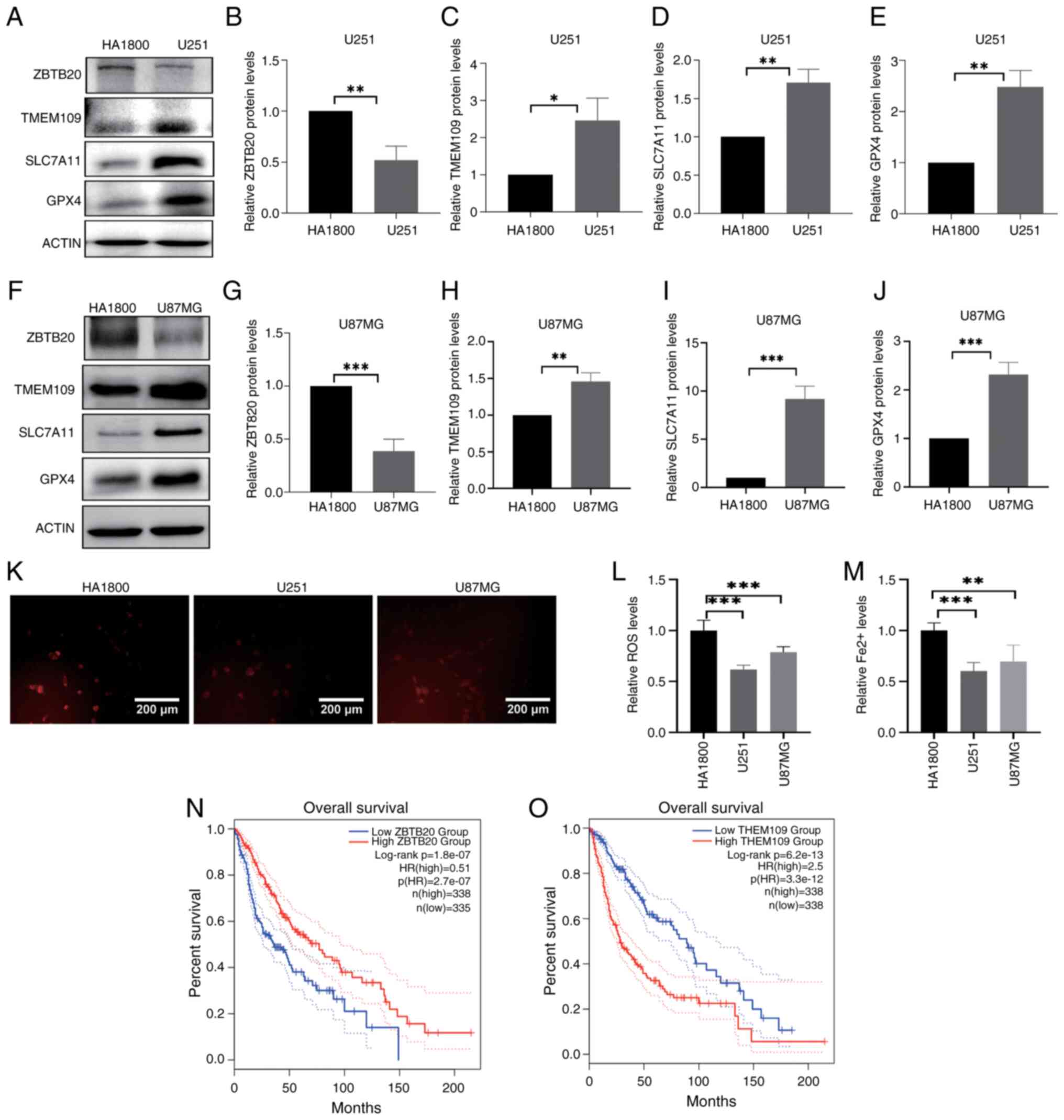
role of ZBTB20 in ferroptosis through gain- and loss-of-function
experiments (Fig. 2A, B, E and
F). U251 GBM cells stably expressing ZBTB20 were obtained by
lentiviral infection and puromycin screening. The results of
western blotting revealed that the expression of SLC7A11 and GPX4
was lower in GBM cells with ZBTB20 overexpression, compared with
that in the vector control (Fig. 2A,
C and D), and higher in GBM cells with ZBTB20 knockdown,
compared with that in the shRNA control (Fig. 2E, G and H). Furthermore, both ROS
(Fig. 2I-K) and Fe2+
(Fig. 2L and M) levels were
higher in GBM cells with ZBTB20 overexpression and lower in GBM
cells with ZBTB20 knockdown compared with wild-type cells. In U87MG
glioma cells, the aforementioned results were consistent (Fig. S1).
 | Figure 2ZBTB20 promotes ferroptosis in U251
GBM cells. (A-D) Expression of ZBTB20, SLC7A11 and GPX4 detected by
(A) western blotting and (B-D) the corresponding histograms in GBM
cells with ZBTB20 overexpression. (E-H) Expression of ZBTB20,
SLC7A11 and GPX4 detected by (E) western blotting and (F-H) the
corresponding histograms in GBM cells with ZBTB20 knockdown. (I-K)
ROS levels detected by (I) fluorescence microscopy and the
corresponding histogram in GBM cells with ZBTB20 (J) overexpression
and (K) knockdown. (L and M) Histograms of Fe2+ content
in GBM cells with ZBTB20 (L) overexpression and (M) knockdown.
*P<0.05, **P<0.01 and
***P<0.001. ZBTB20, zinc finger and BTB domain
containing 20, GBM, glioblastoma; TMEM109, transmembrane protein
109; SLC7A11, solute carrier family 7 member 11, GPX4, glutathione
peroxidase 4; ROS, reactive oxygen species. |
ZBTB20 transcriptionally represses
TMEM109 expression in U251 GBM cells
Although the present study demonstrated that ZBTB20
promotes ferroptosis, the existing literature cannot adequately
explain the mechanisms involved. Genes transcriptionally repressed
by ZBTB20 were validated via RT-qPCR. By integrating Jaspar online
predictions (https://jaspar.genereg.net/) of ZBTB20's promoter
binding regions, TMEM109 was ultimately identified as the target
gene for further investigation, and gain- and loss-of-function
experiments demonstrated that ZBTB20 repressed TMEM109 expression
through transcriptional repression (Fig. 3A-H). ZBTB20 acts as a
transcription factor, and in order to assess whether ZBTB20
directly transcriptionally regulates TMEM109, ChIP (Fig. 3I) and dual-luciferase reporter
gene experiments (Fig. 3J) were
performed. The results revealed that ZBTB20 can directly bind to
the TMEM109 promoter region and inhibit its expression. The results
of PCR (Fig. 1A-D) and western
blotting (Fig. 1E-H) showed that
in U251 and U87MG cells (Fig.
S2), the transcriptional inhibitory effect of ZBTB20 on TMEM109
remained consistent.
ZBTB20 promotes ferroptosis through
repressing TMEM109 expression in U251 GBM cells
TMEM109 has been reported to be a Ca2+
channel located in the membrane of the ER (48). It was hypothesized that TMEM109
alters intracellular calcium homeostasis, and as Ca2+
alteration is one of the inducing factors of ferroptosis (49), TMEM109 may be involved in the
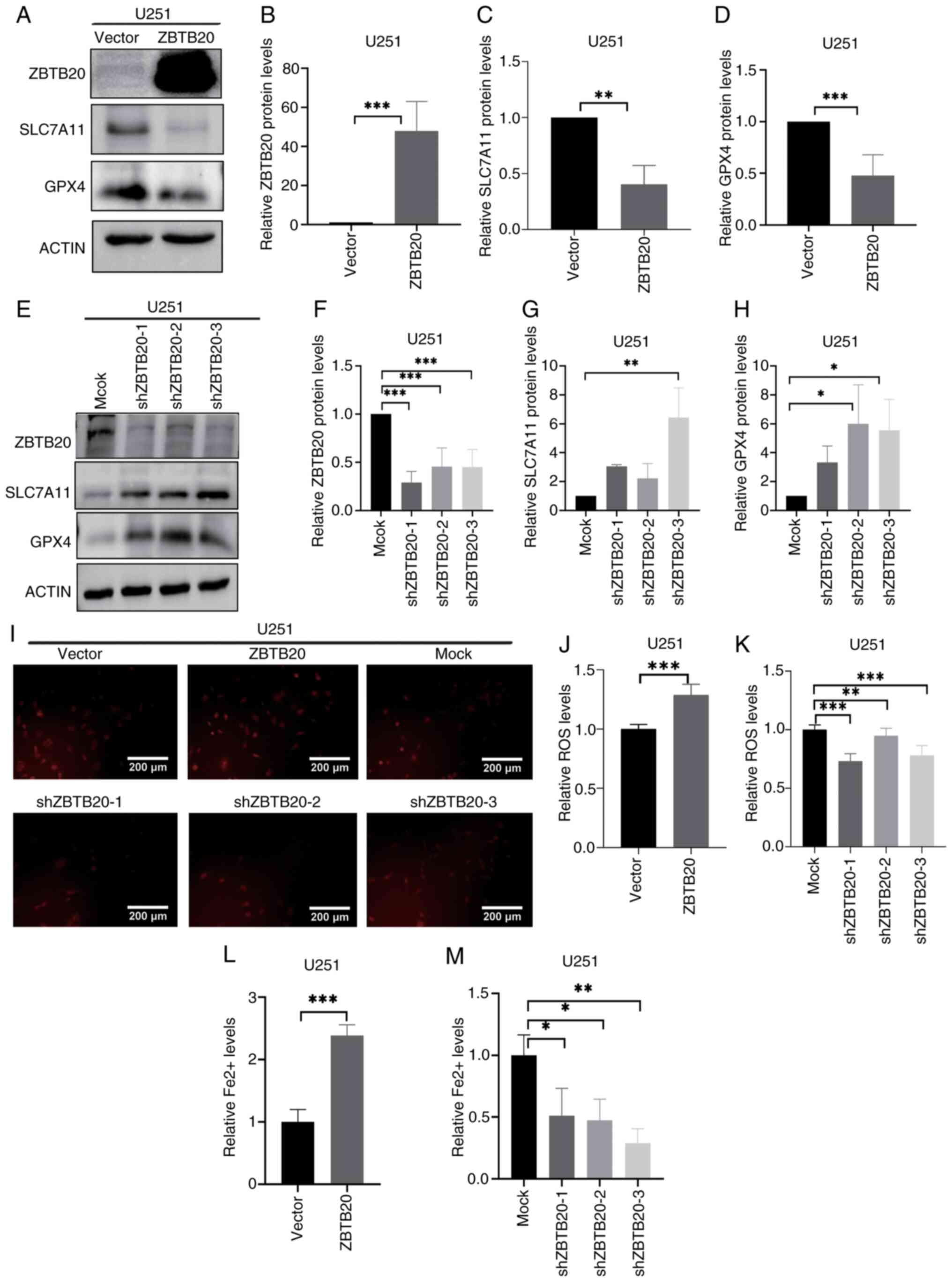
regulation of ferroptosis. To assess this, U251 cells were infected
with lentivirus carrying TMEM109 cDNA, and GBM cells with TMEM109
overexpression and knockdown were obtained. The results of western
blot analysis revealed that the expression of SLC7A11 and GPX4 was
higher in GBM cells with TMEM109 overexpression compared with that
in the vector control cells (Fig.
4A-D), and lower in GBM cells with TMEM109 knockdown compared
with that in the shRNA control cells (Fig. 4E-H). The results also demonstrated
that TMEM109 overexpression decreased ROS levels (Fig. 4I-K) and Fe2+ content
(Fig. 4L and M), whereas TMEM109
knockdown induced the opposite results (Fig. 4I-M). Furthermore, the findings
revealed that patients with high TMEM109 expression had shorter
overall survival (Fig. 1O). The
same result of TMEM109 inhibiting ferroptosis was also demonstrated
in U87MG cells (Fig. S3).
 | Figure 4TMEM109 represses ferroptosis in U251
GBM cells. (A-D) Expression of ZBTB20, SLC7A11 and GPX4 detected by
(A) western blotting and (B-D) the corresponding histograms in GBM
cells with TMEM109 overexpression. (E-H) Expression of ZBTB20,
SLC7A11 and GPX4 detected by (E) western blotting and (F-H) the
corresponding histograms in GBM cells with TMEM109 knockdown. (I-K)
ROS levels detected by (I) fluorescence microscopy and the
corresponding histogram in GBM cells with TMEM109 (J)
overexpression and (K) knockdown. (L and M) Histograms of
Fe2+ content in GBM cells with TMEM109 (L)
overexpression and (M) knockdown. *P<0.05,
**P<0.01 and ***P<0.001. ZBTB20, zinc
finger and BTB domain containing 20, GBM, glioblastoma; TMEM109,
transmembrane protein 109; SLC7A11, solute carrier family 7 member
11, GPX4, glutathione peroxidase 4; ROS, reactive oxygen
species. |
Subsequently, co-transfected cells highly expressing
both TMEM109 and ZBTB20 were constructed. RT-qPCR (Fig. 5A-D) and western blotting (Fig. 5E-I) results revealed that the
expression of both SLC7A11 and GPX4 was significantly higher in
co-transfected cells, whilst ROS levels (Fig. 5J and K) and Fe2+
content (Fig. 5L) were
significantly lower compared with cells overexpressing ZBTB20
alone. Furthermore, the MDA detection experiment revealed that the
content of MDA was significantly higher in co-transfected cells
compared with ZBTB20 overexpression cells (Fig. 5M). These results suggested that
the promoting effect of ZBTB20 on ferroptosis was realized by
inhibiting TMEM109 expression. In U87MG cells, TMEM109 reversed the
iron-depletion effect promoted by ZBTB20, in consistency with the
aforementioned results (Fig.
S4).
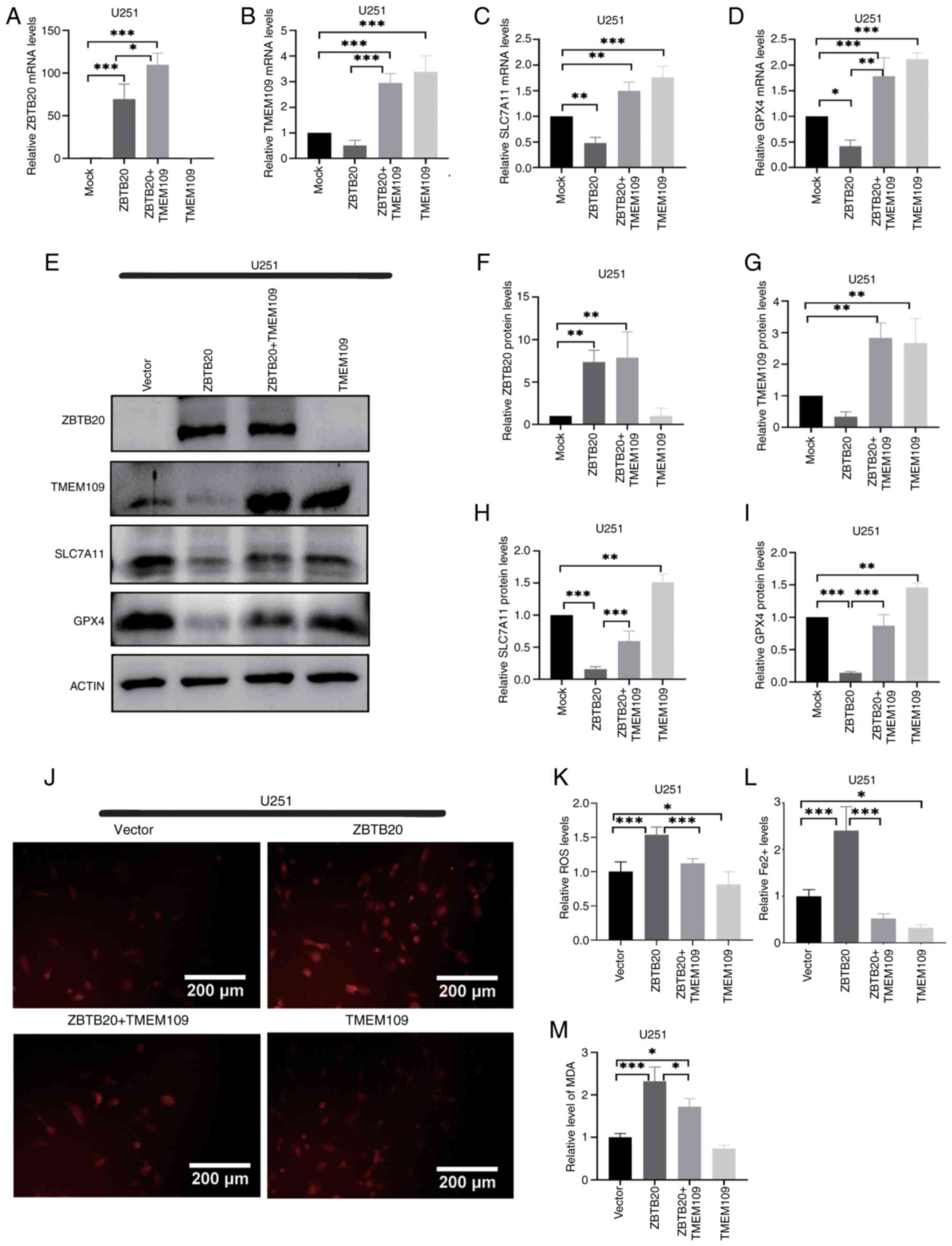 | Figure 5ZBTB20 promotes ferroptosis through
repressing TMEM109 expression in U251 GBM cells. (A-D) Expression
of ZBTB20, TMEM109, SLC7A11 and GPX4 detected by reverse
transcription-quantitative PCR and the corresponding histograms in
GBM cells with ZBTB20 and/or TMEM109 overexpression. (E-I)
Expression of ZBTB20, TMEM109, SLC7A11 and GPX4 detected by (E)
western blotting and (F-I) the corresponding histograms in GBM
cells with ZBTB20 and/or TMEM109 overexpression. (J and K) ROS
levels detected by (J) fluorescence microscopy and (K) the
corresponding histogram. (L) Histograms of Fe2+ content
in GBM cells with ZBTB20 and/or TMEM109 overexpression. (M)
Histograms of MDA content in GBM cells with ZBTB20 and/or TMEM109
overexpression. *P<0.05, **P<0.01 and
***P<0.001. ZBTB20, zinc finger and BTB domain
containing 20, GBM, glioblastoma; TMEM109, transmembrane protein
109; SLC7A11, solute carrier family 7 member 11, GPX4, glutathione
peroxidase 4; ROS, reactive oxygen species; MDA,
malondialdehyde. |
Discussion
The present study demonstrated that ZBTB20 promotes
ferroptosis in GBM cells through transcriptionally inhibiting
TMEM109 expression. Furthermore, the results illustrate the
regulatory mechanism of ferroptosis in GBM and highlight the role
of ZBTB20 and TMEM109 in it, thus providing new ideas for clinical
treatment of GBM. Moreover, whilst the present study demonstrated a
positive effect of ZBTB20 on ferroptosis in glioma cells, other
studies have reported that ZBTB20 can promote the proliferation of
hepatocellular carcinoma (34)
and gastric cancer (30),
suggesting that ZBTB20 serves different roles on progression in
different cancer tissues.
Numerous studies have also reported that ferroptosis
is involved in the oncogenesis and development of several cancers.
For example, cystatin SN regulates the stability of GPX4 protein
through OTU deubiquitinase, ubiquitin aldehyde binding 1, which
inhibits ferroptosis and promotes metastasis of gastric cancer
(50). Additionally, it has been
reported that, compared with paraneoplastic tissue, the expression
level of STAT3 (a ferroptosis inhibitor), is markedly increased in
gliomas (51); this indicates
that the ferroptosis level in gliomas is relatively low. Heat shock
protein 90 induces acyl-CoA synthetase long chain family member
4-dependent ferroptosis in gliomas by dephosphorylating Ser637 at
the Drp1 locus (52). Another
study using the Chinese Glioma Gene Atlas database reported that
ferroptosis-related genes were differentially expressed in GBM and
paraneoplastic tissues (53). The
present study demonstrated that the level of ferroptosis was
significantly reduced in GBM cells compared with in human normal
glial cells.
It has been reported that ZBTB20 can increase ROS
levels in GBM cells by regulating the stress-inducible protein
Sestrin3 (54,55), and that ROS are an important
regulator in ferroptosis (56).
The present study demonstrated that ZBTB20 increased ROS levels and
promoted ferroptosis using gain- and loss-of-function experiments.
The expression of two key genes in ferroptosis (57), SLC7A11 and GPX4, was assessed in
the present study to measure ferroptosis. SLC7A11 is a key gene in
the antioxidant damage pathway in ferroptosis and is located on the
cell membrane. The light chain subunit SLC7A11, a functional
subunit, and the heavy chain subunit SLC3A2, which maintains the
stability of SLC711 (58), form
the cystine-glutamate reverse transporter (Xc-system). SLC7A11 has
a high degree of specificity for cystine and glutamate, and its
main function is to participate in the uptake of cystine and the
release of glutamate (59), which
promotes the synthesis of GSH (60), avoids cellular exposure to
oxidative stress damage, and maintains the redox balance inside the
cell, thus preventing cell death caused by membrane lipid
peroxidation (15). Several
studies have reported that SLC7A11 is highly expressed in several
solid malignant cancers, such as breast cancer, pancreatic cancer,
ovarian cancer and glioma, and has an association with drug
resistance (4,15). Currently, SLC7A11 has become a
research focus in anticancer therapy. The main role of SLC7A11 is
to increase the synthesis of glutathione, promote the activity of
GPX4, and enhance the ability of cells to resist oxidative damage
(61). GPX4 can attenuate lipid
peroxidation toxicity and maintain homeostasis of the membrane
lipid bilayer (5). GPX4 depends
on GSH to catalyze the conversion of peroxides to alcohols, and GSH
deficiency directly causes inactivation of GPX4, which further
triggers ferroptosis (62).
Meanwhile, SLC7A11 can regulate intracellular GSH expression;
therefore, SLC7A11 exists as an upstream regulator of GPX4
(63). The present study
demonstrated that ZBTB20 decreased the expression of SLC7A11 and
GPX4, indicating that ZBTB20 may be involved in the SLC7A11/GPX4
antioxidant damage pathway to regulate ferroptosis.
Furthermore, the present study revealed that the
regulatory effect of ZBTB20 on ferroptosis was partly through
transcriptional repression of TMEM109. Previous research has
reported that TMEM109 overexpression altered the cytoplasmic
Ca2+ concentration and caused calcium imbalance
(64), and calcium imbalance
contributes to ferroptosis (64-66). Downregulation of membrane-spanning
4-domains subfamily A member 15 expression, which is localized in
the ER, promotes luminal Ca2+ accumulation, which
inhibits lipid elongation and desaturation, driving lipid droplet
dispersion and the formation of shorter, more saturated ether
lipids, thereby protecting phospholipids from peroxidation and
preventing ferroptosis in cells (65). Inositol trisphosphate receptor
channel-mediated calcium release promotes ferroptosis in SH-SY5Y
neuroblastoma cells (64). RyR
receptor-mediated calcium release contributes to the inhibition of
GPX4 and thus induces ferroptosis in primary hippocampal neurons
(66). Overall, it was
hypothesized that the regulatory effect of TMEM109 on ferroptosis
may be associated with its regulation of calcium leakage.
However, there are certain limitations of the
present study that need to be improved. First, although the results
demonstrated a regulatory effect of TMEM109 on ferroptosis and
suggested an association with Ca2+ imbalance, altered
Ca2+ levels were not measured to validate this argument.
Second, the way in which the inhibitory effect of ZBTB20 on SLC7A11
and GPX4 expression was realized was not further explored.
In conclusion, in the present study, it was found
that ZBTB20 promotes ferroptosis by repressing the expression of
TMEM109 through transcription. And ferroptosis serves a pivotal
role in cancers, which in recent years has provided new
opportunities for treatment of cancers (9). Therefore, the present study provides
a new therapeutic approach for combating the progression of glioma
and chemotherapy resistance.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XC and ML designed experimental programs and
contributed to molecular biology testing and cell experiments. PD
and HC contributed to analysis and interpretation of data. WW and
XN contributed to the conception of the study and assisted with
performing the analysis with constructive discussions. RW and LZ
contributed to cellular experiments and cell immunofluorescence. XC
and PD confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81171250), and the Education and
teaching reform and practice in Zhengzhou University (grant no.
2023zzujgxm009).
References
|
1
|
Dang Q, Sun Z, Wang Y, Wang L, Liu Z and
Han X: Ferroptosis: A double-edged sword mediating immune tolerance
of cancer. Cell Death Dis. 13:9252022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tang D, Chen X, Kang R and Kroemer G:
Ferroptosis: Molecular mechanisms and health implications. Cell
Res. 31:107–125. 2021. View Article : Google Scholar :
|
|
3
|
Park E and Chung SW: ROS-mediated
autophagy increases intracellular iron levels and ferroptosis by
ferritin and transferrin receptor regulation. Cell Death Dis.
10:8222019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang HH, Fan SQ, Zhan YT, Peng SP and Wang
WY: Suppression of the SLC7A11/glutathione axis causes ferroptosis
and apoptosis and alters the mitogen-activated protein kinase
pathway in nasopharyngeal carcinoma. Int J Biol Macromol.
254:1279762024. View Article : Google Scholar
|
|
5
|
Bersuker K, Hendricks JM, Li Z, Magtanong
L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, et al:
The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit
ferroptosis. Nature. 575:688–692. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su L, Zhang J, Gomez H, Kellum JA and Peng
Z: Mitochondria ROS and mitophagy in acute kidney injury.
Autophagy. 19:401–414. 2023. View Article : Google Scholar :
|
|
7
|
Mou Y, Wang J, Wu J, He D, Zhang C, Duan C
and Li B: Ferroptosis, a new form of cell death: Opportunities and
challenges in cancer. J Hematol Oncol. 12:342019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou C, Yu T, Zhu R, Lu J, Ouyang X, Zhang
Z, Chen Q, Li J, Cui J, Jiang F, et al: Timosaponin AIII promotes
non-small-cell lung cancer ferroptosis through targeting and
facilitating HSP90 mediated GPX4 ubiquitination and degradation.
Int J Biol Sci. 19:1471–1489. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Obrador E, Moreno-Murciano P,
Oriol-Caballo M, López-Blanch R, Pineda B, Gutiérrez-Arroyo JL,
Loras A, Gonzalez-Bonet LG, Martinez-Cadenas C, Estrela JM and
Marqués-Torrejón MÁ: Glioblastoma therapy: Past, present and
future. Int J Mol Sci. 25:25292024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang C, Liu X, Jin S, Chen Y and Guo R:
Ferroptosis in cancer therapy: A novel approach to reversing drug
resistance. Mol Cancer. 21:472022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou Q, Meng Y, Li D, Yao L, Le J, Liu Y,
Sun Y, Zeng F, Chen X and Deng G: Ferroptosis in cancer: From
molecular mechanisms to therapeutic strategies. Signal Transduct
Target Ther. 9:552024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khan AB, Lee S, Harmanci AS, Patel R,
Latha K, Yang Y, Marisetty A, Lee HK, Heimberger AB, Fuller GN, et
al: CXCR4 expression is associated with proneural-to-mesenchymal
transition in glioblastoma. Int J Cancer. 152:713–724. 2023.
View Article : Google Scholar :
|
|
13
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A 'state of the
science' review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khan F, Pang L, Dunterman M, Lesniak MS,
Heimberger AB and Chen P: Macrophages and microglia in
glioblastoma: Heterogeneity, plasticity, and therapy. J Clin
Invest. 133:e1634462023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu T, Zhu C, Chen X, Guan G, Zou C, Shen
S, Wu J, Wang Y, Lin Z, Chen L, et al: Ferroptosis, as the most
enriched programmed cell death process in glioma, induces
immunosuppression and immunotherapy resistance. Neuro Oncol.
24:1113–1125. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yadav N, Xiao A, Zhong Q, Kumar P, Konduru
G, Hart W, Lazzara M and Purow B: Synergistic activity of
simvastatin and irinotecan chemotherapy against glioblastoma
converges on TGF-β signaling. J Neurooncol. 174:621–633. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xing Z, Wei X, Fan Q, Zhao D, He J and
Cheng J: Cryptotanshinone promotes ferroptosis in glioblastoma via
KEAP1/NRF2/HMOX1 signaling pathway. Biochem Biophys Res Commun.
768:1519592025. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nedaeinia R, Dianat-Moghadam H,
Movahednasab M, Khosroabadi Z, Keshavarz M, Amoozgar Z and Salehi
R: Therapeutic and prognostic values of ferroptosis signature in
glioblastoma. Int Immunopharmacol. 155:1145972025. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun J, Xue B, Sun T, Xu X, Zhang D, Shan
Z, Wang Y and Chen B: PTBP1 acts as a tumor suppressor in glioma by
promoting HMOX1-dependent ferroptosis. Biochem Pharmacol.
239:1170412025. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su IC, Su YK, Setiawan SA, Yadav VK, Fong
IH, Yeh CT, Lin CM and Liu HW: NADPH oxidase subunit CYBB confers
chemotherapy and ferroptosis resistance in mesenchymal glioblastoma
via Nrf2/SOD2 modulation. Int J Mol Sci. 24:77062023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu H, Wu Z, Tang J, Gan Y, Li J, Yu Y,
Chen Y, Sui R, Liu J, Zhang Y and Piao H: Ginsenoside F2-modified
liposomes delivering FTY720 enhance glioblastoma targeting and
antitumor activity via ferroptosis. Phytomedicine. 144:1569172025.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng H, Guo Z, Chen F, Zhong Q, Hu Y, Du
C, Wang H, Wei P, Huang W, Wang D, et al: Engineering charge
density in s-block potassium single-atom nanozyme for amplified
ferroptosis in glioblastoma therapy. Mater Today Bio.
32:1018892025. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qiao Y, Liu M, Zhang Y, Ni F, Yu L, Chen
Z, Dai X and Wang X: Gambogic acid-iron nanozymes as effective
carriers for enhanced chemotherapy by inducing excessive autophagy
and oxidative stress. J Nanobiotechnology. 23:4352025. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yin N, Wang B, Wang Y, Tian L, Han S,
Zheng B, Feng F, Song S and Zhang H: A metal-phenolic network
nanoresensitizer overcoming glioblastoma drug resistance through
the metabolic adaptive strategy and targeting drug-tolerant cells.
Nano Lett. 25:9570–9580. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang H, Feng K, Han M, Shi Y, Zhang Y, Wu
J, Yang W, Wang X, Di L and Wang R: Homologous magnetic targeted
immune vesicles for amplifying immunotherapy via ferroptosis
activation augmented photodynamic therapy against glioblastoma. J
Control Release. 383:1138162025. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang Y, Zhao J, Li R, Liu Y, Zhou L, Wang
C, Lv C, Gao L and Cui D: CircLRFN5 inhibits the progression of
glioblastoma via PRRX2/GCH1 mediated ferroptosis. J Exp Clin Cancer
Res. 41:3072022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Janczarek M: The Ros/MucR zinc-finger
protein family in bacteria: Structure and functions. Int J Mol Sci.
23:155362022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li F, Du M, Yang Y, Wang Z, Zhang H, Wang
X and Li Q: Zinc finger and BTB domain-containing protein 20
aggravates angiotensin II-induced cardiac remodeling via the
EGFR-AKT pathway. J Mol Med (Berl). 100:427–438. 2022. View Article : Google Scholar
|
|
29
|
Zhang W, Mi J, Li N, Sui L, Wan T, Zhang
J, Chen T and Cao X: Identification and characterization of DPZF, a
novel human BTB/POZ zinc finger protein sharing homology to BCL-6.
Biochem Biophys Res Commun. 282:1067–1073. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Zhou X, Zhang M, Cheng L, Zhang Y
and Wang X: ZBTB20 promotes cell migration and invasion of gastric
cancer by inhibiting IκBα to induce NF-κB activation. Artif Cells
Nanomed Biotechnol. 47:3862–3872. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Juven A, Nambot S, Piton A, Jean-Marçais
N, Masurel A, Callier P, Marle N, Mosca-Boidron AL, Kuentz P,
Philippe C, et al: Primrose syndrome: A phenotypic comparison of
patients with a ZBTB20 missense variant versus a 3q13.31
microdeletion including ZBTB20. Eur J Hum Genet. 28:1044–1055.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han Q, Yan X, Ye Y, Han L, Ma X, Wang T,
Cao D and Zhang WJ: ZBTB20 regulates prolactin expression and
lactotrope function in adult mice. Endocrinology. 163:bqac1812022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cao D, Ma X, Cai J, Luan J, Liu AJ, Yang
R, Cao Y, Zhu X, Zhang H, Chen YX, et al: ZBTB20 is required for
anterior pituitary development and lactotrope specification. Nat
Commun. 7:111212016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
To JC, Chiu AP, Tschida BR, Lo LH, Chiu
CH, Li XX, Kuka TP, Linden MA, Amin K, Chan WC, et al: ZBTB20
regulates WNT/CTNNB1 signalling pathway by suppressing PPARG during
hepatocellular carcinoma tumourigenesis. JHEP Rep. 3:1002232020.
View Article : Google Scholar
|
|
35
|
Kan H, Huang Y, Li X, Liu D, Chen J and
Shu M: Zinc finger protein ZBTB20 is an independent prognostic
marker and promotes tumor growth of human hepatocellular carcinoma
by repressing FoxO1. Oncotarget. 7:14336–14349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nagao M, Ogata T, Sawada Y and Gotoh Y:
Zbtb20 promotes astrocytogenesis during neocortical development.
Nat Commun. 7:111022016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li F, Yang Y, Xue C, Tan M, Xu L, Gao J,
Xu L, Zong J and Qian W: Zinc finger protein ZBTB20 protects
against cardiac remodelling post-myocardial infarction via
ROS-TNFα/ASK1/JNK pathway regulation. J Cell Mol Med.
24:13383–13396. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang B, Wang Y, Zhang J, Hu C, Jiang J, Li
Y and Peng Z: ROS-induced lipid peroxidation modulates cell death
outcome: Mechanisms behind apoptosis, autophagy, and ferroptosis.
Arch Toxicol. 97:1439–1451. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang S, Liu Q, Chang M, Pan Y, Yahaya BH,
Liu Y and Lin J: Chemotherapy impairs ovarian function through
excessive ROS-induced ferroptosis. Cell Death Dis. 14:3402023.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tirincsi A, O'Keefe S, Nguyen D, Sicking
M, Dudek J, Förster F, Jung M, Hadzibeganovic D, Helms V, High S,
et al: Proteomics identifies substrates and a novel component in
hSnd2-dependent ER protein targeting. Cells. 11:29252022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Takeshima H, Venturi E and Sitsapesan R:
New and notable ion-channels in the sarcoplasmic/endoplasmic
reticulum: Do they support the process of intracellular
Ca2+ release? J Physiol. 593:3241–3251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nishi M, Komazaki S, Iino M, Kangawa K and
Takeshima H: Mitsugumin23, a novel transmembrane protein on
endoplasmic reticulum and nuclear membranes. FEBS Lett.
432:191–196. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Venturi E, Mio K, Nishi M, Ogura T, Moriya
T, Pitt SJ, Okuda K, Kakizawa S, Sitsapesan R, Sato C and Takeshima
H: Mitsugumin 23 forms a massive bowl-shaped assembly and
cation-conducting channel. Biochemistry. 50:2623–2632. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yamashita A, Taniwaki T, Kaikoi Y and
Yamazaki T: Protective role of the endoplasmic reticulum protein
mitsugumin23 against ultraviolet C-induced cell death. FEBS Lett.
587:1299–1303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liang D, Feng Y, Zandkarimi F, Wang H,
Zhang Z, Kim J, Cai Y, Gu W, Stockwell BR and Jiang X: Ferroptosis
surveillance independent of GPX4 and differentially regulated by
sex hormones. Cell. 186:2748–2764.e22. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen X, Yu C, Kang R, Kroemer G and Tang
D: Cellular degradation systems in ferroptosis. Cell Death Differ.
28:1135–1148. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Duan P, Li B, Zhou Y, Cao H, Chen S and
Xing Y: ZBTB20 suppresses tumor growth in glioblastoma through
activating the TET1/FAS/caspase-3 pathway. Oncol Lett. 28:3582024.
View Article : Google Scholar
|
|
48
|
Watanabe D, Nishi M, Liu F, Bian Y and
Takeshima H: Ca2+ storage function is altered in the
sarcoplasmic reticulum of skeletal muscle lacking mitsugumin 23. Am
J Physiol Cell Physiol. 326:C795–C809. 2024. View Article : Google Scholar
|
|
49
|
Wang S, Li W, Zhang P, Wang Z, Ma X, Liu
C, Vasilev K, Zhang L, Zhou X, Liu L, et al: Mechanical overloading
induces GPX4-regulated chondrocyte ferroptosis in osteoarthritis
via Piezo1 channel facilitated calcium influx. J Adv Res. 41:63–75.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li D, Wang Y, Dong C, Chen T, Dong A, Ren
J, Li W, Shu G, Yang J, Shen W, et al: CST1 inhibits ferroptosis
and promotes gastric cancer metastasis by regulating GPX4 protein
stability via OTUB1. Oncogene. 42:83–98. 2023. View Article : Google Scholar :
|
|
51
|
Sun Q, Lu H, Yuan F, Zhao Q, Wei Y, Wang
R, Chen Q and Liu B: SLC10A3 regulates ferroptosis of glioblastoma
through the STAT3/GPX4 pathway. Sci Rep. 15:212592025. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Miao Z, Tian W, Ye Y, Gu W, Bao Z, Xu L,
Sun G, Li C, Tu Y, Chao H, et al: Hsp90 induces Acsl4-dependent
glioma ferroptosis via dephosphorylating Ser637 at Drp1. Cell Death
Dis. 13:5482022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wan RJ, Peng W, Xia QX, Zhou HH and Mao
XY: Ferroptosis-related gene signature predicts prognosis and
immunotherapy in glioma. CNS Neurosci Ther. 27:973–986. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li S, Lu Z, Sun R, Guo S, Gao F, Cao B and
Aa J: The role of SLC7A11 in cancer: Friend or foe? Cancers
(Basel). 14:30592022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lin Y, Dong Y, Liu W, Fan X and Sun Y:
Pan-cancer analyses confirmed the ferroptosis-related gene SLC7A11
as a prognostic biomarker for cancer. Int J Gen Med. 15:2501–2513.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen Y, Mi Y, Zhang X, Ma Q, Song Y, Zhang
L, Wang D, Xing J, Hou B, Li H, et al: Dihydroartemisinin-induced
unfolded protein response feedback attenuates ferroptosis via
PERK/ATF4/HSPA5 pathway in glioma cells. J Exp Clin Cancer Res.
38:4022019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen X, Wang Z, Li C, Zhang Z, Lu S, Wang
X, Liang Q, Zhu X, Pan C, Wang Q, et al: SIRT1 activated by AROS
sensitizes glioma cells to ferroptosis via induction of NAD+
depletion-dependent activation of ATF3. Redox Biol. 69:1030302024.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Koppula P, Zhuang L and Gan B: Cystine
transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient
dependency, and cancer therapy. Protein Cell. 12:599–620. 2021.
View Article : Google Scholar :
|
|
59
|
Ye Y, Chen A, Li L, Liang Q, Wang S, Dong
Q, Fu M, Lan Z, Li Y, Liu X, et al: Repression of the antiporter
SLC7A11/glutathione/glutathione peroxidase 4 axis drives
ferroptosis of vascular smooth muscle cells to facilitate vascular
calcification. Kidney Int. 102:1259–1275. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang L, Liu Y, Du T, Yang H, Lei L, Guo M,
Ding HF, Zhang J, Wang H, Chen X and Yan C: ATF3 promotes
erastin-induced ferroptosis by suppressing system Xc. Cell Death
Differ. 27:662–675. 2020. View Article : Google Scholar
|
|
61
|
Liu Y, Lu S, Wu LL, Yang L, Yang L and
Wang J: The diversified role of mitochondria in ferroptosis in
cancer. Cell Death Dis. 14:5192023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu S, Wu W, Chen Q, Zheng Z, Jiang X, Xue
Y and Lin D: TXNRD1: A key regulator involved in the ferroptosis of
CML cells induced by cysteine depletion in vitro. Oxid Med Cell
Longev. 2021:76745652021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang X, Wang Y, Huang D, Shi S, Pei C, Wu
Y, Shen Z, Wang F and Wang Z: Astragaloside IV regulates the
ferroptosis signaling pathway via the Nrf2/SLC7A11/GPX4 axis to
inhibit PM2.5-mediated lung injury in mice. Int Immunopharmacol.
112:1091862022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Campos J, Gleitze S, Hidalgo C and Núñez
MT: IP3R-mediated calcium release promotes ferroptotic
death in SH-SY5Y neuroblastoma cells. Antioxidants (Basel).
13:1962024. View Article : Google Scholar
|
|
65
|
Xin S, Mueller C, Pfeiffer S, Kraft VAN,
Merl-Pham J, Bao X, Feederle R, Jin X, Hauck SM, Schmitt-Kopplin P
and Schick JA: MS4A15 drives ferroptosis resistance through
calcium-restricted lipid remodeling. Cell Death Differ. 29:670–686.
2022. View Article : Google Scholar :
|
|
66
|
Gleitze S, Ramírez OA, Vega-Vásquez I, Yan
J, Lobos P, Bading H, Núñez MT, Paula-Lima A and Hidalgo C:
Ryanodine receptor mediated calcium release contributes to
ferroptosis induced in primary hippocampal neurons by GPX4
inhibition. Antioxidants (Basel). 12:7052023. View Article : Google Scholar : PubMed/NCBI
|