Introduction
With an aging population and the continuation of
organized cancer screening programs, gastric cancer (GC) has shown
a consistent trend of increasing morbidity and mortality (1,2).
GC has a significant demographic disparity, with male prevalence
rates nearly twice that of females (1,3).
Geographical variations are also evident, with disproportionately
high incidence rates in East Asia (particularly Japan and Mongolia)
and Eastern Europe, which together account for 87% of GC cases
worldwide (1,3). GC has thus become a global public
health challenge and even surgically treated patients, such as
those with gastric stump cancer, still face a considerable disease
burden and poor outcomes (4). The
low detection rate of early-stage GC can be attributed to the lack
of specific clinical symptoms (5). GC screening can detect precancerous
lesions and early-stage GC in asymptomatic patients, reducing
mortality and improving treatment efficacy (6). The majority (>70%) of patients
are diagnosed at advanced stages, mainly due to the absence of
clinical symptoms during early pathogenesis (7). This delay in diagnosis contributes
to a poor prognosis and the clinical management of GC presents
obvious challenges. Despite the availability of chemotherapy and
radiotherapy, treatment outcomes for advanced gastrointestinal
malignancies, such as pancreatic and GC, remain unsatisfactory
(8). Patients diagnosed at a late
stage have a poor prognosis without targeted treatment (2). Traditional Chinese Medicine
(TCM)-derived polysaccharides, such as Rhizoma Coptidis
polysaccharides and Anemarrhena asphodeloides
polysaccharides, have been explored as adjuncts in GC management
(9,10). To address these clinical
challenges, there is a pressing need to improve our understanding
of the pathogenesis of GC. Therefore, it is necessary to elucidate
the mechanisms associated with the pathogenesis and progression of
GC and to explore potential targets for effective treatment. Such
discoveries are essential for the development of more effective
treatment strategies and improved survival outcomes.
Mitochondria are vital for cellular homeostasis
concerning energy production, calcium regulation and apoptosis.
Their dysfunction has been widely reported to be associated with
neurodegenerative diseases, diabetes and chronic pulmonary
conditions (11-14). Mitophagy, the selective
degradation of defective mitochondria, is essential for maintaining
cellular homeostasis and influencing various cancer-related
pathways. In various types of cancer, including bladder and
colorectal, mitophagy has been linked to survival and therapy
resistance. The influence of mitophagy on tumor microenvironments
and immune responses has the potential to facilitate tumor growth
(15-17). Conversely, the progression of
breast cancer is inhibited by Urolithin A, which activates
transcription factor EB (TFEB)-mediated mitophagy in tumor
macrophages (18). Esophageal
cancer was associated with a reduction in mitophagy, which has been
linked to an increased potential for metastasis (19). These results indicate that
mitophagy inhibition may facilitate tumor spread. Consequently, the
manipulation of mitophagy pathways may present a novel therapeutic
strategy with the potential to enhance treatment outcomes in cancer
management.
The Ras homolog family member T1 (RHOT1) gene
encodes a mitochondrial GTPase that plays a crucial role in various
processes within mitochondria, including mitochondrial transport,
mitochondrial calcium buffering and mitophagy. Its role in disease
pathogenesis, particularly neurodegenerative disorders and
metabolic conditions, has been well-documented. For instance, the
dysfunction of RHOT1 has been demonstrated to disrupt
Parkin-mediated mitophagy, leading to the accumulation of damaged
mitochondria and exacerbating neuronal loss in Parkinson's disease
(20). Additionally, the
dysfunction of RHOT1 has been observed to impede calcium handling
and mitochondrial transport, thereby contributing to metabolic
dysregulation in diabetes (11).
RHOT1 plays a pivotal role in maintaining mitochondrial
quality control and has been identified as essential for preserving
mitochondrial integrity (20). In
GC, increasing research has identified mitochondrial dysfunction as
a pivotal factor driving tumor progression, therapy resistance and
metabolic reprogramming (21-24). Peng et al (25) investigated the correlation between
the expression of RHOT1 and the clinicopathological features of
tumor-node-metastasis staging and lymph node metastasis. RHOT1 has
been thought to serve as a biomarker for GC. However, to date, no
studies have reported the specific role of RHOT1 in the regulation
of mitophagy in GC. Its conserved function in mitochondrial quality
control suggests that aberrant RHOT1 activity may influence tumors
by regulating mitophagy. Consequently, the investigation of RHOT1
in GC cells has the potential to reveal novel associations between
mitochondrial dynamics and oncogenesis, thus providing potential
therapeutic targets for diseases associated with mitochondrial
dysfunction.
The present study hypothesized that RHOT1 may affect
the behavior of GC cells through mitophagy. It aimed to investigate
the role of RHOT1 in regulating mitochondrial quality control,
energy metabolism and mitophagy-related signaling in GC.
Furthermore, its effect on the proliferation, invasion and
migration of GC cells in vitro was examined. These findings
may provide a theoretical basis for considering RHOT1 as a
potential alternative therapeutic target for GC.
Materials and methods
Cell lines and cell culture
The cell lines used were HGC-27, MKN-45, AGS, SNU-1
and GES-1 (human gastric cancer cell lines and a human gastric
epithelial cell line, respectively) obtained from Wuhan Servicebio
Technology Co., Ltd. (cell batch: IM-H084202303). They were
cultured in Roswell Park Memorial Institute (RPMI) 1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Clark Bioscience), 100 U/ml penicillin and 100
μg/ml streptomycin and maintained at 37°C in a humidified
atmosphere with 5% CO2.
Total RNA extraction and reverse
transcription-quantitative (RT-q) PCR
RNA Easy Fast Tissue/Cell Kit (Tiangen Biotech Co.,
Ltd.) was prepared for total RNA extraction according to the
manufacturer's instructions under RNase-free conditions. Total RNA
was extracted from GC cell lines (HGC-27, MKN-45, AGS, SNU-1) and
GES-1 after transfection or treatment, depending on the
experimental group. PrimeScript RT Master Mix Kit (Takara
Biotechnology Co., Ltd.) was used for reverse transcription and the
SYBR Green Master Mix Kit (Takara Biotechnology Co., Ltd.) was
employed to detect the relative expression of mRNAs. The reaction
conditions were performed under the following conditions: 95°C for
2 min, 95°C for 15 sec, 60°C for 1 min and 72°C for 30 sec for 40
cycles. Primer sequences are presented in Table I. All primers were designed based
on Homo sapiens RefSeq/NCBI sequences and targeted exon-exon
junctions to ensure specificity for mRNA. GAPDH was used to
normalize the gene being tested. All primers were synthesized by
Sangon Biotech Co., Ltd. and validated by melt curve analysis and
agarose gel electrophoresis to confirm specificity. Each sample was
run in triplicate using the 7500 Fast (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Gene expression results were expressed as
fold changes relative to GAPDH using the 2-ΔΔCq method
(26).
 | Table IPrimer information. |
Table I
Primer information.
| Gene | Forward
(5'-3') | Reverse
(5'-3') | Amplicon size
(bp) | RefSeq/NCBI |
|---|
| RHOT1 |
CTGATTTCTGCAGGAAACACAA |
GCAAAAACAGTAGCACCAAAAC | 142 | NM_001033566.3 |
| PINK1 |
GTGGACCATCTGGTTCAACAGG |
GCAGCCAAAATCTGCGATCACC | 110 | NM_032409.4 |
| Parkin |
CCAGAGGAAAGTCACCTGCGAA |
CTGAGGCTTCAAATACGGCACTG | 125 | NM_004562.3 |
| Tomm20 |
CGACCGCAAAAGACGAAGTGAC |
GCTTCAGCATCTTTAAGGTCAGG | 130 | NM_007019.5 |
| Timm23 |
ACACGAGGTGCAGAAGATGACC |
CTGTCAGACCACCTCGTGCTAT | 115 | NM_006991.3 |
| GAPDH |
GAGTCAACGGATTTGGTCGT |
TTGATTTTGGAGGGATCTCG | 101 | NM_002046.7 |
Short interfering (si)RNA
transfection
Target-specific siRNA (si-RHOT1) (Table II) and a negative control siRNA
(si-NC) were transfected into HGC-27 cells cultured in six-well
plates. Cells were transfected with 20 μM siRNA using Lipo
6000 transfection reagent (Beyotime Institute of Biotechnology) and
then cultured for 48 h at 37°C. The silencing efficiency was
evaluated by qPCR. The si-NC used in the present study was a
non-targeting control siRNA purchased from Shanghai GenePharma Co.,
Ltd. and its sequence was not disclosed by the manufacturer.
 | Table IIsiRNA sequences. |
Table II
siRNA sequences.
| siRNA | Sense (5'-3') | Antisense
(5'-3') |
|---|
| RHOT1-homo-393 |
CCAACACACAUUGUAGAUUTT |
AAUCUACAAUGUGUGUUGGTT |
|
RHOT1-homo-1384 |
GCUAUCUAGGCUAUUCAAUTT |
AUUGAAUAGCCUAGAUAGCTT |
|
RHOT1-homo-1804 |
GCUUAAUCGUAGCUGCAAATT |
UUUGCAGCUACGAUUAAGCTT |
|
RHOT1-homo-1174 |
CCUUUGACAAGCAUGAUUUTT |
AAAUCAUGCUUGUCAAAGGTT |
CCK-8 proliferation assay
Cell counting kit 8 (CCK-8; MedChemExpress) was used
for the detection of proliferation of HGC-27 cells. Cells
(~2×103) were seeded in 100 μl of medium in a
96-well plate and incubated at 37°C. CCK-8 reagent (10 μl)
was added to each well at 0, 24, 48, 72 and 96 h. Incubation was
performed at 37°C for 2 h. A microplate reader (Molecular Devices,
LLC) was used to read OD values at 450 nm. Blank control wells
containing medium and CCK-8 reagent without cells were included and
their absorbance values were subtracted from sample readings to
eliminate background interference. Cell proliferation results were
expressed as OD values at 450 nm, normalized to control wells.
Wound healing assay
A wound-healing assay was used to evaluate the
migration ability of HGC-27 cells. Cells were cultured at
3×105/ml per well in a 12-well plate. After 6 h of
incubation, 1 ml of cell medium was added to the 12-well plate. A
20 μl pipette tip was used to create a wound and PBS was
used to wash the slide 2-3 times. Then serum-free medium was
prepared and the images were captured under the microscope at 0, 24
and 48 h. The experiment was performed three times.
Transwell assay
For the Transwell migration assay, 200 μl of
cell suspension was transferred to a Transwell upper chamber
(Corning, Inc.) and 500 μl of medium with 20% FBS was
transferred to a Transwell lower chamber and incubated with 5%
CO2 in a 37°C incubator for 48 h. Finally, 50 μl
of the diluted cell suspension was added to the upper chamber of
the Transwell (Corning, Inc.) and incubated at 37°C for 1 h. For
the invasion assay, a layer of Matrigel matrix glue (Corning, Inc.;
ratio of serum-free medium:matrix glue 8:1) was coated within the
lower chamber, and the cells were incubated for 24 h at 37°C. After
incubation, a 4% paraformaldehyde solution was used to fix the
cells in the Transwell. The Transwell was subsequently stained with
0.1% crystal violet for 20 min. Cell counts for migration and
invasion were recorded by bright-field microscopy. Three biological
replicates were performed. The migration and invasion abilities
were expressed as the number of stained cells counted under the
microscope.
Flow cytometry
Flow cytometry was performed to evaluate the cell
cycle and apoptosis of the cells. HGC-27 cells were stained with
propidium iodide (PI; cat. Y267501; Beyotime Institute of
Biotechnology) for the cell cycle assay and Annexin Ⅴ and PI
(Annexin Ⅴ; cat. C1062M; Beyotime Institute of Biotechnology) were
used for the apoptosis assay. Cells were stained with PI for cell
cycle and Annexin Ⅴ and PI for apoptosis at 37°C for 30 min.
FACSCalibur and FACSCelesta (BD Biosciences) were used for cell
cycle and apoptosis detection, respectively. The flow cytometer was
operated with a 488 nm excitation laser at standard power. For cell
cycle analysis, gating was based on PI fluorescence intensity to
distinguish populations with different DNA content, corresponding
to G0/G1, S and G2/M phases. For apoptosis
analysis, gating was performed on Annexin V/PI two-parameter dot
plots to classify cells as viable (Annexin V−/PI−), early apoptotic
(Annexin V+/PI-), or late apoptotic/necrotic (Annexin V+/PI+).
Three biological replicates were performed. Cell cycle distribution
was expressed as the percentage of cells in
G0/G1, S and G2/M phases and
apoptosis results were expressed as the percentage of apoptotic
cells.
Reactive oxygen species (ROS) and
mitochondrial membrane potential (MMP)
ROS were detected using the Reactive Oxygen Specific
Test Kit (Beyotime Institute of Biotechnology). ROS levels were
measured using 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA).
HGC-27 cells (1×106) were divided equally. In the
positive control group, 1 μl of Rosup stimulating drug was
added and incubated at 37°C for 30 min. The treatment group was
directly treated with 10 μM DCFH-DA dissolved in serum-free
culture medium (1 ml) and incubated at 37°C for 30 min.
Fluorescence was detected by flow cytometry at an excitation
wavelength of 488 nm and an emission wavelength of 530 nm and
changes in MMP were detected using the Mitochondrial Membrane
Potential Detection Kit (Beyotime Institute of Biotechnology). The
HGC-27 cells were harvested by centrifugation at 1,000 × g for 5
min at 37°C and resuspended in 50 mM phosphate-buffered saline (pH
7.0). A total of 5×104 cells collected by centrifugation
were resuspended in 188 μl Annexin V-FITC conjugate. Then 2
μl Mito-Tracker Red CMXRos was added and incubated at 25°C
for 30 min and the cells placed in an ice bath. Fluorescence was
detected by flow cytometry using an excitation wavelength of 579 nm
and an emission wavelength of 599 nm. ROS levels were expressed as
mean fluorescence intensity and MMP was expressed as fluorescence
intensity. The experiment was performed three times.
Bioinformatics analysis
The String website (https://cn.string-db.org/) was used to retrieve target
genes for analysis of protein-protein interactions (PPI). The
species was limited to Homo sapiens and the parameters were
set as follows: Choice of significance term for network edges;
evidence: Choice of active interaction source; experimental: Choice
of minimum required interaction score; confidence: 0.2.
After downloading the enriched gene set,
de-duplication of the gene set was carried out to obtain the set of
interacting genes. The final PPI network map was calculated by
selecting the top 10 pivotal genes in the Hubba node using the
Cytoscape (https://cytoscape.org/; version 3.8.0)
plugin cyto-Hubba. R software (https://www.r-project.org/; version 3.6.3) was used to
analyze the RHOT1 mRNA expression levels of STAD RNA-seq based on
The Cancer Genome Atlas (TCGA) database and a single-gene
co-expression heatmap was constructed. Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of
selected data were performed using the clusterProfiler package
(https://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html;
version 3.14.3) and visualized using the ggplot2 package
(https://cran.r-project.org/web/packages/ggplot2/index.html;
version 3.3.3). The intersection of mitophagy genes obtained from
the Reactome database (https://curator.reactome.org; version 87) with those
obtained from the KEGG database was taken and plotted as a Venn
diagram. The expression matrix of 20 mitophagy pathway core genes
was screened based on the intersection of mitophagy pathways from
KEGG and Reactome. Principal component analysis (PCA) was performed
(after normalization) using Omicstudio (https://www.omicstudio.cn/tool) and the correlation
heatmap of these core genes was plotted.
Western blotting
Total proteins from transfected HGC-27 cells were
isolated by RIPA buffer (cat. no. R0010; Beijing Solarbio Science
& Technology Co., Ltd.) and then separated by 8-11% SDS-PAGE.
Protein concentration was determined using the BCA protein assay
kit (cat. no. WLA019; Wanleibio Co., Ltd.). The 20 μg
protein samples were dissolved and transferred to 0.45 μm
polyvinylidene fluoride (PVDF) membranes (cat. no. IPVH00010;
MilliporeSigma). The PVDF membranes were blocked with 5% skimmed
milk powder for 2 h at room temperature and then incubated with
primary antibodies at 4°C overnight. The antibodies were RHOT1
(cat. no. A5838; 1:500; ABclonal Biotech Co., Ltd.), PINK1 (cat.
no. WL04963; 1:500; Wanleibio Co., Ltd.), Parkin (cat. no. WL02512;
1:500; Wanleibio Co., Ltd.), GAPDH (cat. no. 5174, 1:1,000; Cell
Signaling Technology, Inc.). The PVDF membranes were then washed
with tris-buffered saline-Tween 20 (0.1% TBST-20; cat. GC204002;
Wuhan Servicebio Technology Co., Ltd.) and incubated with
horseradish peroxidase-conjugated goat anti-rabbit IgG (cat. no.
WLA023; 1:5,000; Wanleibio Co., Ltd.) for 1.5 h at room
temperature. The membranes were scanned and analyzed using the
Gel-Pro Analyzer 4.5 program for Windows (Media Cybernetics, Inc.).
Three biological replicates were performed. Protein expression
levels were expressed as band intensities normalized to GAPDH.
Statistical analysis
The data were presented as the mean ± standard
deviation. The findings from each experiment were corroborated
through independent replicates. Normality of data distribution was
assessed using the Shapiro-Wilk test prior to parametric analyses.
For comparisons between two groups, a two-tailed unpaired Student's
t-test was applied. For comparisons among multiple groups with a
single variable, one-way ANOVA followed by the Tukey-Kramer post
hoc test was used. For datasets involving more than one variable,
two-way ANOVA with appropriate post-hoc analyses was performed.
SPSS (version 29.0; IBM Corp.) and GraphPad Prism (version 9.5.0;
Dotmatics) were used for statistical analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
RHOT1 is involved in mitophagy via
PINK1/Parkin pathway
To investigate the function of RHOT1, the
interacting proteins of RHOT1 were investigated from the STRING
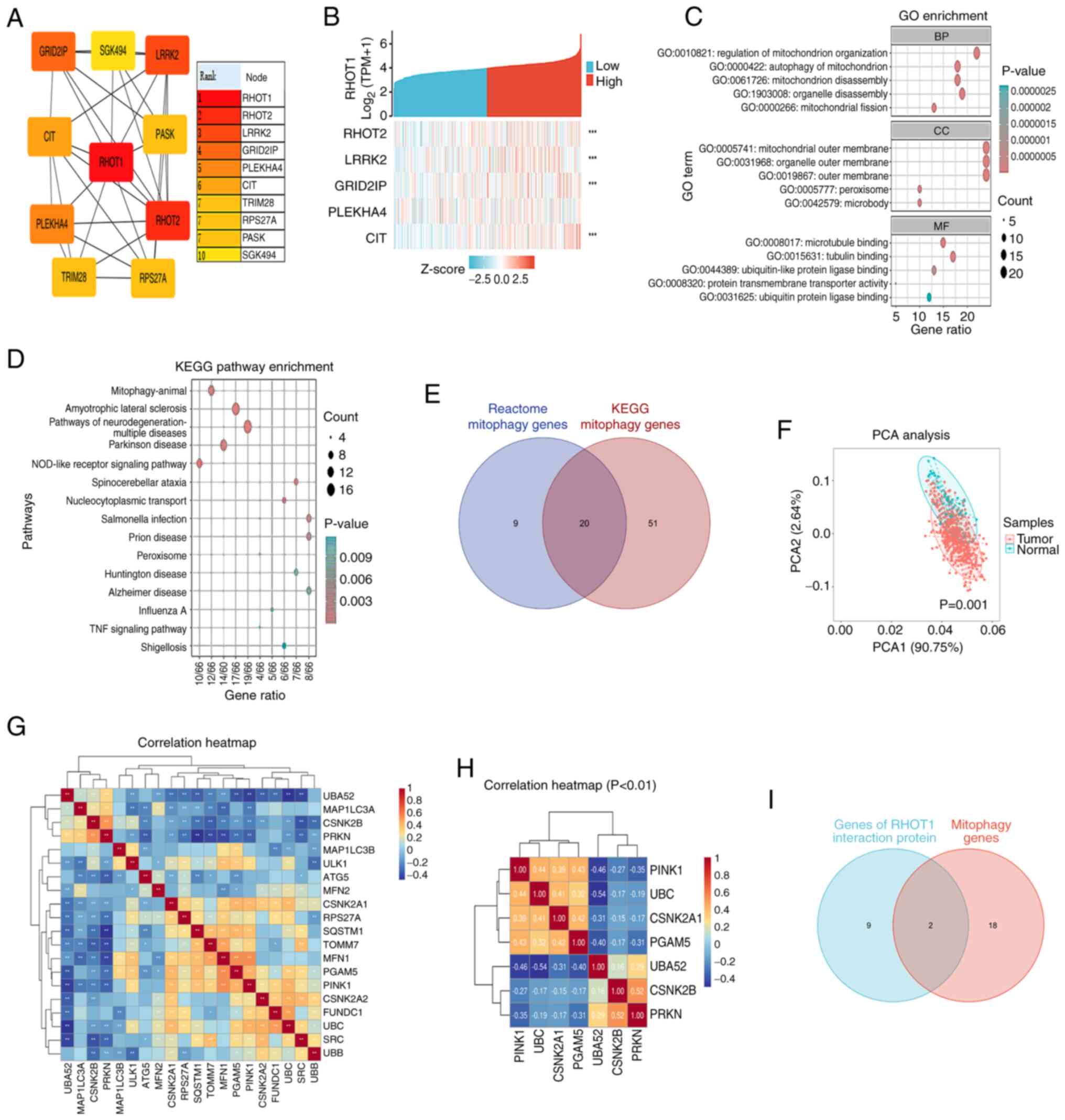
search sets. Analysis of RHOT1-interacting proteins identified
RHOT2, LRRK2, GRID2IP, PLEKHA4 and CIT as hub genes
(Fig. 1A). Single-gene
co-expression analysis using TCGA-STAD RNA-seq data showed that
RHOT2, LRRK2, GRID2IP and CIT were significantly
co-expressed with RHOT1 (P<0.001; Fig. 1B). GO and KEGG enrichment analysis
indicated involvement in mitochondrion organization, mitophagy,
microtubule binding and pathways including Parkinson's disease and
neurodegeneration (Fig. 1C and
D).
A Venn diagram identified 20 mitophagy-related genes
from Reactome and KEGG databases (Fig. 1E) and PCA showed significant
differences between tumor and normal groups (Fig. 1F). Next, a correlation heatmap was
constructed from the 20 mitophagy genes (Fig. 1G). The correlation heatmap
displayed the genes with a statistically significant difference
P<0.01 (Fig. 1H). Intersection
of RHOT1-interacting genes and mitophagy genes revealed
PINK1 and Parkin (Fig.
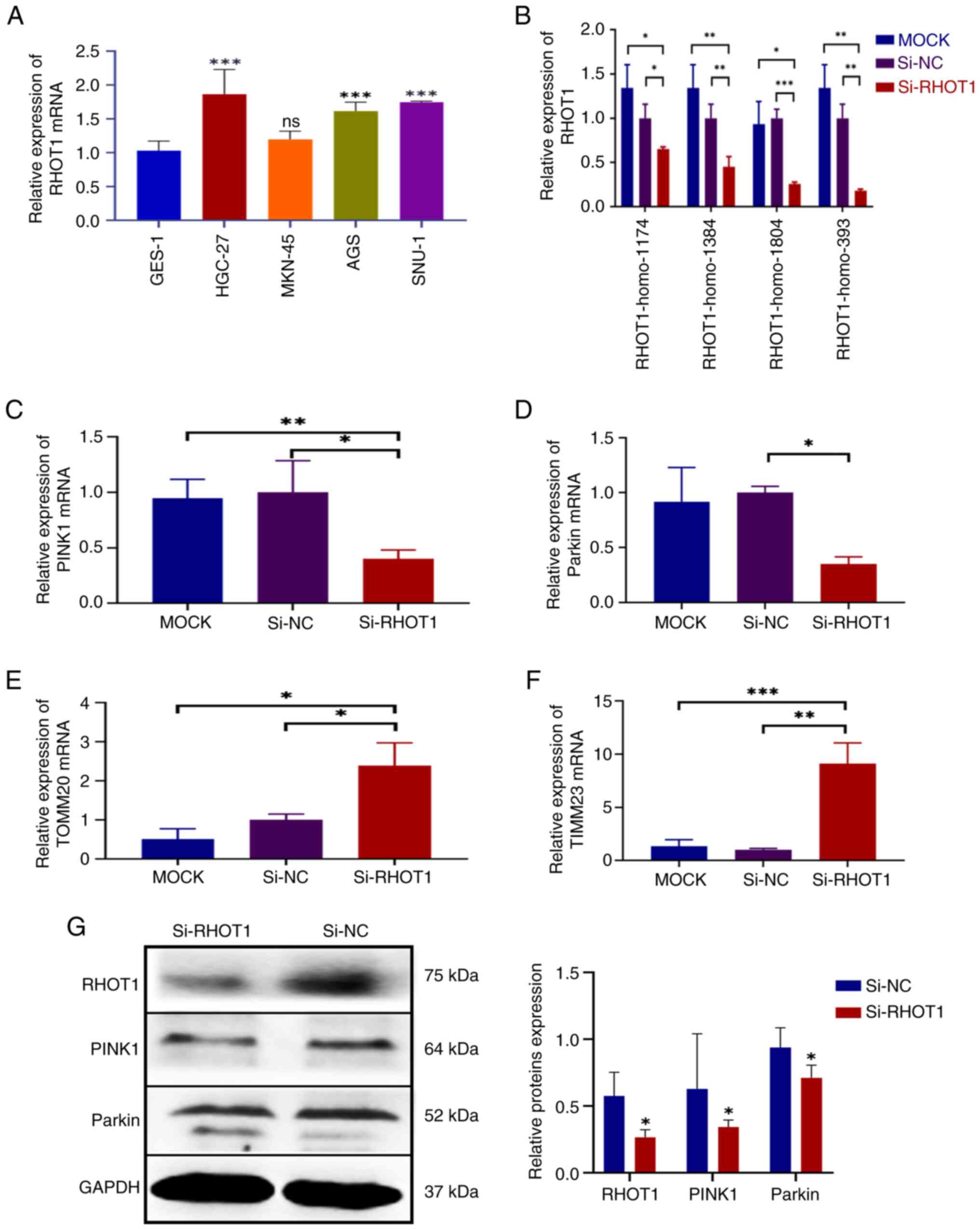
1I). RHOT1 expression was upregulated in GC cell lines
HGC-27, MKN-45, AGS and SNU-1 compared with GES-1, with HGC-27
showing the highest expression (Fig.
2A). Among four siRHOT1 constructs, siRNA RHOT1-homo-393
achieved the highest silencing efficiency (Fig. 2B). Subsequently, the mRNA
expression levels of the genes were examined. The result showed
that the mRNA expression of PINK1 and Parkin were
both downregulated after silencing RHOT1, with PINK1
decreased by 59.75% (P=0.025) and Parkin decreased by 65.12%
compared with the si-NC group (P=0.0189) (Fig. 2C and D). Additionally, the mRNA
expression of mitochondrial membrane-related genes TOMM20
(translocase of the outer mitochondrial membrane 20) and
TIMM23 (translocase of the inner mitochondrial membrane 23)
were increased after silencing RHOT1 (Fig. 2E and F). The protein expression of
PINK1 and Parkin were downregulated after silencing RHOT1 (Fig. 2G).
These results indicated that silencing RHOT1 leads
to decreased PINK1 and Parkin expression and inhibition of
mitophagy.
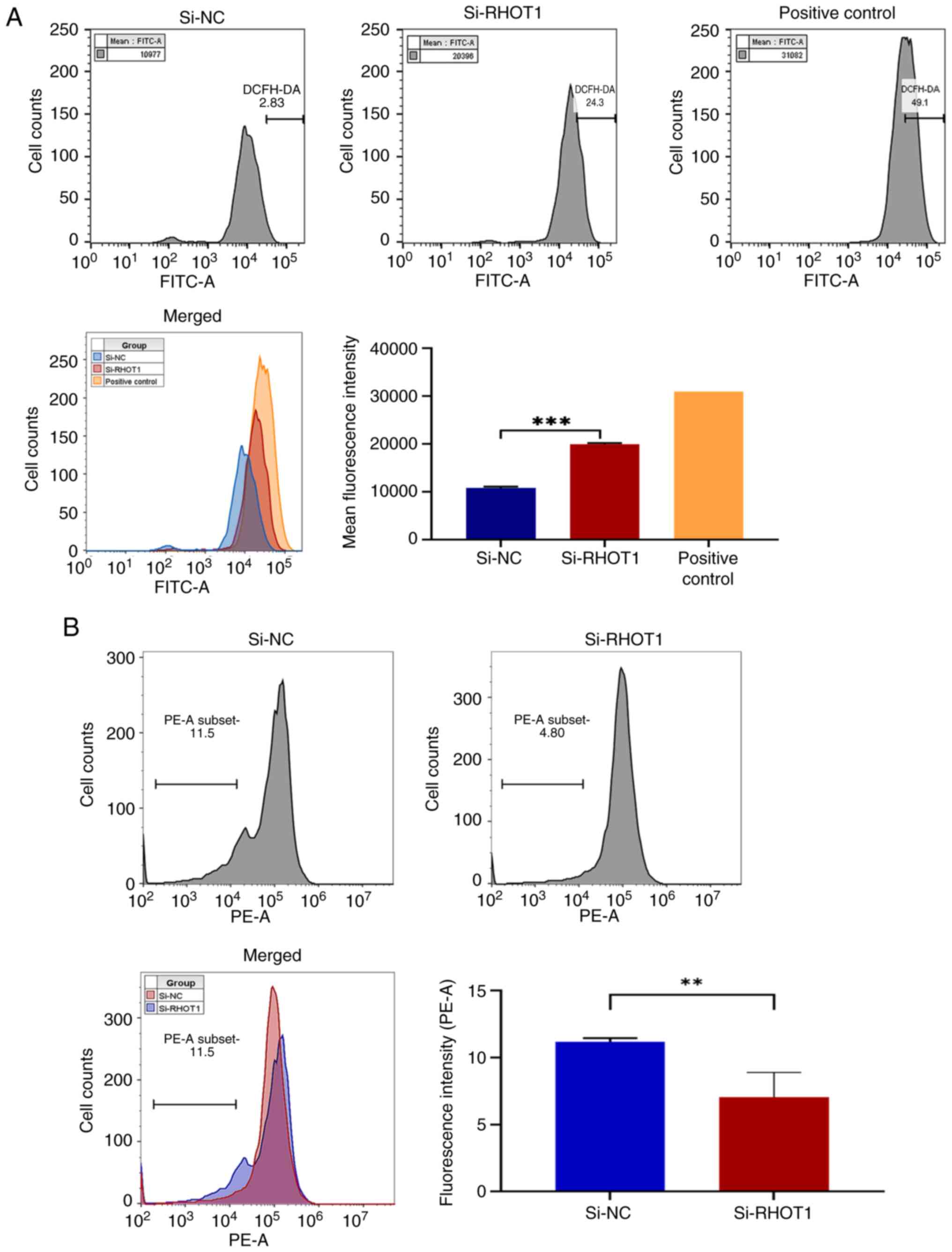
Silencing RHOT1 affects ROS and MMP in GC
cells
Based on the aforementioned results for PINK1,
Parkin and mitochondrial membrane-related genes, mitochondrial
function was further examined to assess mitophagy. Flow cytometry
analysis showed that the average ROS fluorescence intensity in
HGC-27 cells was increased by 84.73% (P<0.001) after silencing
RHOT1 compared with the negative control, while it remained lower
than the positive control (Fig.
3A). MMP assessment indicated that silencing RHOT1 caused a
36.94% decrease in MMP compared with the negative control
(P=0.0061), reflecting mitochondrial depolarization (Fig. 3B).
These results demonstrate that silencing RHOT1
elevates ROS levels and depolarizes the mitochondrial membrane,
indicating mitochondrial damage and dysfunction in HGC-27
cells.
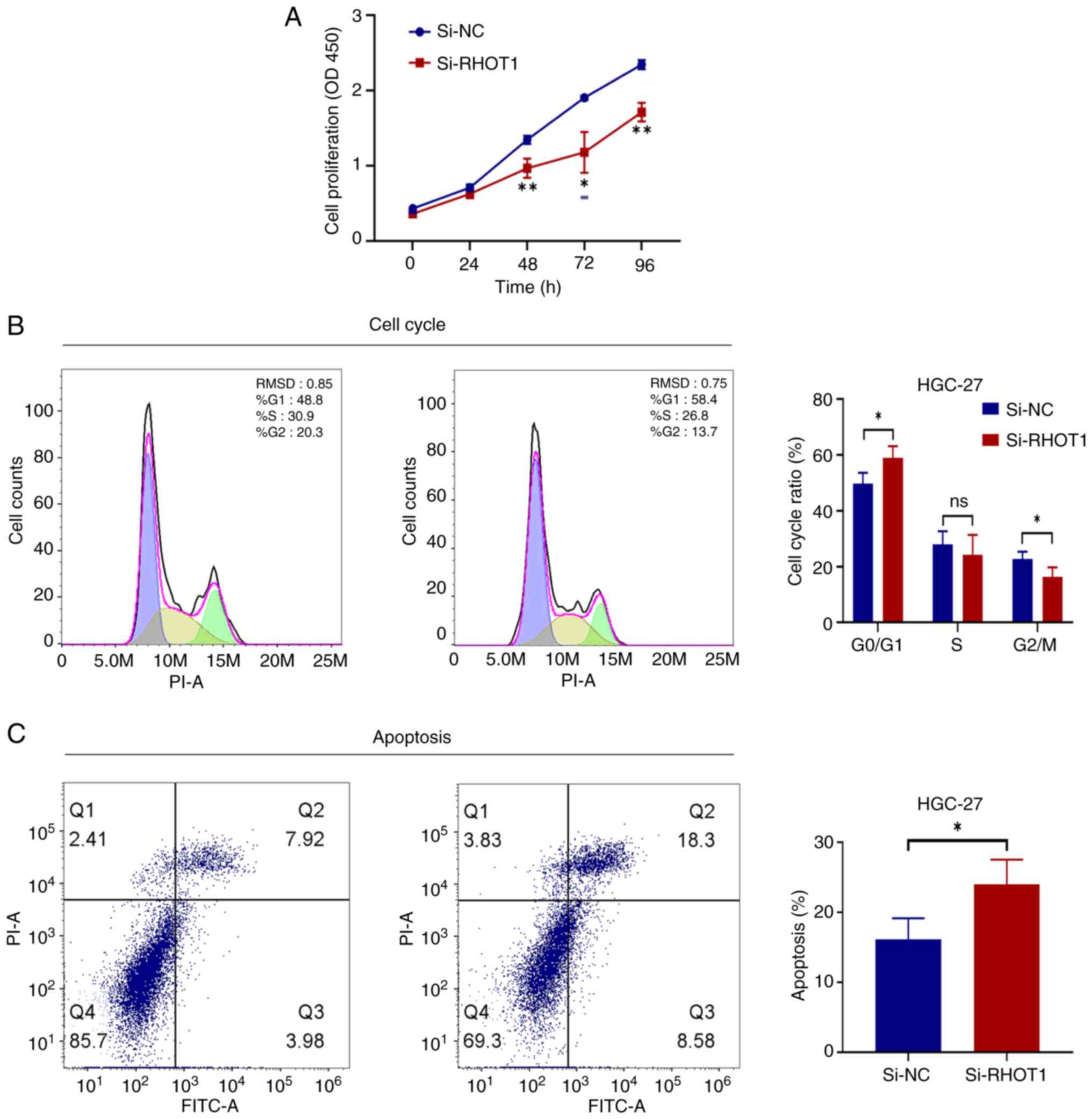
Silencing RHOT1 inhibits the
proliferation of GC cells arrests the cell cycle and promotes cell
apoptosis
Mitochondria were damaged and dysfunctional, which
would cause abnormal cellular energy metabolism and affect cell
growth. The effect of silencing RHOT1 on the growth state and
apoptosis of GC cells was assessed in HGC-27 cells. CCK-8 assay
showed that silencing RHOT1 markedly slowed HGC-27 cell
proliferation from 48 h to 96 h compared with the si-NC group
(Fig. 4A). Flow cytometry
analysis revealed that silencing RHOT1 increased the proportion of
cells in G0/G1 phase, causing cell cycle
arrest. Specifically, the G0/G1 phase
proportion was markedly higher than the si-NC group (P<0.05)
(Fig. 4B). Apoptosis assessment
demonstrated that silencing RHOT1 elevated the percentage of
apoptotic cells compared with the si-NC group (P<0.05) (Fig. 4C).
These results indicated that silencing RHOT1
affected mitophagy to reduce the proliferation of GC cells, arrest
the cell cycle in the G0/G1 phase and promote
cell apoptosis.
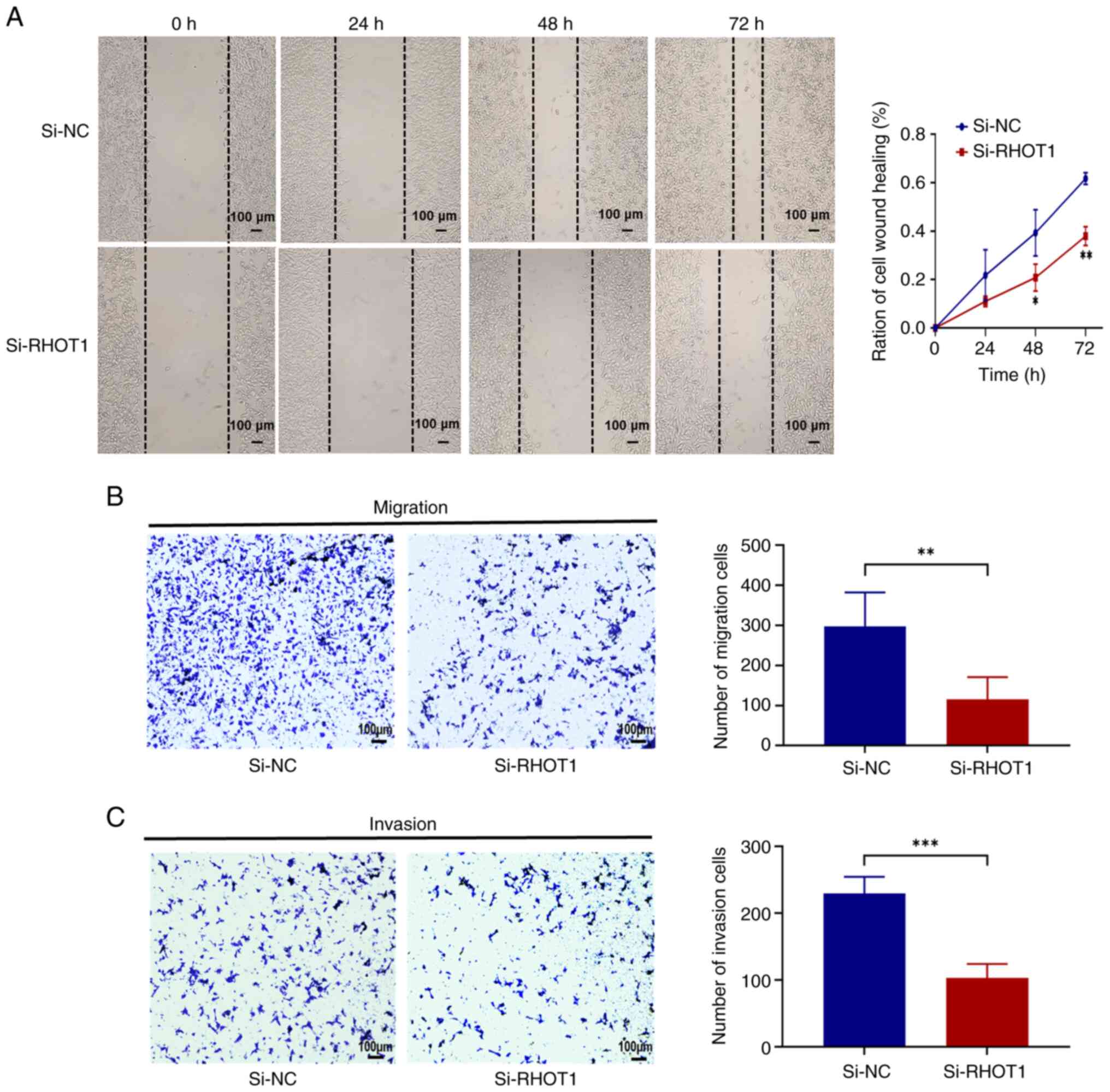
Silencing RHOT1 inhibits the migration
and invasion of GC cells
The present results showed that the expression of
PINK1 and Parkin was downregulated after silencing
RHOT1, so RHOT1 may affect mitophagy, leading to the accumulation
of damaged mitochondria, which further increased ROS and depleted
energy in the cells and consequently affected the invasion and
migration phenotypes of the cells. The effects of RHOT1 on the
invasion and migration of GC cells were subsequently investigated.
Wound healing and Transwell migration assays were employed to
evaluate the migration ability of HGC-27 cells. Wound healing assay
showed that silencing RHOT1 markedly reduced the migration of
HGC-27 cells compared with the si-NC group (Fig. 5A); Transwell migration assay
confirmed that the number of migrating cells in the si-RHOT1 group
was markedly lower than in the si-NC group (P<0.05) (Fig. 5B). Transwell invasion assay
further demonstrated that silencing RHOT1 markedly reduced the
number of invasive HGC-27 cells through Matrigel compared with the
control group (P<0.05; Fig.
5C).
These results indicated that silencing RHOT1
affected mitophagy to reduce the migration and invasion of GC
cells.
Discussion
GC is the fifth most common cause of cancer-related
death worldwide, contributing to 8.8% of all such deaths (27). Screening programs have reduced
mortality in some cancers by detecting early-stage and precancerous
lesions in asymptomatic patients, but organized GC screening has
only been implemented in a few high-prevalence countries (1). The etiology of GC is complex and
multifactorial, with numerous molecular alterations influencing
tumorigenesis and progression through aberrant gene expression or
protein dysfunction (28). At
present, there remains a necessity to explore treatment targets and
effective biomarkers.
Mitochondrial health is critical for cellular
function and mitophagy, the selective degradation of damaged
mitochondria, is central to maintaining mitochondrial quality
(29,30). Mitophagy exerts dual roles in
cancer, either prompting or suppressing carcinogenesis (31). RHOT1, an outer mitochondrial
membrane protein, serves as a molecular switch in the mitochondrial
dynamics and mitophagy (11,20,32). Mutations in RHOT1 disrupt
mitochondria transport, impair quality control and have been linked
to neurodegenerative diseases such as Parkinson's (11,33-35). RHOT1 is upregulated in several
cancers: Jiang et al (36)
predicted its prognostic value in pancreatic cancer and Li et
al (37) found that silencing
RHOT1 inhibited pancreatic cancer cell proliferation and migration.
In mice, RHOT1 knockdown reduced mitochondrial motility and
impaired quality control (38).
It was also found that RHOT1 interacted with LRRK2, consistent with
findings that LRRK2 knockdown or mutation impaired basal mitophagy
(39). Collectively, these data
position RHOT1 as a critical regulator of mitochondrial dynamics
and mitophagy. Database analyses revealed strong associations
between RHOT1 and PINK1/Parkin, the canonical ubiquitin-dependent
mitophagy pathway (40). In the
present study, silencing RHOT1 altered PINK1 and Parkin expression,
suggesting that RHOT1 modulates mitophagy via this axis. GO and
KEGG enrichment of RHOT1-interacting genes further supported
predominant effects on mitochondrial function and the involvement
of the PINK1/Parkin pathway.
Silencing RHOT1 also induced mitochondrial
dysfunction, characterized by increased ROS production and MMP
depolarization. ROS are central regulators of proliferation,
differentiation and metabolism, but excessive ROS promotes
oxidative stress, mtDNA mutations and mitochondrial damage
(41,42). RHOT1 has been implicated in
peroxisomal transport, which contributes to ROS clearance (43,44). Thus, the present findings of ROS
accumulation and MMP depolarization aligned with reports that RHOT1
deficiency aggravates oxidative stress and compromises
mitochondrial homeostasis. Consequently, dysfunctional mitochondria
accumulate, reinforcing oxidative stress and energy imbalance.
In addition, silencing RHOT1 upregulated TOMM20 and
TIMM23, proteins essential for mitochondrial protein import. This
upregulation probably reflects a compensatory response to
mitochondrial stress (45-47),
consistent with previous studies showing increased import machinery
and UPRmt components to preserve organelle integrity (48-50). Thus, RHOT1 deficiency appears to
couple impaired mitophagy with a UPRmt-like compensatory mechanism,
which will be a subgoal for future research.
Mitochondrial dysfunction also influenced cell cycle
progression and apoptosis. Oxidative stress stabilizes p53,
upregulates p21, inhibits CDK2/Cyclin E and blocks G1-S
transition (48,51-53). Disruption of MMP triggers
cytochrome c release, caspase activation and PARP cleavage,
promoting apoptosis (54-56). In agreement, silencing RHOT1 in GC
cells induced G0/G1 arrest and apoptosis,
linking mitochondrial quality control to cell-cycle regulation and
survival. Furthermore, RHOT1 loss impaired migration and invasion.
Reduced ATP production and ROS accumulation inhibit cytoskeletal
remodeling and pseudopodia formation (57,58), while ROS modulates EMT gene
expression via NF-κB and HIF-1α signaling (59). Myc has been shown to
transcriptionally regulate RHOT1, coordinating mitochondrial
trafficking with cytoskeletal dynamics (60). Dysregulated mitophagy and
accumulation of damaged mitochondria further promote tumor
progression (61).
In summary, RHOT1 deficiency in GC cells perturbs
mitophagy, exacerbates oxidative stress, disrupts mitochondrial
homeostasis and impairs key cellular processes, including cell
cycle progression, apoptosis and invasion. These findings
highlighted RHOT1 as a potential therapeutic target in GC. A
potential limitation of the present study is that functional assays
were mainly conducted in the HGC-27 cell line, which showed the
highest RHOT1 expression among the GC cell lines examined. While
this strengthens the rationale for its selection as the primary
model, further validation in additional GC subtypes would help to
generalize the findings. Addressing this limitation will be an
important focus of future research. In addition, the results
remained correlative, as no additional experiments were designed to
directly validate the causal relationship. This limitation was
acknowledged and the findings were therefore interpreted with
caution. Future studies will be required to provide more definitive
evidence of causality.
Silencing RHOT1 induced mitochondrial dynamics
disorder, resulting in the inhibition of the PINK1/Parkin mitophagy
pathway, which impaired mitophagy and caused the accumulation of
intracellular ROS and depolarization of MMP. The aforementioned
multiple ways triggered cell cycle arrest and promoted apoptosis,
resulting in decreased proliferation of GC cells and weakened
invasion and migration ability. Therefore, suppression of RHOT1 may
become a new strategy for the treatment of GC and provide a new
direction for the study of targeted treatment and drug resistance
mechanisms in GC.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YQP and XC conceived and designed the research; YQP,
XC, FK, JZ, SY were responsible for data analysis. RF, JW and YYP
were provided supervision and project administration. XC provided
supervision. YQP wrote the first draft and YYP reviewed and edited
the manuscript. FK, JZ and YYP confirm the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Doctoral Start-up
Foundation of Liaoning Province, China (grant no. 2022-BS-340); the
Science and Technology Research Project of Department of Education
of Liaoning Province (grant no. JYTZD2023145), the Science and
Technology Key Research Project of Liaoning Province (grant no.
2024JH2/102500059) and the Research Project on Integrated
Traditional Chinese and Western Medicine for Chronic Disease
Management (grant no. CXZH2024134).
References
|
1
|
Conti CB, Agnesi S, Scaravaglio M,
Masseria P, Dinelli ME, Oldani M and Uggeri F: Early gastric
cancer: Update on prevention, diagnosis and treatment. Int J
Environ Res Public Health. 20:21492023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rocken C: Predictive biomarkers in gastric
cancer. J Cancer Res Clin Oncol. 149:467–481. 2023. View Article : Google Scholar :
|
|
3
|
Smyth EC, Nilsson M, Grabsch HI, Van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Drizlionoks E, Tercioti V Junior, Coelho
Neto JS, Andreollo NA and Lopes LR: Surgical treatment of gastric
stump cancer: A cohort study of 51 patients. Arq Bras Cir Dig.
37:e18502025. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang WJ, Zhao HP, Yu Y, Wang JH, Guo L,
Liu JY, Pu J and Lv J: Updates on global epidemiology, risk and
prognostic factors of gastric cancer. World J Gastroenterol.
29:2452–2468. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu H, Cao S and Xu R: Cancer incidence,
mortality, and burden in China: A time-trend analysis and
comparison with the United States and United Kingdom based on the
global epidemiological data released in 2020. Cancer Commun (Lond).
41:1037–1048. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Veitch AM, Uedo N, Yao K and East JE:
Optimizing early upper gastrointestinal cancer detection at
endoscopy. Nat Rev Gastroenterol Hepatol. 12:660–667. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Haggstrom L, Chan WY, Nagrial A, Chantrill
LA, Sim HW, Yip D and Chin V: Chemotherapy and radiotherapy for
advanced pancreatic cancer. Cochrane Database Syst Rev.
12:CD0110442024.PubMed/NCBI
|
|
9
|
Gao S, Wang Y, Shan Y, Wang W, Li J and
Tan H: Rhizoma Coptidis polysaccharides: Extraction, separation,
purification, structural characteristics and bioactivities. Int J
Biol Macromol. 320(Pt 1): 1456772025. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao S, Xu T, Wang W, Li J, Shan Y, Wang Y
and Tan H: Polysaccharides from Anemarrhena asphodeloides Bge, the
extraction, purification, structure characterization, biological
activities and application of a traditional herbal medicine. Int J
Biol Macromol. 311(Pt 1): 1434972025. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kavyashree S, Harithpriya K and Ramkumar
KM: Miro1-a key player in β-cell function and mitochondrial
dynamics under diabetes mellitus. Mitochondrion. 84:1020392025.
View Article : Google Scholar
|
|
12
|
Li JM, Li X, Chan LWC, Hu R, Zheng T, Li H
and Yang S: Lipotoxicity-polarised macrophage-derived exosomes
regulate mitochondrial fitness through Miro1-mediated mitophagy
inhibition and contribute to type 2 diabetes development in mice.
Diabetologia. 66:2368–2386. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeong YY, Jia N, Ganesan D and Cai Q:
Broad activation of the PRKN pathway triggers synaptic failure by
disrupting synaptic mitochondrial supply in early tauopathy.
Autophagy. 18:1472–1474. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maremanda KP, Sundar IK and Rahman I:
Protective role of mesenchymal stem cells and mesenchymal stem
cell-derived exosomes in cigarette smoke-induced mitochondrial
dysfunction in mice. Toxicol Appl Pharmacol. 385:1147882019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li D, Su H, Deng X, Huang Y, Wang Z, Zhang
J, Chen C, Zheng Z, Wang Q, Zhao S, et al: DARS2 promotes bladder
cancer progression by enhancing PINK1-mediated mitophagy. Int J
Biol Sci. 21:1530–1544. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deng X, Huang Y, Zhang J, Chen Y, Jiang F,
Zhang Z, Li T, Hou L, Tan W and Li F: Histone lactylation regulates
PRKN-Mediated mitophagy to promote M2 Macrophage polarization in
bladder cancer. Int Immunopharmacol. 148:1141192025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Yu C, Xie G, Tao K, Yin Z and Lv
Q: USP14 inhibits mitophagy and promotes tumorigenesis and
chemosensitivity through deubiquitinating BAG4 in microsatellite
instability-high colorectal cancer. Mol Med. 31:1632025. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng B, Wang Y, Zhou B, Qian F, Liu D, Ye
D, Zhou X and Fang L: Urolithin A inhibits breast cancer
progression via activating TFEB-mediated mitophagy in tumor
macrophages. J Adv Res. 69:125–138. 2025. View Article : Google Scholar :
|
|
19
|
Zheng S, Zhang Y, Gong X, Teng Z and Chen
J: CREB1 regulates RECQL4 to inhibit mitophagy and promote
esophageal cancer metastasis. J Clin Biochem Nutr. 75:102–110.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chemla A, Arena G, Sacripanti G, Barmpa K,
Zagare A, Garcia P, Gorgogietas V, Antony P, Ohnmacht J, Baron A,
et al: Parkinson's disease mutant Miro1 causes mitochondrial
dysfunction and dopaminergic neuron loss. Brain. Feb 6–2025.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cavinato M: Mitochondrial dysfunction and
cisplatin sensitivity in gastric cancer: GDF15 as a master player.
FEBS J. 291:1111–1114. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lo YL, Wang TY, Chen CJ, Chang YH and Lin
AM: Two-in-one nanoparticle formulation to deliver a tyrosine
kinase inhibitor and microRNA for targeting metabolic reprogramming
and mitochondrial dysfunction in gastric cancer. Pharmaceutics.
14:17592022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen W, Zou P, Zhao Z, Weng Q, Chen X,
Ying S, Ye Q, Wang Z, Ji J and Liang G: Selective killing of
gastric cancer cells by a small molecule via targeting TrxR1 and
ROS-mediated ER stress activation. Oncotarget. 7:16593–16609. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rong Y, Teng Y and Zhou X: Advances in the
study of metabolic reprogramming in gastric cancer. Cancer Med.
14:e709482025. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng YY, Sun D and Xin Y: Hsa_circ_0005230
is up-regulated and promotes gastric cancer cell invasion and
migration via regulating the miR-1299/RHOT1 axis. Bioengineered.
13:5046–5063. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Li M, Pi H, Yang Z, Reiter RJ, Xu S, Chen
X, Chen C, Zhang L, Yang M, Li Y, et al: Melatonin antagonizes
cadmium-induced neurotoxicity by activating the transcription
factor EB-dependent autophagy-lysosome machinery in mouse
neuroblastoma cells. J Pineal Res. 61:353–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Procaccio L, Schirripa M, Fassan M,
Vecchione L, Bergamo F, Prete AA, Intini R, Manai C, Dadduzio V,
Boscolo A, et al: Immunotherapy in gastrointestinal cancers. Biomed
Res Int. 2017:43465762017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Leites EP and Morais VA: Mitochondrial
quality control pathways: PINK1 acts as a gatekeeper. Biochem
Biophys Res Commun. 500:45–50. 2018. View Article : Google Scholar
|
|
30
|
Yao RQ, Ren C, Xia ZF and Yao YM:
Organelle-specific autophagy in inflammatory diseases: A potential
therapeutic target underlying the quality control of multiple
organelles. Autophagy. 17:385–401. 2021. View Article : Google Scholar :
|
|
31
|
Wang Z, Chen C, Ai J, Shu J, Ding Y, Wang
W, Gao Y, Jia Y and Qin Y: Identifying mitophagy-related genes as
prognostic biomarkers and therapeutic targets of gastric carcinoma
by integrated analysis of single-cell and bulk-RNA sequencing data.
Comput Biol Med. 163:1072272023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Eberhardt EL, Ludlam AV, Tan Z and
Cianfrocco MA: Miro: A molecular switch at the center of
mitochondrial regulation. Protein Sci. 29:1269–1284. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lopez-Domenech G, Howden JH, Covill-Cooke
C, Morfill C, Patel JV, Burli R, Crowther D, Birsa N, Brandon NJ
and Kittler JT: Loss of neuronal Miro1 disrupts mitophagy and
induces hyperactivation of the integrated stress response. EMBO J.
40:e1007152021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Diaz-Moreno I and De La Rosa MA: IUBMB
focused meeting/FEBS workshop: Crosstalk between nucleus and
mitochondria in human disease (CrossMitoNus). IUBMB Life.
73:489–491. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grossmann D, Berenguer-Escuder C, Chemla
A, Arena G and Kruger R: The Emerging role of RHOT1/Miro1 in the
pathogenesis of parkinson's disease. Front Neurol. 11:5872020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang H, He C, Geng S, Sheng H, Shen X,
Zhang X, Li H, Zhu S, Chen X, Yang C and Gao H: RhoT1 and Smad4 are
correlated with lymph node metastasis and overall survival in
pancreatic cancer. PLoS One. 7:e422342012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Q, Yao L, Wei Y, Geng S, He C and Jiang
H: Role of RHOT1 on migration and proliferation of pancreatic
cancer. Am J Cancer Res. 5:1460–1470. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lopez-Domenech G, Higgs NF, Vaccaro V, Ros
H, Arancibia-Carcamo IL, Macaskill AF and Kittler JT: Loss of
dendritic complexity precedes neurodegeneration in a mouse model
with disrupted mitochondrial distribution in mature dendrites. Cell
Rep. 17:317–327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Singh F, Prescott AR, Rosewell P, Ball G,
Reith AD and Ganley IG: Pharmacological rescue of impaired
mitophagy in Parkinson's disease-related LRRK2 G2019S knock-in
mice. Elife. 10:e676042021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cao Y, Zheng J, Wan H, Sun Y, Fu S, Liu S,
He B, Cai G, Cao Y, Huang H, et al: A mitochondrial SCF-FBXL4
ubiquitin E3 ligase complex degrades BNIP3 and NIX to restrain
mitophagy and prevent mitochondrial disease. EMBO J.
42:e1130332023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lu Y, Li Z, Zhang S, Zhang T, Liu Y and
Zhang L: Cellular mitophagy: Mechanism, roles in diseases and small
molecule pharmacological regulation. Theranostics. 13:736–766.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lemasters JJ: Selective mitochondrial
autophagy, or mitophagy, as a targeted defense against oxidative
stress, mitochondrial dysfunction, and aging. Rejuvenation Res.
8:3–5. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Covill-Cooke C, Toncheva VS, Drew J, Birsa
N, Lopez-Domenech G and Kittler JT: Peroxisomal fission is
modulated by the mitochondrial Rho-GTPases, Miro1 and Miro2. EMBO
Rep. 21:e498652020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Costello JL, Castro IG, Camoes F, Schrader
TA, Mcneall D, Yang J, Giannopoulou EA, Gomes S, Pogenberg V,
Bonekamp NA, et al: Predicting the targeting of tail-anchored
proteins to subcellular compartments in mammalian cells. J Cell
Sci. 130:1675–1687. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Raiff A, Zhao S, Bekturova A, Zenge C,
Mazor S, Chen X, Ru W, Makaros Y, Ast T, Ordureau A, et al:
TOM20-driven E3 ligase recruitment regulates mitochondrial dynamics
through PLD6. Nat Chem Biol. Apr 22–2025.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Saleem A, Iqbal S, Zhang Y and Hood DA:
Effect of p53 on mitochondrial morphology, import, and assembly in
skeletal muscle. Am J Physiol Cell Physiol. 308:C319–C329. 2015.
View Article : Google Scholar :
|
|
47
|
Zha J, Li J, Yin H, Shen M and Xia Y:
TIMM23 overexpression drives NSCLC cell growth and survival by
enhancing mitochondrial function. Cell Death Dis. 16:1742025.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kumar N, Shukla A, Kumar S, Ulasov I,
Singh RK, Kumar S, Patel A, Yadav L, Tiwari R, Paswan R, et al: FNC
(4'-azido-2'-deoxy-2'-fluoro(arbino)cytidine) as an effective
therapeutic agent for NHL: ROS generation, cell cycle arrest, and
mitochondrial-mediated apoptosis. Cell Biochem Biophys. 82:623–639.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tian X, Yuan M, Li L, Chen D, Liu B, Zou
X, He M and Wu Z: Enterovirus 71 induces mitophagy via PINK1/Parkin
signaling pathway to promote viral replication. FASEB J.
39:e706592025. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lu J, Zhang Y, Song H, Wang F, Wang L,
Xiong L and Shen X: A novel polysaccharide from tremella fuciformis
alleviated high-fat diet-induced obesity by promoting
AMPK/PINK1/PRKN-Mediated mitophagy in mice. Mol Nutr Food Res.
69:e2024006992025. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bai LY, Su JH, Chiu CF, Lin WY, Hu JL,
Feng CH, Shu CW and Weng JR: Antitumor effects of a sesquiterpene
derivative from marine sponge in human breast cancer cells. Mar
Drugs. 19:2442021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bhavya K, Mantipally M, Roy S, Arora L,
Badavath VN, Gangireddy M, Dasgupta S, Gundla R and Pal D: Novel
imidazo[1,2-a]pyridine derivatives induce apoptosis and cell cycle
arrest in non-small cell lung cancer by activating NADPH oxidase
mediated oxidative stress. Life Sci. 294:1203342022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Minella AC, Swanger J, Bryant E, Welcker
M, Hwang H and Clurman BE: p53 and p21 form an inducible barrier
that protects cells against cyclin E-cdk2 deregulation. Curr Biol.
12:1817–1827. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yu C, Sun J, Lai X, Tan Z, Wang Y, Du H,
Pan Z, Chen T, Yang Z, Ye S, et al: Gefitinib induces apoptosis in
Caco-2 cells via ER stress-mediated mitochondrial pathways and the
IRE1alpha/JNK/p38 MAPK signaling axis. Med Oncol. 42:712025.
View Article : Google Scholar
|
|
55
|
Guefack MF, Talukdar D, Mukherjee R, Guha
S, Mitra D, Saha D, Das G, Damen F, Kuete V and Murmu N: Hypericum
roeperianum bark extract suppresses breast cancer proliferation via
induction of apoptosis, downregulation of PI3K/Akt/mTOR signaling
cascade and reversal of EMT. J Ethnopharmacol. 319(Pt 1):
1170932024. View Article : Google Scholar
|
|
56
|
Huang J, Zhang Y, Cheng A, Wang M, Liu M,
Zhu D, Chen S, Zhao X, Yang Q, Wu Y, et al: Duck Circovirus
genotype 2 ORF3 protein induces apoptosis through the mitochondrial
pathway. Poult Sci. 102:1025332023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Peng J, Yang Z, Li H, Hao B, Cui D, Shang
R, Lv Y, Liu Y, Pu W, Zhang H, et al: Quercetin reprograms
immunometabolism of macrophages via the SIRT1/PGC-1alpha signaling
pathway to ameliorate lipopolysaccharide-induced oxidative damage.
Int J Mol Sci. 24:55422023. View Article : Google Scholar
|
|
58
|
Chen JT, Wei L, Chen TL, Huang CJ and Chen
RM: Regulation of cytochrome P450 gene expression by ketamine: A
review. Expert Opin Drug Metab Toxicol. 14:709–720. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Qin W, Li C, Zheng W, Guo Q, Zhang Y, Kang
M, Zhang B, Yang B, Li B, Yang H and Wu Y: Inhibition of autophagy
promotes metastasis and glycolysis by inducing ROS in gastric
cancer cells. Oncotarget. 6:39839–39854. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Agarwal E, Altman BJ, Ho Seo J, Bertolini
I, Ghosh JC, Kaur A, Kossenkov AV, Languino LR, Gabrilovich DI,
Speicher DW, et al: Myc regulation of a mitochondrial trafficking
network mediates tumor cell invasion and metastasis. Mol Cell Biol.
39:e00109–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Panigrahi DP, Praharaj PP, Bhol CS,
Mahapatra KK, Patra S, Behera BP, Mishra SR and Bhutia SK: The
emerging, multifaceted role of mitophagy in cancer and cancer
therapeutics. Semin Cancer Biol. 66:45–58. 2020. View Article : Google Scholar
|