Introduction
Lung cancer accounts for 18% of global cancer
deaths, with non-small-cell lung cancer (NSCLC) comprising 85% of
cases (1). Despite advances in
targeted agents and immune checkpoint blockade (ICB), the 5-year
survival rate remains at ≤25% (2). Notably, primary or acquired
resistance occurs in 50-60% of patients with EGFR mutations within
12-18 months (3) and in ≥30% of
patients treated with ICB (4).
Intratumoral heterogeneity and clonal evolution are recognized
drivers of resistance; however, the tumor microenvironment (TME) is
increasingly appreciated as an active factor that imposes
metabolic, physical and immunological barriers to therapy (5).
The heterogeneity of NSCLC further complicates
treatment. Genomic analyses have revealed distinct molecular
subtypes of NSCLC, such as KRAS, BRAF and RET, each requiring
tailored therapies (6). However,
even within the same subtype, intratumoral heterogeneity drives
clonal evolution and therapeutic escape (5). For example, KRAS G12C inhibitors,
such as sotorasib, achieve objective response rates (ORRs) of 37%;
however, resistance emerges through alternative pathway activation
or phenotypic switching (7).
Immunotherapy resistance is equally complex, with TME-mediated
mechanisms such as T-cell exhaustion, myeloid-derived suppressor
cells (MDSCs) and collagen-rich physical barriers impeding immune
cell infiltration (8).
The TME has emerged as a critical factor in cancer
progression and therapeutic resistance, as supported by both
preclinical models and clinical trial data (9-14).
Comprising tumor cells, immune cells, stromal components,
extracellular matrix (ECM) and bioactive molecules, the TME is a
dynamic ecosystem that actively promotes tumor evolution, immune
evasion and therapeutic resistance (10). Chronic inflammation within the
TME, for example, has been linked to the development of an
immunosuppressive environment, which facilitates tumor progression
and resistance to immunotherapy (11). Studies have shown that components
of the TME, such as tumor-associated macrophages (TAMs), MDSCs and
regulatory T cells (Tregs), serve key roles in suppressing
antitumor immune responses (12,13). Additionally, the ECM and its
degradative enzymes contribute to tumor invasion and metastasis by
remodeling the physical structure of the TME (14). Understanding these interactions is
crucial for developing novel therapeutic strategies that target the
TME to overcome resistance and improve patient outcomes.
The present review aims to provide a comprehensive
analysis of the role of the TME in lung cancer progression and
therapeutic resistance. Firstly, the molecular mechanisms
underlying TME-mediated immune evasion and resistance to
chemotherapy, radiotherapy and immunotherapy were explored.
Furthermore, emerging therapeutic strategies targeting the TME,
such as immunotherapy combinations, anti-angiogenic therapies and
nanoparticle-based drug delivery systems, were critically
evaluated, and their potential to overcome resistance and enhance
treatment efficacy discussed. The current review will highlight the
importance of integrating TME profiling into personalized medicine
approaches, and emphasize the need for further research to address
the challenges posed by TME heterogeneity and plasticity. By
bridging gaps in the current knowledge, the review seeks to inform
future research directions and clinical applications in the
management of lung cancer.
Heterogeneity of the lung cancer
microenvironment
The heterogeneity of the lung cancer
microenvironment arises from the complex interplay of cellular and
non-cellular components, as well as emerging factors such as
microbiota and neuronal crosstalk (15,16). This heterogeneity poses notable
challenges for therapeutic intervention, necessitating a deeper
understanding of the dynamic interactions within the TME to develop
effective treatment strategies.
Cellular components
The TME in lung cancer is characterized by a complex
array of immune cells, including TAMs, MDSCs and Tregs, which
collectively contribute to an immunosuppressive environment
(5) (Fig. 1). TAMs, particularly the
M2-polarized subset, secrete interleukin (IL)-10 and transforming
growth factor-β (TGF-β) to inhibit antitumor immunity and support
tumor progression (5). Similarly,
MDSCs impair cytotoxic T-cell function via reactive oxygen species
and arginase production (17),
and Tregs further reinforce immunosuppression through secretion of
immunosuppressive cytokines, such as IL-10 and TGF-β (18). However, studies have highlighted
the heterogeneity of these immune cells within the TME (19,20). For example, it has been suggested
that TAMs can exhibit both protumorigenic and antitumorigenic
functions depending on their spatial distribution and functional
orientation within the tumor (21,22). This heterogeneity complicates
therapeutic strategies targeting TAMs, as interventions may need to
account for their dual roles (23).
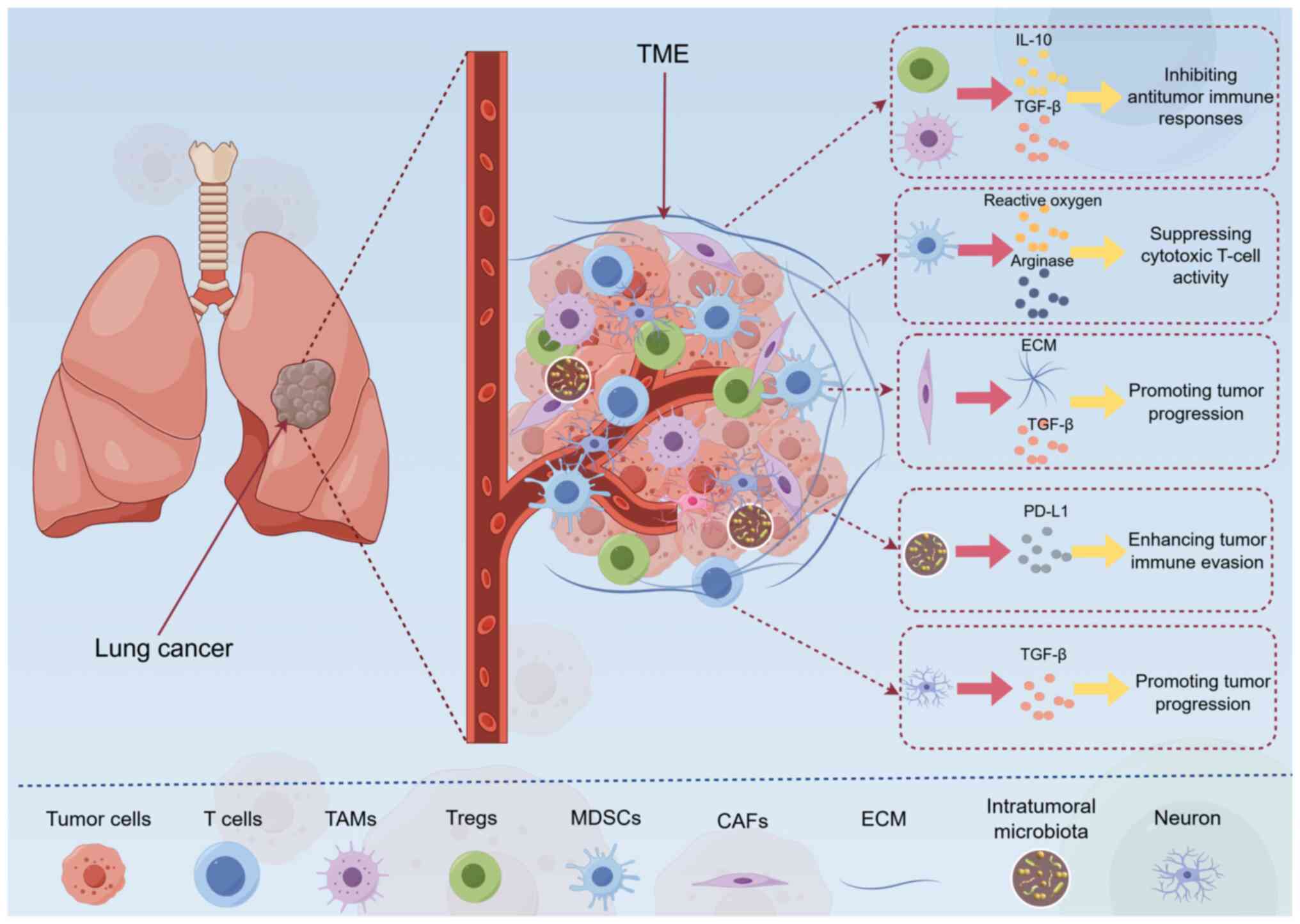 | Figure 1TME composition and interactions in
lung cancer. Components of the TME, including immune cells (such as
TAMs, MDSCs and Tregs), stromal cells (for example, CAFs and
endothelial cells), ECM and soluble factors (cytokines, chemokines
and growth factors), are shown, as are the interaction between
these components. For example, TAMs secrete IL-10 and TGF-β, thus
inhibiting antitumor immune responses, and CAFs promote tumor
progression by secreting ECM components and growth factors. This
figure was created using Figdraw (www.figdraw.com, ID: YTYOW4a999). CAFs,
cancer-associated fibroblasts; ECM, extracellular matrix; IL-10,
interleukin-10; MDSCs, myeloid-derived suppressor cells; PD-L1,
programmed death-ligand 1; TAMs, tumor-associated macrophages; TME,
tumor microenvironment; TGF-β, transforming growth factor-β; Tregs,
regulatory T cells. |
Cancer-associated fibroblasts (CAFs) contribute to
tumor progression and therapeutic resistance by remodeling the ECM
and secreting growth factors such as TGF-β and fibroblast growth
factor (FGF) (24) (Fig. 1). Through ECM deposition and
cross-linking, CAFs create a physical barrier that impedes drug
penetration and immune cell infiltration, while also promoting
angiogenesis and metastatic dissemination (25,26). Additionally, the presence of other
immune cell subsets, such as natural killer (NK) cells and γδ T
cells, has been shown to influence tumor outcomes. NK cells, known
for their ability to recognize and lyse tumor cells without prior
sensitization, can be impaired in the TME due to factors such as
TGF-β and programmed death-ligand 1 (PD-L1) expression (27). γδ T cells, which are involved in
both innate and adaptive immunity, have also been shown to serve a
role in shaping the TME and affecting therapeutic responses
(28).
Non-cellular components
The ECM undergoes dynamic remodeling in the TME,
driven by matrix stiffness, collagen crosslinking and protease
activity (29). Matrix
metalloproteinases (MMPs) and other proteases degrade the ECM,
facilitating tumor invasion and metastasis (30). Additionally, increased matrix
stiffness can enhance tumor cell proliferation and resistance to
therapy by promoting mechanotransduction pathways (30). These changes in the ECM create a
physical barrier that limits drug delivery and contributes to
therapeutic resistance.
Soluble factors, such as cytokines (for example,
IL-6 and TNF-α), chemokines and growth factors, are critical
drivers of TME heterogeneity. IL-6, for example, promotes tumor
progression by activating the STAT3 pathway, which enhances cell
survival and proliferation (31).
TNF-α, while often associated with inflammation, can also
contribute to immunosuppression by upregulating PD-L1 expression on
tumor cells (31). Growth factors
such as vascular endothelial growth factor (VEGF) drive
angiogenesis and create an immunosuppressive microenvironment,
further complicating therapeutic strategies (32).
Emerging factors
Recent studies have identified the presence of
intratumoral microbiota within the lung cancer microenvironment,
with microbial metabolites influencing oncogenic signaling and
immune evasion (33,34). For example, certain bacterial
species have been shown to modulate the expression of immune
checkpoint molecules, such as PD-L1, thereby enhancing tumor immune
evasion (35) (Fig. 1). Furthermore, the role of the
gut-lung axis in shaping the lung TME has garnered attention. The
gut microbiota can influence systemic immunity and inflammation,
which in turn affects the composition and function of the lung
microbiota and TME (36). This
interplay between the gut and lung may offer novel targets for
therapeutic intervention.
Neuronal interactions within the TME have also
emerged as a novel area of research (37). Neurotrophic factors and
axonogenesis promote tumor innervation, which can enhance tumor
growth and metastasis (38).
Studies have suggested that neural signaling may influence the
secretion of protumorigenic factors, such as TGF-β, and create a
permissive environment for tumor progression (39,40) (Fig.
1). Additionally, the involvement of neuronal-derived exosomes
in transmitting signals that promote tumor cell survival and
therapeutic resistance has been revealed (41), highlighting the importance of
understanding the bidirectional communication between neurons and
tumor cells in shaping the TME.
The heterogeneity of the lung cancer
microenvironment, driven by diverse cellular interactions and
dynamic non-cellular components, presents a notable barrier to
effective therapy. Understanding these complex interactions is
essential for developing targeted approaches that can modulate the
TME to enhance treatment efficacy. By addressing the dual roles of
immune cells, the fibrotic networks created by CAFs and the
physical barriers of the ECM, future therapeutic strategies may be
better designed to overcome resistance and improve patient
outcomes.
Dynamic crosstalk in the TME: Mechanisms of
progression
The intricate crosstalk within the TME facilitates
immune evasion, metabolic reprogramming and epigenetic modulation,
thereby creating a permissive niche for tumor growth and
therapeutic resistance (12,42). Understanding these dynamic
interactions is crucial for developing novel therapeutic strategies
that target the TME to enhance treatment efficacy and overcome
resistance in lung cancer.
Immune evasion and checkpoint
dysregulation
The TME facilitates immune evasion through various
mechanisms, including the upregulation of immune checkpoint
molecules and the polarization of immune cells (11). PD-L1, a well-known immune
checkpoint protein, is often upregulated in lung cancer cells,
enabling them to suppress T-cell activity and evade immune
detection (43,44) (Fig.
2). Previous studies have also highlighted the role of the
lymphocyte-activation gene 3 (LAG-3)/fibrinogen-like protein 1
(FGL1) axis and V-type immunoglobulin domain-containing suppressor
of T-cell activation (VISTA) in immune evasion. LAG-3, expressed on
exhausted T cells, interacts with FGL1, leading to T-cell
dysfunction (45). VISTA, another
immune checkpoint molecule, contributes to the immunosuppressive
environment by inhibiting T-cell activation (46). TAMs serve a notable role in immune
evasion by polarizing toward an M2 phenotype. This polarization is
driven by cytokines such as colony stimulating factor 1 (CSF-1),
which activates CSF-1 receptor signaling in macrophages (47). However, the extent to which these
mechanisms contribute to immune evasion may vary among different
lung cancer subtypes and patient populations, necessitating further
investigation to clarify their roles and potential therapeutic
targets.
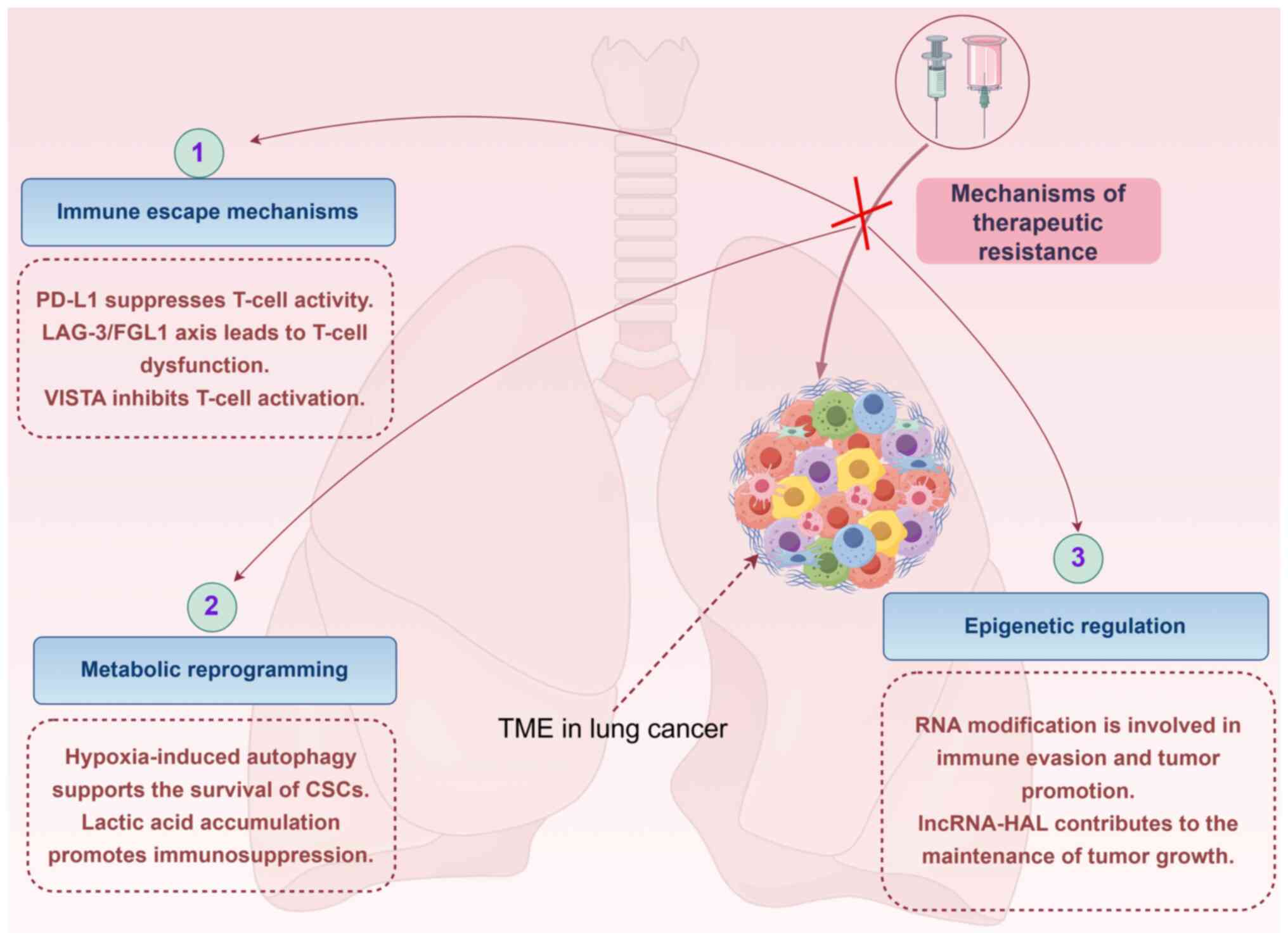 | Figure 2Mechanisms of therapeutic resistance
in the TME. The mechanism by which the TME drives treatment
resistance is shown, including immune escape mechanisms (such as
PD-L1 upregulation, the LAG-3/FGL1 axis and the role of VISTA),
metabolic reprogramming (for example, hypoxia-induced autophagy and
immunosuppression due to lactic acid accumulation) and epigenetic
regulation (such as RNA modification and the role of lncRNAs). This
figure was created using Figdraw (www.figdraw.com, ID: STSWAa4006). CSCs, cancer stem
cells; FGL1, fibrinogen-like protein 1; LAG-3,
lymphocyte-activation gene 3; lncRNA, long non-coding RNA; PD-L1,
programmed death-ligand 1; TME, tumor microenvironment; VISTA,
V-type immunoglobulin domain-containing suppressor of T-cell
activation. |
Metabolic reprogramming and hypoxia
Hypoxia is a common feature of the TME, and has
marked effects on tumor progression and therapeutic resistance
(48) (Fig. 2). Hypoxia-induced autophagy,
mediated by hypoxia-inducible factor-1α and BCL-2/adenovirus E1B 19
kDa protein-interacting protein 3, supports the survival of cancer
stem cells (CSCs). CSCs are known for their ability to self-renew
and differentiate, contributing to tumor heterogeneity and
resistance to therapy (49). By
maintaining CSC survival, hypoxia-induced autophagy ensures a
reservoir of cells capable of repopulating the tumor after
treatment.
Lactate, a byproduct of anaerobic metabolism,
accumulates in the TME under hypoxic conditions and serves a role
in promoting immunosuppression (50) (Fig.
2). Lactate-mediated signaling in stromal cells, such as
Notch/C-C motif chemokine ligand 5, fosters an immunosuppressive
environment by attracting Tregs and inhibiting the function of
cytotoxic T cells (51). This
metabolic reprogramming not only supports tumor growth but also
creates a barrier to effective immune responses, highlighting the
complex interplay between metabolic changes and immune modulation
in the TME.
Epigenetic modulation in the TME
Epigenetic changes within the TME markedly influence
tumor progression and therapeutic resistance (42) (Fig.
2). RNA modifications, including m6A, m5C and ac4C, have
emerged as critical regulators of mRNA stability and translation.
These modifications can affect the expression of immune-related
genes, such as PD-L1, STAT1 and IRF7, thereby
modulating immune cell function and the overall immune response
within the TME (52). For
example, m6A methylation of specific mRNA transcripts can enhance
their stability and translation efficiency, leading to increased
production of proteins involved in immune evasion and tumor
promotion (53).
Long non-coding RNAs (lncRNAs) also serve a role in
shaping the TME through epigenetic mechanisms (54). Hypoxia-induced lncRNA-HAL has been
shown to promote stemness and therapeutic resistance by interacting
with chromatin remodeling complexes (55). By altering the epigenetic
landscape, lncRNA-HAL contributes to the maintenance of a favorable
environment for tumor growth and resistance to treatment (56). Understanding the specific roles of
these epigenetic modulators and their interactions within the TME
is crucial for developing novel therapeutic strategies aimed at
reversing epigenetic changes and enhancing treatment efficacy in
lung cancer.
The intricate crosstalk within the TME, involving
immune evasion, metabolic reprogramming and epigenetic modulation,
creates a complex landscape of resistance mechanisms in lung
cancer. By targeting key nodes in these networks, such as immune
checkpoint regulation, hypoxia-driven pathways and epigenetic
modifiers, novel therapies can be developed to disrupt the
supportive role of the TME in tumor progression. These approaches
hold promise for enhancing the effectiveness of existing treatments
and addressing the challenge of therapeutic resistance.
Resistance to targeted therapy
Resistance to targeted therapies, such as
EGFR-tyrosine kinase inhibitors (TKIs), is a notable challenge in
lung cancer treatment. One of the key mechanisms underlying this
resistance is the activation of alternative signaling pathways.
TAMs contribute to resistance against targeted therapies such as
EGFR-TKIs by secreting hepatocyte growth factor, which activates
the c-MET pathway and bypasses EGFR inhibition (57). Moreover, TAMs and CAFs
collaboratively foster a fibrotic niche via ECM remodeling, which
physically restricts drug access and activates alternative survival
pathways (58,59).
Genetic alterations, including secondary EGFR
mutations (such as T790M) and MET amplification, remain key drivers
of acquired resistance (60).
These changes are often facilitated by a pro-inflammatory TME,
which promotes genomic instability and enriches for resistant
clones (61).
Resistance to ICB
Immune checkpoint inhibitors (ICIs) have
revolutionized the treatment landscape for lung cancer, but
resistance to these therapies remains a key issue. Resistance to
ICIs can be influenced by various factors within the TME (62). For example, the presence of
immunosuppressive cells, such as MDSCs and Tregs, can inhibit the
activation and function of cytotoxic T cells. These
immunosuppressive cells can be recruited and activated by cytokines
such as TGF-β and IL-10, which are often upregulated in the TME.
The dense infiltration of MDSCs and Tregs creates a suppressive
milieu that limits the efficacy of ICB (63).
Moreover, physical barriers within the TME, such as
a dense ECM and poor vascularization, can prevent immune cells from
infiltrating the tumor (64). A
recent study demonstrated that the ECM stiffness, mediated by
collagen crosslinking and MMPs, can physically impede T-cell
migration and reduce the delivery of ICIs to their targets. This
structural barrier is further exacerbated by the presence of
immunosuppressive cytokines, which collectively contribute to the
resistance of tumors to immunotherapy (64).
The interplay between targeted therapeutic
resistance and immune evasion is complex and can be influenced by
the TME. Metabolic changes induced by targeted therapies can alter
the TME and contribute to immune resistance (65). For example, hypoxia resulting from
tumor growth can lead to the upregulation of PD-L1 and the
recruitment of immunosuppressive cells. Additionally, the release
of damage-associated molecular patterns from dying cancer cells
during targeted therapy can activate innate immune responses that
paradoxically promote immunosuppression.
TME-driven therapeutic resistance in lung
cancer
Comprehending TME-mediated resistance mechanisms is
essential to devise new lung cancer therapies that restore drug
sensitivity and prolong patient survival. Future research should
focus on assessing the molecular and cellular interactions within
the TME and exploring combination therapies that target multiple
resistance pathways simultaneously.
Immune evasion and resistance
mechanisms
Previous studies have highlighted the role of immune
evasion mechanisms in therapeutic resistance (Table I). Utsumi et al (66) demonstrated that AXL-mediated drug
resistance in ALK-rearranged NSCLC was enhanced by growth-arrest
specific protein 6 from macrophages and MMP11-positive fibroblasts,
underscoring the importance of cellular interactions within the
TME. Moreover, Peyraud et al (67) utilized spatially resolved
transcriptomics to reveal determinants of primary resistance to
immunotherapy in NSCLC with mature tertiary lymphoid structures,
suggesting that the spatial organization of immune cells may impact
therapy outcomes. However, Nishinakamura et al (68) showed that coactivation of innate
immune suppressive cells induced acquired resistance against
combined Toll-like receptor 7/8 agonist treatment and programmed
cell death protein 1 (PD-1) blockade, further illustrating the
dynamic nature of immune resistance mechanisms. Notably, large
clinical trials, such as KEYNOTE-189, have validated the impact of
TME features on immunotherapy response, reinforcing the clinical
relevance of these mechanisms (69).
 | Table IStudies on the TME driving drug
resistance in lung cancer. |
Table I
Studies on the TME driving drug
resistance in lung cancer.
| First author,
year | Treatment
measures | Study type | Model | Resistance
mechanisms | (Refs.) |
|---|
| Utsumi, 2025 | AXL inhibition +
GAS6/MMP11 blockade | Preclinical |
ALK-rearranged
NSCLC cell lines | Macrophage-derived
GAS6 and fibroblast MMP11 activated AXL to bypass ALK
inhibition | (66) |
| Peyraud, 2025 | Spatially resolved
transcriptomics | Clinical | NSCLC with tertiary
lymphoid structures | Spatial exclusion
of CD8+ T cells by stromal barriers and
immunosuppressive cytokine gradients | (67) |
| Nishinakamura,
2025 | Toll-like receptor
agonist + PD-1 blockade | Preclinical | Syngeneic murine
models | MDSC recruitment
via CCL2/CCR2 axis and Treg activation through TGF-β/IL-10
signaling | (68) |
| Ebid, 2025 | Cisplatin +
fibroblast crosstalk | In
vitro | NSCLC cell lines +
fibroblast co-culture | CAF-secreted IL-6
activated STAT3 to upregulate anti-apoptotic BCL-2 family
proteins | (78) |
| Zhang, 2024 | LDHA-targeted
inhibition | Pan-cancer
analysis | NSCLC clinical
datasets | Lactate-driven
acidosis induced PD-L1 upregu lation and impaired T-cell
cytotoxicity | (79) |
| Wang, 2024 | POSTN CAF/ACKR1 EC
interaction |
Single-cell
RNA-sequencing |
TKI-resistant
NSCLC xenografts | CAF-derived POSTN
activated endothelial ACKR1 to recruit immunosuppressive
neutrophils | (80) |
| Wang, 2024 | SMARCA4 mutation
analysis | Preclinical | NSCLC
organoids | Chromatin
remodeling defects reduced neoantigen presentation and
CD8+ T-cell infiltration | (81) |
| Huang, 2024 | EGFR-TKI + TGF-β
blockade | In vivo | EGFR-mutant PDX
models | ERK1/2-p90RSK axis
enhanced TGF-β secretion, promoting T-cell exhaustion and Treg
expansion | (82) |
| Kobayashi,
2024 | Bevacizumab +
miR-200c delivery | Organoid
models | EGFR-mutant NSCLC
organoids | EMT-mediated
VEGF-independent angiogenesis and ECM remodeling via ZEB1/MMP9
activation | (83) |
| Tan, 2024 | Lung-on-a-chip drug
screening | Multicellular
model |
EGFR-TKI-resistant
NSCLC | Fibroblast-mediated
paracrine HGF/c-MET signaling drove bypass survival pathways | (84) |
| Pan, 2024 | Disulfidptosis gene
targeting | Radiogenomics | Lung adenocarcinoma
cohorts | Cysteine metabolism
rewiring protected against radiation-induced ferroptosis | (85) |
| Han, 2024 | Osimertinib + anti
angiogenic therapy | Phase II trial |
Osimertinib-resistant
NSCLC | VEGFR2/PDGFRβ
crosstalk induced CAF activation and hyaluronan-rich ECM
deposition | (86) |
| Shen, 2023 | T1-mapping MRI
nanoprobe | Diagnostic
study | Patients with
multidrug-resistant
NSCLC | Hypoxia-induced
collagen crosslinking reduced drug permeability and enhanced efflux
pumps | (87) |
| Lu, 2023 | STAT3/CD47-SIRPα
axis inhibition | Preclinical |
EGFR-TKI-resistant
PDX models | TAM phagocytosis
evasion via CD47 upregulation and STAT3-driven immunosuppressive
niche | (88) |
The involvement of specific T-cell subsets in lung
cancer resistance mechanisms has garnered attention in previous
research. γδ T cells, which are part of the innate-like T-cell
population, have been shown to serve a dual role in the TME
(70). Some studies have
indicated that γδ T cells can mediate antitumor responses through
the secretion of IFN-γ and the killing of tumor cells (71,72). However, other research has
suggested that γδ T cells may also contribute to immunosuppression
by producing immunosuppressive cytokines, such as IL-10 and TGF-β,
in the TME, thereby promoting tumor progression and resistance to
therapy (73). For example, Liu
et al (28) demonstrated
that the frequency and function of γδ T cells were altered in
patients with lung cancer, and these cells may influence the
efficacy of immunotherapy. Additionally, tissue-resident memory T
cells (Trm) have been identified as key players in local immune
responses. Trm cells persist in tissues long-term and can provide
immediate protection against tumor recurrence (74). However, in the context of chronic
inflammation and an immunosuppressive TME, Trm cells may lose their
function or even promote tumor growth. It has been shown that the
expression of checkpoint molecules, such as PD-1 and LAG-3, on Trm
cells can limit their antitumor activity, contributing to
therapeutic resistance (75).
In addition to T-cell subsets, other immune cells
such as innate lymphoid cells (ILCs) have been implicated in
shaping the TME and influencing therapeutic outcomes. ILCs,
including ILC1s, ILC2s and ILC3s, can regulate tumor inflammation
and immune responses through the secretion of cytokines. For
example, ILC3s have been reported to promote tumor progression by
secreting IL-22, which can enhance tumor cell survival and
resistance to therapy (76).
Furthermore, the interaction between ILCs and other immune cells,
such as macrophages and dendritic cells, can modulate the overall
immune response in the TME, affecting the efficacy of immunotherapy
(77).
Fibroblast-induced resistance and
metabolic reprogramming
Additionally, paracrine signaling mechanisms between
tumor cells and CAFs can synergize with the TME to drive resistance
(Table I). Ebid et al
(78) investigated the cross-talk
signaling between NSCLC cell lines and fibroblasts, demonstrating
that this interaction can attenuate the cytotoxic effect of
cisplatin. This previous study highlighted the role of CAFs in
mediating chemoresistance. Additionally, Zhang et al
(79) identified lactate
dehydrogenase A as a novel predictor for immunotherapy resistance,
linking metabolic reprogramming within the TME to therapy outcomes.
Wang et al (80) conducted
single-cell transcriptomics analysis and revealed an
immunosuppressive network between periostin (POSTN) CAFs and
atypical chemokine receptor 1 (ACKR1) endothelial cells (ECs) in
TKI-resistant lung cancer, providing insights into the cellular and
molecular mechanisms underlying resistance to targeted
therapies.
Genetic alterations and resistance
Additionally, genetic alterations within tumor cells
can synergize with the TME to drive resistance (Table I). Wang et al (81) showed that SMARCA4 mutations
induced tumor cell-intrinsic defects and resistance to
immunotherapy, suggesting that genetic alterations within tumor
cells may synergize with the TME to drive resistance. Huang et
al (82) demonstrated that
EGFR mutations can induce suppression of CD8+ T cells
and anti-PD-1 resistance via the ERK1/2-p90RSK-TGF-β axis, linking
genetic alterations to immune evasion mechanisms. Kobayashi et
al (83) explored the impact
of bevacizumab and microRNA (miR)-200c on epithelial-mesenchymal
transition (EMT) and EGFR-TKI resistance in EGFR-mutant lung cancer
organoids, suggesting that targeting EMT-related pathways may offer
a promising strategy to overcome resistance.
Multicellular models and combination
therapies
Moreover, advanced multicellular models and
combination therapies are being explored to better understand and
overcome TME-driven resistance (Table
I). Tan et al (84)
evaluated drug resistance for EGFR-TKIs in lung cancer using a
multicellular lung-on-a-chip model, allowing for a more accurate
simulation of the TME and providing valuable data on resistance
mechanisms. Pan et al (85) investigated the role of
disulfidptosis-related genes in radiotherapy resistance of lung
adenocarcinoma, emphasizing the importance of understanding
TME-driven resistance across different therapeutic modalities. Han
et al (86) showed that
osimertinib in combination with anti-angiogenesis therapy may be a
promising option for osimertinib-resistant NSCLC, highlighting the
potential of combination therapies in overcoming resistance
mediated by the TME.
Nanotechnology and resistance
management
Finally, nanotechnology offers innovative approaches
to monitor and manage resistance within the TME (Table I). Shen et al (87) developed an adaptable nanoprobe
integrated with quantitative T1-mapping MRI for accurate
differential diagnosis of multidrug-resistant lung cancer, offering
a novel option for monitoring and managing resistance within the
TME. Lu et al (88) showed
that reprogramming of TAMs via the STAT3/CD47-signal regulatory
protein α axis promoted acquired resistance to EGFR-TKIs in lung
cancer, emphasizing the role of immune cell reprogramming in
resistance.
Challenges and future directions
The heterogeneity of the TME presents notable
challenges for the development of effective biomarkers for lung
cancer. Spatial transcriptomics has emerged as a powerful tool for
mapping immune-stromal interactions among different lung cancer
subtypes (89). This technology
allows for the simultaneous analysis of multiple cell types and
their spatial configurations, providing insights into how immune
cells, such as CD8+ T cells and TAMs, interact with
stromal components including CAFs and ECM proteins (90). Studies have shown that the spatial
distribution of immune cells within the TME can influence
therapeutic responses, with immune-excluded or immune-desert tumors
exhibiting poor responses to immunotherapy (90,91). By leveraging spatial
transcriptomics, researchers can better characterize the TME
landscape and identify key biomarkers that predict treatment
outcomes (91). This approach may
not only enhance the understanding of TME heterogeneity but also
aid in the development of personalized therapeutic strategies.
The integration of single-cell sequencing and
artificial intelligence (AI) offers promising options for
developing personalized TME-targeted therapies (92). However, translation into clinical
practice requires validation through large-scale trials, as
exemplified by the ongoing efforts in biomarker-driven studies.
Single-cell RNA sequencing (scRNA-seq) has revolutionized the
understanding of cellular heterogeneity within the TME, revealing
distinct subpopulations of immune cells and their functional states
(92). AI algorithms can analyze
vast datasets generated from scRNA-seq to predict therapeutic
vulnerabilities and optimize treatment combinations. For example,
AI models can identify specific immune cell subsets or signaling
pathways that are critical for tumor progression and resistance,
enabling the design of targeted therapies that disrupt these
interactions (93). This approach
has shown potential in improving the efficacy of immunotherapy and
overcoming resistance in patients with lung cancer (93). However, challenges remain in
translating these findings into clinical practice, including the
need for standardized protocols and the integration of multiomics
data to capture the full complexity of the TME.
Translating TME-targeted therapies into clinical
practice faces several barriers, particularly related to off-target
effects and delivery systems (19,94,95). TME-modulating agents, such as ICIs
and anti-angiogenic drugs, often exhibit off-target effects that
can limit their therapeutic efficacy and cause adverse events
(96). For example, the secretion
of FGL1 by hepatocytes and tumor cells can blunt the efficacy of
anti-PD-1 immunotherapy, highlighting the need for strategies to
enhance treatment specificity (97). Additionally, optimizing delivery
systems to ensure targeted drug delivery and minimize systemic
toxicity remains a critical challenge. Nanoparticle-based drug
delivery systems and targeted conjugates are being explored as
potential solutions to overcome these barriers (98). These approaches aim to enhance the
precision of TME-targeted therapies, ensuring that drugs reach
their intended targets while minimizing off-target effects. Further
research is needed to refine these technologies and evaluate their
clinical feasibility in patients with lung cancer.
The complexity of TME heterogeneity poses notable
hurdles in developing effective therapeutic strategies. The dynamic
nature of the TME, with its diverse cell types and signaling
pathways, makes it difficult to identify consistent biomarkers for
patient stratification and treatment monitoring (99). For example, the coexistence of
immunosuppressive and immunostimulatory signals within the TME can
lead to variable responses to immunotherapy, complicating the
prediction of treatment outcomes. Recent studies have highlighted
the need for a deeper understanding of TME plasticity and the
identification of stable biomarkers that can reliably predict
therapeutic responses across different patient populations
(9,100).
Another critical challenge lies in the technical
limitations of advanced imaging and sequencing technologies. While
spatial transcriptomics and single-cell sequencing have markedly
advanced the understanding of TME heterogeneity, these techniques
require highly specialized equipment and expertise, limiting their
widespread adoption in clinical settings. Furthermore, the
integration of multiomics data remains a complex task, as it
necessitates sophisticated bioinformatics tools and standardized
protocols to ensure data comparability and reproducibility.
In addition, addressing the challenges posed by TME
heterogeneity and developing personalized TME-targeted therapies
requires a multidisciplinary approach that integrates advanced
technologies, including spatial transcriptomics, single-cell
sequencing and AI. Overcoming clinical translation barriers will
necessitate innovative strategies to enhance drug specificity and
delivery. By addressing these challenges, more effective and
personalized treatments may be developed for patients with lung
cancer.
Therapeutic strategies and clinical
application of the TME
Therapeutic strategies targeting the TME offer
innovative approaches to overcome resistance and improve outcomes
in lung cancer treatment (100).
By modulating the immune microenvironment (Table II) and targeting metabolic and
epigenetic pathways (Table
III), researchers and clinicians are developing more effective
and personalized treatment regimens. These strategies hold the
promise of enhancing the efficacy of existing therapies and
addressing the challenges posed by the dynamic and heterogeneous
nature of the TME.
 | Table IIStudies on immune
microenvironment-modulating therapeutic strategies in lung cancer
treatment. |
Table II
Studies on immune
microenvironment-modulating therapeutic strategies in lung cancer
treatment.
| First author,
year |
Intervention/Target | Study type | Model | Clinical
value/Treatment outcome | (Refs.) |
|---|
| Forde, 2018 | Neoadjuvant
PD-1 | Phase II trial | Resectable
NSCLC | 45% major
pathological response; enhanced T-cell clonality and reduced
immunosuppressive cells | (104) |
| Niemeijer,
2018 | PD-1/PD-L1 PET
imaging | Observational | Advanced NSCLC | Identified spatial
heterogeneity of PD-1/PD-L1; associated with ICI response | (105) |
| Zhang, 2018 | Anti-PD-1 vs.
anti-PD-L1 + chemo | Retrospective | Squamous NSCLC | Comparable efficacy
(ORR: 40-45%); higher pneumonitis risk with anti-PD-L1 | (106) |
| Bozorgmehr,
2019 | Nivolumab +
radiotherapy | Phase II trial | Advanced NSCLC | Synergistic effect:
ORR 45 vs. 29% (monotherapy); increased CD8+ T-cell
infiltration | (107) |
| Zhao, 2019 | Apatinib + PD-1
blockade | Phase Ib/II | NSCLC | Optimized TME via
VEGF inhibition; improved PFS (7.1 vs. 4.2 months) | (108) |
| Leighl, 2021 | Durvalumab +
tremelimumab | Phase II trial | PD-1-resistant
NSCLC | Modest activity
(ORR: 9%); grade 3-4 toxicity in 35% of patients | (109) |
| Ott, 2020 | Neoantigen vaccine
+ anti-PD-1 | Phase Ib trial | Advanced NSCLC | Enhanced
tumor-specific T-cell responses; ORR 50% in NSCLC cohort | (110) |
| Awad, 2022 | NEO-PV-01 vaccine +
chemotherapy/ICI | Phase I/II | Non-squamous
NSCLC | Feasibility
confirmed; 2-year OS rate 75% in responders | (111) |
| Li, 2015 | TAM reprogramming
(Fuzheng Sanjie) | Preclinical | Lewis lung
cancer | Reduced tumor
growth via M2-to-M1 polarization; improved CD8+ T-cell
infiltration | (112) |
| Li, 2018 | Hydroxychloroquine
+ chemotherapy | Preclinical | NSCLC | Enhanced
chemosensitivity; reduced M2-TAMs and increased M1-like
macrophages | (113) |
| Zhang, 2021 |
EGFR-specific
CAR-T cells | Phase I trial | Relapsed NSCLC | Manageable safety;
median PFS 4.1 months; 30% disease control rate | (121) |
 | Table IIIStudies on therapeutic strategy that
target tumor microenvironment components in lung cancer
treatment. |
Table III
Studies on therapeutic strategy that
target tumor microenvironment components in lung cancer
treatment.
| First author,
year |
Intervention/Target | Study type | Model | Clinical
value/Treatment outcome | (Refs.) |
|---|
| Li, 2020 | FUT8 inhibition in
CAFs | Preclinical | NSCLC | Reduced EGFR core
fucosylation; suppressed CAF-mediated tumor proliferation | (128) |
| Yang, 2020 | CAF-derived
exosomal miR-210 | Preclinical | NSCLC | Promoted metastasis
via the PTEN/PI3K/AKT pathway; reversed by miR-210 inhibition | (129) |
| Chen, 2024 | CAF-secreted
SERPINE2 | Preclinical | NSCLC | Enhanced tumor
resistance via exosomal transfer; SERPINE2 knockdown restored
chemosensitivity | (130) |
| Sun, 2024 | PRRX1-OLR1 axis in
CAFs | Preclinical | NSCLC | Promoted immune
suppression; dual targeting reduced MDSC infiltration | (131) |
| Wang, 2013 | Integrin β1
inhibition | Preclinical | NSCLC | Suppressed
metastasis via ERK1/2 pathway inhibition; reduced ECM adhesion | (133) |
| Wang, 2013 | miR-29c targeting
ECM | Preclinical | NSCLC | Inhibited integrin
β1/MMP2; reduced lung metastasis in xenografts | (134) |
| Shie, 2023 | Acidosis-induced
ECM remodeling | Preclinical | NSCLC | Promoted
vasculogenic mimicry; acidosis blockade suppressed metastatic
colonization | (138) |
| Abdel-Hafez,
2024 | Inhalable
ECM-modulating nanoparticles | Preclinical | NSCLC | Enhanced drug
delivery via ECM degradation; improved tumor penetration | (140) |
| Cai, 2024 | Apatinib +
chemotherapy/ICI | Phase II trial |
KRAS-mutant
NSCLC | Improved PFS (8.2
vs. 5.1 months); tumor cavitation linked to anti-angiogenic therapy
response | (145) |
| Zhang, 2024 | Anti-angiogenic
therapy + RT/ICI | Retrospective | NSCLC brain
metastasis | Intracranial ORR
45%; median OS 14.7 months | (146) |
Immune microenvironment modulation
ICIs
ICIs have revolutionized the treatment landscape of
NSCLC by targeting key pathways such as PD-1, PD-L1 and cytotoxic
T-lymphocyte associated protein 4 (CTLA-4) (101). Large clinical trials, including
CHECKMATE-227 and KEYNOTE-024, have established the efficacy of
ICIs in improving overall survival, underscoring their clinical
importance (102,103). These therapies aim to enhance
the antitumor immune response by blocking inhibitory signals that
suppress T-cell activity within the TME. PD-1 and PD-L1 inhibitors
are the most extensively studied ICIs in NSCLC. Forde et al
(104) demonstrated the
potential of neoadjuvant PD-1 blockade (pembrolizumab) in
resectable lung cancer, showing notable tumor regression and
increased major histological response rates. In addition, grade ≥3
adverse events were reported in <10% of patients, with the most
common being fatigue and elevated transaminases. Similarly,
Niemeijer et al (105)
utilized PET imaging to identify PD-1 and PD-L1 expression patterns
in patients with NSCLC, providing insights into the spatial
distribution of these targets within the TME. However,
discrepancies in response rates across studies highlight the need
for biomarker-guided patient selection. Zhang et al
(106) compared the efficacy of
anti-PD-1 and anti-PD-L1 therapies in combination with chemotherapy
for advanced squamous NSCLC, concluding that both approaches were
effective but with varying toxicity profiles. This finding
underscores the importance of optimizing treatment combinations
based on tumor subtype and patient-specific factors.
Combining ICIs with other therapies, such as
radiotherapy or anti-angiogenic agents, has shown promise in
enhancing therapeutic efficacy. Nivolumab plus radiotherapy has
been reported to yield an ORR of 45%, with grade ≥3 toxicities in
18% of patients (primarily radiation pneumonitis and lymphopenia)
(107). By contrast, low-dose
apatinib combined with PD-1 blockade resulted in grade ≥3 adverse
events in 15% of patients, mainly hypertension and hand-foot
syndrome (108). These data
facilitate comparative risk-benefit assessment across TME-targeted
strategies.
Targeting multiple immune checkpoints, such as PD-1
and CTLA-4, has emerged as a strategy to amplify immune responses;
however, this approach is often limited by substantial toxicity.
Leighl et al (109)
evaluated the combination of durvalumab (anti-PD-L1) and
tremelimumab (anti-CTLA-4) in patients with
anti-PD-1/PD-L1-resistant NSCLC, showing only modest activity (ORR:
9%) but grade 3-4 toxicities in 35% of patients. Similarly, the
CHECKMATE-227 trial, while demonstrating survival benefit, reported
treatment-related adverse events in 76% of patients receiving
nivolumab plus ipilimumab, with 33% experiencing grade 3-4 events
(103). These findings highlight
the challenging risk-benefit balance of dual checkpoint blockade,
particularly in heavily pretreated patients, and underscore the
need for better patient stratification and toxicity management
strategies.
Previous studies have explored innovative
combinations, such as personalized neoantigen vaccines. Ott et
al (110) reported promising
results with a neoantigen vaccine combined with anti-PD-1 therapy
in patients with advanced melanoma and NSCLC. Similarly, Awad et
al (111) demonstrated the
feasibility of integrating neoantigen vaccines with chemotherapy
and anti-PD-1 therapy in non-squamous NSCLC. These approaches
leverage tumor antigens to enhance immune recognition, potentially
overcoming resistance mechanisms within the TME.
Immune cells
Immune cells within the TME serve a critical role in
modulating tumor progression and therapeutic resistance in lung
cancer. This section focuses on the therapeutic strategies and
clinical applications of targeting specific immune cell
populations, such as TAMs, tumor-infiltrating lymphocytes (TILs)
and chimeric antigen receptor T (CAR-T) cells.
Natural product-based interventions, such as herbal
extracts, have shown immunomodulatory potential in preclinical
models. Li et al (112)
demonstrated that the Fuzheng Sanjie recipe could reprogram TAMs
and reduce tumor growth in Lewis lung cancer mice. Similarly, Gao
et al (113) reported
that ginseng extract altered the behavior of A549 lung cancer cells
and TAMs in co-culture systems. However, the translational
potential of these natural products is limited by several factors,
including undefined active components, batch-to-batch variability,
poor bioavailability and a lack of rigorous clinical trial data
(114). While these studies
provide valuable insights into TME modulation, their clinical
applicability remains uncertain without standardized formulations
and validation in human trials. Li et al (115) showed that hydroxychloroquine
could enhance chemosensitivity and promote the transition of
M2-TAMs to M1-like macrophages, thereby suppressing tumor growth in
NSCLC. However, discrepancies exist in the effectiveness of
TAM-targeted therapies, as some studies highlight the challenges of
achieving consistent reprogramming of TAMs across different tumor
models (47,115).
TILs are a diverse population of immune cells that
infiltrate the tumor site and serve a pivotal role in antitumor
immune responses (116).
CD8+ T cells, a major subset of TILs, are particularly
important for their ability to recognize and kill tumor cells;
however, the function of TILs is often suppressed within the
immunosuppressive TME (116).
Mechanistically, TILs can be inhibited through several pathways.
For example, the upregulation of immune checkpoint molecules such
as PD-1 and PD-L1 on TILs can lead to T-cell exhaustion, reducing
their cytotoxic activity against tumor cells (117). Furthermore, Tregs and MDSCs
within the TME can secrete immunosuppressive cytokines, including
IL-10 and TGF-β, which further suppress the activation and
proliferation of TILs (118).
Additionally, the metabolic environment of the TME, characterized
by hypoxia and high levels of adenosine, can impair TIL function by
promoting the expression of inhibitory receptors and reducing the
availability of essential nutrients (119).
The spatial distribution and density of TILs within
tumors can predict response to immunotherapy. For example, tumors
with a high density of CD8+ TILs in the tumor core tend
to respond better to ICIs compared with those with a peripheral
distribution of TILs (119).
This highlights the importance of understanding not only the
presence but also the localization and functional state of TILs in
the TME. Sumitomo et al (120) investigated the association
between PD-L1/PD-L2 expression and TILs in NSCLC, revealing that
M2-TAMs and TILs interact to create an immunosuppressive TME. This
previous study underscored the importance of targeting both TAMs
and TILs to enhance therapeutic outcomes. Furthermore, CAR-T cells
have shown promise in hematological malignancies but face notable
challenges in solid tumors such as lung cancer. Zhang et al
(121) performed a Phase I trial
of EGFR-specific CAR-T cells in relapsed/refractory NSCLC,
demonstrating manageable safety and preliminary efficacy. However,
the therapeutic potential of CAR-T in solid tumors is limited by
several factors, including on-target/off-tumor toxicity, inadequate
tumor infiltration and the immunosuppressive TME (122). Additionally, manufacturing
complexity, high costs and the risk of cytokine release syndrome
further constrain their widespread clinical application (123). Current research focuses on
improving CAR-T design to overcome these barriers, but their role
in lung cancer remains investigational.
Emerging evidence has suggested that targeting other
immune cells, such as MDSCs, may also enhance therapeutic efficacy.
For example, Kong et al (124) showed that the Modified Bushen
Yiqi formula reduced the chemotactic recruitment of MDSCs in Lewis
lung cancer-bearing mice, thereby enhancing antitumor immunity.
Therapies that target TME components
CAFs
CAFs are a critical component of the TME and serve a
multifaceted role in promoting tumor progression and therapeutic
resistance in lung cancer (125). CAFs contribute to tumor
progression through various mechanisms, including promoting cancer
cell proliferation, enhancing metastasis and inducing therapeutic
resistance (126). Li et
al (127) demonstrated that
α1,6-fucosyltransferase regulates the cancer-promoting capacity of
CAFs by modifying EGFR core fucosylation in NSCLC. Similarly, Yang
et al (128) showed that
exosomes derived from CAFs containing miR-210 promoted NSCLC
migration and invasion through the PTEN/PI3K/AKT pathway.
CAFs are also implicated in therapeutic resistance.
Chen et al (129)
reported that CAFs can transfer SERPINE2 via exosomes, enhancing
tumor progression and resistance to treatment in lung cancer.
Additionally, Sun et al (130) highlighted the role of the paired
related homeobox 1-oxLDL receptor 1 axis in supporting
CAFs-mediated immune suppression and tumor progression, suggesting
potential therapeutic targets.
CAFs influence tumor metabolism and signaling
pathways to promote resistance. Wang et al (80) revealed an immunosuppressive
network between POSTN CAFs and ACKR1 ECs in TKI-resistant lung
cancer, emphasizing the role of CAFs in mediating resistance to
targeted therapies. Furthermore, studies have shown that CAFs
promote glycolysis in NSCLC cells, enhancing DNA damage repair and
radioresistance (126,128,129).
ECM
The ECM is a critical component of the TME, and
serves an important role in lung cancer progression and therapeutic
resistance. Targeting the ECM offers promising therapeutic
strategies to enhance treatment outcomes. Integrins and MMPs are
key mediators of cancer cell adhesion and invasion. Wang et
al (131) demonstrated that
shikonin attenuated lung cancer cell adhesion to the ECM and
metastasis by inhibiting integrin β1 expression and the ERK1/2
signaling pathway. Similarly, Wang et al (132) showed that miR-29c suppressed
lung cancer cell adhesion to the ECM and metastasis by targeting
integrin β1 and MMP2. These studies highlight the potential of
targeting integrins and MMPs to reduce metastasis and improve
patient outcomes.
ECM degradation is a critical step in tumor
invasion and metastasis. Bi et al (133) reported that PRDM14 could promote
the migration of human NSCLC cells through ECM degradation in
vitro. Additionally, Zhang et al (134) demonstrated that protein arginine
methyltransferase 1 small hairpin RNA inhibited NSCLC cell
migration by suppressing EMT, ECM degradation, and Src
phosphorylation. These findings underscore the importance of
targeting ECM degradation pathways to prevent tumor
progression.
ECM remodeling contributes to therapeutic
resistance in lung cancer. Wang et al (135) revealed that stromal ECM is a
microenvironmental cue that can promote resistance to EGFR-TKIs in
lung cancer cells. Furthermore, Shie et al (136) showed that acidosis promoted the
metastatic colonization of lung cancer via remodeling of the ECM
and vasculogenic mimicry. These studies emphasize the role of ECM
remodeling in mediating therapeutic resistance and the need for
strategies to modulate ECM composition.
Previous studies have explored innovative
approaches to target the ECM. Peláez et al (137) demonstrated that sterculic acid
can alter the expression of adhesion molecules and ECM compounds to
regulate the migration of lung cancer cells. Additionally,
Abdel-Hafez et al (138)
investigated inhalable nano-structured microparticles for ECM
modulation as a potential delivery system for lung cancer. These
approaches offer promising avenues to enhance therapeutic efficacy
by modulating the ECM.
Immune cells
Emerging evidence has underscored the therapeutic
value of targeting immune cell-TME crosstalk. For example, beyond
herbal compounds, previous studies have highlighted that modulation
of TAM polarity via STAT3 inhibition or CD47-SIRPα axis blockade
can resensitize EGFR-TKI-resistant tumors by restoring phagocytic
clearance and enhancing T-cell infiltration (88,115). Furthermore, the interaction
between immune cells and other TME components, such as CAFs and
ECM, can modulate the overall immune response, affecting the
efficacy of immunotherapy.
Anti-angiogenic therapy
Anti-angiogenic therapy has emerged as a promising
strategy in the treatment of lung cancer, targeting the VEGF
pathway and other angiogenic factors to normalize tumor blood
vessels, alleviate hypoxia and enhance drug delivery (139). The VEGF pathway is a critical
target for anti-angiogenic therapy (140). The phase III IMpower150 trial
demonstrated that combining atezolizumab, bevacizumab and
chemotherapy markedly improved survival in non-squamous NSCLC,
providing robust clinical validation (141). Similarly, Qiang et al
(142) demonstrated the efficacy
of first-line chemotherapy combined with immunotherapy or
anti-angiogenic therapy in advanced KRAS-mutant NSCLC, showing
improved progression-free survival. Similarly, Cai et al
(143) reported tumor cavitation
in patients with NSCLC receiving anti-angiogenic therapy with
apatinib, highlighting the potential of this approach in specific
patient populations.
Combining anti-angiogenic therapy with other
modalities has shown promise in enhancing therapeutic efficacy.
Zhang et al (146)
evaluated the combination of anti-angiogenic therapy, radiotherapy
and PD-1 inhibitors in patients with driver gene-negative NSCLC
brain metastases, demonstrating improved outcomes. Additionally,
Song et al (145)
reported the efficacy of PD-1 inhibitors combined with
anti-angiogenic therapy in NSCLC with brain metastases, further
supporting the benefits of multi-modal approaches.
Despite initial success, resistance to
anti-angiogenic therapy remains a challenge. Studies have
identified mechanisms, such as upregulation of compensatory
pro-angiogenic factors (for example, basic FGF, platelet-derived
growth factor and VEGF-C) and recruitment of bone marrow-derived
endothelial progenitor cells as key contributors to resistance
(146,147). Furthermore, tumor heterogeneity
and the TME serve notable roles in mediating resistance,
necessitating strategies to overcome these limitations.
Novel therapeutic approaches:
Antibody-drug conjugates (ADCs) and bispecific antibodies
In addition to the aforementioned therapies, novel
approaches such as ADCs and bispecific antibodies are emerging as
promising strategies in the treatment of lung cancer. ADCs are
engineered molecules that combine the specificity of antibodies
with the potency of cytotoxic drugs. They selectively deliver
chemotherapy agents to cancer cells expressing specific antigens,
thereby minimizing off-target effects (141). For example, enfortumab vedotin,
an ADC targeting Nectin-4, has shown notable efficacy in patients
with advanced NSCLC. In a phase I/II trial, an ORR of 41% and a
median duration of response of 10.5 months was demonstrated
(141). Another ADC, sacituzumab
govitecan, which targets Trop-2, has also exhibited promising
results in early-phase trials for NSCLC (6). These ADCs represent a novel frontier
in personalized lung cancer therapy by leveraging antigen
specificity to enhance treatment efficacy and reduce systemic
toxicity. Bispecific antibodies, which can simultaneously bind to
two different antigens, offer unique advantages in cancer
immunotherapy. They can redirect immune cells to tumor cells or
modulate immune checkpoints more effectively. For example,
bispecific antibodies targeting PD-L1 and TGF-β have been developed
to overcome immunosuppression in the TME (148). Preclinical studies have shown
that these bispecific antibodies can enhance T-cell activation and
tumor cell killing (148,149).
Additionally, bispecific T-cell engagers are being explored to
redirect T cells to cancer cells expressing specific antigens, such
as EGFR or HER2, which are frequently upregulated in lung cancer
(149,150). Early clinical trials have
indicated that bispecific antibodies can achieve tumor regression
in a subset of patients with refractory NSCLC, highlighting their
potential as next-generation immunotherapies (148-150).
These emerging therapies, including ADCs and
bispecific antibodies, are expected to markedly impact the
treatment landscape of lung cancer by addressing the limitations of
current targeted therapies and immunotherapies. Further clinical
research is warranted to optimize their application and combination
strategies in the context of the TME.
Conclusion
In conclusion, the TME drives drug resistance in
lung cancer through immune evasion, metabolic reprogramming and
epigenetic modulation. While targeting TME components shows
promise, several challenges remain. Immune-based strategies, such
as dual checkpoint blockade and CAR-T therapy, are limited by
toxicity and poor efficacy in solid tumors. Natural product
interventions face translational hurdles due to undefined
mechanisms and a lack of clinical validation. Emerging
technologies, such as spatial transcriptomics and nanodrug
delivery, offer potential solutions but require further
optimization for clinical application. Future research should
prioritize not only multiomics integration but also rigorous
preclinical models and well-designed clinical trials to distinguish
promising research possibilities from preliminary findings.
Availability of data and materials
Not applicable.
Authors' contributions
LiL contributed to the literature review on the
heterogeneity of the lung cancer microenvironment. LY wrote the
part about the role of the tumor microenvironment in treatment
resistance. HL and TS were involved in the literature search and
analysis, as well as in the writing of the manuscript. LihL, as the
corresponding author, oversaw the entire review process, provided
critical guidance and revised the manuscript. Data authentication
is not applicable. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
This work was supported by the First People's Hospital of Baiyin
College Scientific Research Project (grant no. 2021YK-01; project
name: Clinical research on thoracoscopic lobectomy and
segmentectomy in the treatment of early lung cancer).
References
|
1
|
Wéber A, Morgan E, Vignat J, Laversanne M,
Pizzato M, Rumgay H, Singh D, Nagy P, Kenessey I, Soerjomataram I
and Bray F: Lung cancer mortality in the wake of the changing
smoking epidemic: A descriptive study of the global burden in 2020
and 2040. BMJ Open. 13:e0653032023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
3
|
Oxnard GR, Arcila ME, Sima CS, Riely GJ,
Chmielecki J, Kris MG, Pao W, Ladanyi M and Miller VA: Acquired
resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung
cancer: Distinct natural history of patients with tumors harboring
the T790M mutation. Clin Cancer Res. 17:1616–1622. 2011. View Article : Google Scholar
|
|
4
|
Miyata H, Shigeto H, Ikeya T, Ashizawa T,
Iizuka A, Kikuchi Y, Maeda C, Kanematsu A, Yamashita K, Urakami K,
et al: Localization of epidermal growth factor receptor-mutations
using PNA: DNA probes in clinical specimens from patients with
non-small cell lung cancer. Sci Rep. 15:113142025. View Article : Google Scholar
|
|
5
|
Son B, Lee S, Youn H, Kim E, Kim W and
Youn B: The role of tumor microenvironment in therapeutic
resistance. Oncotarget. 8:3933–3945. 2017. View Article : Google Scholar :
|
|
6
|
Yu H, Zhang W, Xu XR and Chen S: Drug
resistance related genes in lung adenocarcinoma predict patient
prognosis and influence the tumor microenvironment. Sci Rep.
13:96822023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Skoulidis F, Li BT, Dy GK, Price TJ,
Falchook GS, Wolf J, Italiano A, Schuler M, Borghaei H, Barlesi F,
et al: Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl
J Med. 384:2371–2381. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Said SS and Ibrahim WN: Cancer resistance
to immunotherapy: Comprehensive insights with future perspectives.
Pharmaceutics. 15:11432023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Glaviano A, Lau HS, Carter LM, Lee EHC,
Lam HY, Okina E, Tan DJJ, Tan W, Ang HL, Carbone D, et al:
Harnessing the tumor microenvironment: Targeted cancer therapies
through modulation of epithelial-mesenchymal transition. J Hematol
Oncol. 18:62025. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tian Y, Yang Y, He L, Yu X, Zhou H and
Wang J: Exploring the tumor microenvironment of breast cancer to
develop a prognostic model and predict immunotherapy responses. Sci
Rep. 15:125692025. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tan Z, Xue H, Sun Y, Zhang C, Song Y and
Qi Y: The role of tumor inflammatory microenvironment in lung
cancer. Front Pharmacol. 12:6886252021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu Y, Liang J, Zhang Y and Guo Q: Drug
resistance and tumor immune microenvironment: An overview of
current understandings (Review). Int J Oncol. 65:962024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heydenreich B, Bellinghausen I, Lorenz S,
Henmar H, Strand D, Würtzen PA and Saloga J: Reduced in vitro
T-cell responses induced by glutaraldehyde-modified allergen
extracts are caused mainly by retarded internalization of dendritic
cells. Immunology. 136:208–217. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mai Z, Lin Y, Lin P, Zhao X and Cui L:
Modulating extracellular matrix stiffness: A strategic approach to
boost cancer immunotherapy. Cell Death Dis. 15:3072024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen K, Luo L, Li Y and Yang G:
Reprogramming the immune microenvironment in lung cancer. Volume.
16:16848892025.
|
|
16
|
Chandra R, Ehab J, Hauptmann E, Gunturu
NS, Karalis JD, Kent DO, Heid CA, Reznik SI, Sarkaria IS, Huang H,
et al: The current state of tumor Microenvironment-specific
therapies for Non-small cell lung cancer. Cancers (Basel).
17:17322025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He ZN, Zhang CY, Zhao YW, He SL, Li Y, Shi
BL, Hu JQ, Qi RZ and Hua BJ: Regulation of T cells by
myeloid-derived suppressor cells: Emerging immunosuppressor in lung
cancer. Discov Oncol. 14:1852023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lim JU, Lee E, Lee SY, Cho HJ, Ahn DH,
Hwang Y, Choi JY, Yeo CD, Park CK and Kim SJ: Current literature
review on the tumor immune micro-environment, its heterogeneity and
future perspectives in treatment of advanced non-small cell lung
cancer. Transl Lung Cancer Res. 12:857–876. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Genova C, Dellepiane C, Carrega P,
Sommariva S, Ferlazzo G, Pronzato P, Gangemi R, Filaci G, Coco S
and Croce M: Therapeutic implications of tumor microenvironment in
lung cancer: Focus on immune checkpoint blockade. Front Immunol.
12:7994552021. View Article : Google Scholar
|
|
20
|
Cao Q, Li C, Li Y, Kong X, Wang S and Ma
J: Tumor microenvironment and drug resistance in lung
adenocarcinoma: Molecular mechanisms, prognostic implications, and
therapeutic strategies. Discov Oncol. 16:2382025. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chu X, Tian Y and Lv C: Decoding the
spatiotemporal heterogeneity of Tumor-associated macrophages. Mol
Cancer. 23:1502024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mei S, Zhang H, Hirz T, Jeffries NE, Xu Y,
Baryawno N, Wu S, Wu CL, Patnaik A, Saylor PJ, et al: Single-cell
and spatial transcriptomics reveal a Tumor-associated macrophage
subpopulation that mediates prostate cancer progression and
metastasis. Mol Cancer Res. 23:653–665. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashrafi A, Akter Z, Modareszadeh P,
Modareszadeh P, Berisha E, Alemi PS, Chacon Castro MDC, Deese AR
and Zhang L: Current landscape of therapeutic resistance in lung
cancer and promising strategies to overcome resistance. Cancers
(Basel). 14:45622022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo T and Xu J: Cancer-associated
fibroblasts: A versatile mediator in tumor progression, metastasis,
and targeted therapy. Cancer Metastasis Rev. 43:1095–1116. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu C, Gu J, Gu H, Zhang X, Zhang X and Ji
R: The recent advances of cancer associated fibroblasts in cancer
progression and therapy. Front Oncol. 12:10088432022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Papavassiliou KA, Sofianidi AA, Gogou VA
and Papavassiliou AG: Drugging the tumor microenvironment epigenome
for therapeutic interventions in NSCLC. J Cancer. 16:1832–1835.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tong L, Jiménez-Cortegana C, Tay AHM,
Wickström S, Galluzzi L and Lundqvist A: NK cells and solid tumors:
Therapeutic potential and persisting obstacles. Mol Cancer.
21:2062022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu J, Wu M, Yang Y, Wang Z, He S, Tian X
and Wang H: γδ T cells and the PD-1/PD-L1 axis: A love-hate
relationship in the tumor microenvironment. J Transl Med.
22:5532024. View Article : Google Scholar
|
|
29
|
Mancini A, Gentile MT, Pentimalli F,
Cortellino S, Grieco M and Giordano A: Multiple aspects of matrix
stiffness in cancer progression. Front Oncol. 14:14066442024.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Henke E, Nandigama R and Ergün S:
Extracellular matrix in the tumor microenvironment and its impact
on cancer therapy. Front Mol Biosci. 6:1602019. View Article : Google Scholar
|
|
31
|
Marrugal Á, Ojeda L, Paz-Ares L,
Molina-Pinelo S and Ferrer I: Proteomic-based approaches for the
study of cytokines in lung cancer. Dis Markers. 2016:21386272016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang A, Miao K, Sun H and Deng CX: Tumor
heterogeneity reshapes the tumor microenvironment to influence drug
resistance. Int J Biol Sci. 18:3019–3033. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li X, Shang S, Wu M, Song Q and Chen D:
Gut microbial metabolites in lung cancer development and
immunotherapy: Novel insights into gut-lung axis. Cancer Lett.
598:2170962024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li J, Shi B, Ren X, Hu J, Li Y, He S,
Zhang G, Maolan A, Sun T, Qi X, et al: Lung-intestinal axis,
Shuangshen granules attenuate lung metastasis by regulating the
intestinal microbiota and related metabolites. Phytomedicine.
132:1558312024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ankudavicius V, Nikitina D, Lukosevicius
R, Tilinde D, Salteniene V, Poskiene L, Miliauskas S, Skieceviciene
J, Zemaitis M and Kupcinskas J: Detailed characterization of the
Lung-gut microbiome axis reveals the link between PD-L1 and the
microbiome in Non-Small-cell lung cancer patients. Int J Mol Sci.
25:23232024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dong Q, Chen ES, Zhao C and Jin C:
Host-Microbiome interaction in lung cancer. Front Immunol.
12:6798292021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hunt PJ, Andújar FN, Silverman DA and Amit
M: Mini-review: Trophic interactions between cancer cells and
primary afferent neurons. Neurosci Lett. 746:1356582021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hernandez S, Serrano AG and Solis Soto LM:
The role of nerve fibers in the tumor Immune microenvironment of
solid tumors. Adv Biol (Weinh). 6:22000462022. View Article : Google Scholar
|
|
39
|
Li X, Peng X, Yang S, Wei S, Fan Q, Liu J,
Yang L and Li H: Targeting tumor innervation: Premises, promises,
and challenges. Cell Death Discov. 8:1312022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang Y, Ye WL, Zhang RN, He XS, Wang JR,
Liu YX, Wang Y, Yang XM, Zhang YJ and Gan WJ: The role of TGF-β
signaling pathways in cancer and its potential as a therapeutic
target. Evid Based Complement Alternat Med. 2021:66752082021.
|
|
41
|
Jiang C, Zhang N, Hu X and Wang H:
Tumor-associated exosomes promote lung cancer metastasis through
multiple mechanisms. Mol Cancer. 20:1172021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang J, Xu J, Wang W, Zhang B, Yu X and
Shi S: Epigenetic regulation in the tumor microenvironment:
Molecular mechanisms and therapeutic targets. Signal Transduct
Target Ther. 8:2102023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu W, Hua Y, Qiu H, Hao J, Zou K, Li Z, Hu
S, Guo P, Chen M, Sui S, et al: PD-L1 promotes tumor growth and
progression by activating WIP and β-catenin signaling pathways and
predicts poor prognosis in lung cancer. Cell Death Dis. 11:5062020.
View Article : Google Scholar
|
|
44
|
Xiao K, Zhang S, Peng Q, Du Y, Yao X, Ng
II and Tang H: PD-L1 protects tumor-associated dendritic cells from
ferroptosis during immunogenic chemotherapy. Cell Rep.
43:1148682024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shi AP, Tang XY, Xiong YL, Zheng KF, Liu
YJ, Shi XG, Lv Y, Jiang T, Ma N and Zhao JB: Immune checkpoint LAG3
and its ligand FGL1 in cancer. Front Immunol. 12:7850912021.
View Article : Google Scholar
|
|
46
|
Villarroel-Espindola F, Yu X, Datar I,
Mani N, Sanmamed M, Velcheti V, Syrigos K, Toki M, Zhao H, Chen L,
et al: Spatially resolved and quantitative analysis of VISTA/PD-1H
as a novel immunotherapy target in human Non-small cell lung
cancer. Clin Cancer Res. 24:1562–1573. 2018. View Article : Google Scholar :
|
|
47
|
Wang S, Wang J, Chen Z, Luo J, Guo W, Sun
L and Lin L: Targeting M2-like tumor-associated macrophages is a
potential therapeutic approach to overcome antitumor drug
resistance. NPJ Precis Oncol. 8:312024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li Y, Zhao L and Li XF: Hypoxia and the
tumor microenvironment. Technol Cancer Res Treat.
20:153303382110363042021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zaarour RF, Azakir B, Hajam EY, Nawafleh
H, Zeinelabdin NA, Engelsen AST, Thiery J, Jamora C and Chouaib S:
Role of Hypoxia-mediated autophagy in tumor cell death and
survival. Cancers (Basel). 13:5332021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sasidharan Nair V, Saleh R, Toor SM,
Cyprian FS and Elkord E: Metabolic reprogramming of T regulatory
cells in the hypoxic tumor microenvironment. Cancer Immunol
Immunother. 70:2103–2121. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li X, Yan X, Wang Y, Kaur B, Han H and Yu
J: The Notch signaling pathway: A potential target for cancer
immunotherapy. J Hematol Oncol. 16:452023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Luo H, Liu L, Liu X, Xie Y, Huang X, Yang
M, Shao C and Li D: Interleukin-33 (IL-33) promotes DNA
damage-resistance in lung cancer. Cell Death Dis. 16:2742025.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Boo SH and Kim YK: The emerging role of
RNA modifications in the regulation of mRNA stability. Exp Mol Med.
52:400–408. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Guo Y, Xie Y and Luo Y: The role of Long
Non-coding RNAs in the tumor immune microenvironment. Front
Immunol. 13:8510042022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jiang J, Lu Y, Zhang F, Huang J, Ren XL
and Zhang R: The emerging roles of long noncoding RNAs as hallmarks
of lung cancer. Front Oncol. 11:7615822021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Entezari M, Ghanbarirad M, Taheriazam A,
Sadrkhanloo M, Zabolian A, Goharrizi MASB, Hushmandi K, Aref AR,
Ashrafizadeh M, Zarrabi A, et al: Long non-coding RNAs and exosomal
lncRNAs: Potential functions in lung cancer progression, drug
resistance and tumor microenvironment remodeling. Biomed
Pharmacother. 150:1129632022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tian Z, Cen L, Wei F, Dong J, Huang Y, Han
Y, Wang Z, Deng J and Jiang Y: EGFR mutations in non-small cell
lung cancer: Classification, characteristics and resistance to
third-generation EGFR-tyrosine kinase inhibitors (Review). Oncol
Lett. 30:3752025. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Parente P, Parcesepe P, Covelli C,
Olivieri N, Remo A, Pancione M, Latiano TP, Graziano P, Maiello E
and Giordano G: Crosstalk between the tumor microenvironment and
immune system in pancreatic ductal adenocarcinoma: Potential
targets for new therapeutic approaches. Gastroenterol Res Pract.
2018:75306192018. View Article : Google Scholar
|
|
59
|
Reichel D, Tripathi M and Perez JM:
Biological effects of nanoparticles on macrophage polarization in
the tumor microenvironment. Nanotheranostics. 3:66–88. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Coelho MA, Strauss ME, Watterson A, Cooper
S, Bhosle S, Illuzzi G, Karakoc E, Dinçer C, Vieira SF, Sharma M,
et al: Base editing screens define the genetic landscape of cancer
drug resistance mechanisms. Nature Genetics. 56:2479–2492. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wen Y, Zhu Y, Zhang C, Yang X, Gao Y, Li
M, Yang H, Liu T and Tang H: Chronic inflammation, cancer
development and immunotherapy. Front Pharmacol. 13:10401632022.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Qian FF and Han BH: Mechanisms of
resistance to immune checkpoint inhibitors and strategies to
reverse drug resistance in lung cancer. Chin Med J (Engl).
133:2444–2455. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Bagchi S, Yuan R and Engleman EG: Immune
checkpoint inhibitors for the treatment of cancer: Clinical impact
and mechanisms of response and resistance. Annu Rev Pathol.
16:223–249. 2021. View Article : Google Scholar
|
|
64
|
Zhang M and Zhang B: Extracellular matrix
stiffness: Mechanisms in tumor progression and therapeutic
potential in cancer. Exp Hematol Oncol. 14:542025. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Benvenuto M and Focaccetti C: Tumor
microenvironment: Cellular interaction and metabolic adaptations.
Int J Mol Sci. 25:36422024. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Utsumi T, Mizuta H, Seto Y, Uchibori K,
Nishio M, Okamoto I and Katayama R: AXL-Mediated drug resistance in
ALK-rearranged NSCLC enhanced by GAS6 from macrophages and MMP11
positive fibroblasts. Cancer Sci. 116:1034–1047. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Peyraud F, Guégan JP, Rey C, Lara O, Odin
O, Del Castillo M, Vanhersecke L, Coindre JM, Clot E, Brunet M, et
al: Spatially resolved transcriptomics reveal the determinants of
primary resistance to immunotherapy in NSCLC with mature tertiary
lymphoid structures. Cell Rep Med. 6:1019342025. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Nishinakamura H, Shinya S, Irie T,
Sakihama S, Naito T, Watanabe K, Sugiyama D, Tamiya M, Yoshida T,
Hase T, et al: Coactivation of innate immune suppressive cells
induces acquired resistance against combined TLR agonism and PD-1
blockade. Sci Transl Med. 17:adk31602025. View Article : Google Scholar
|
|
69
|
Gandhi L, Rodríguez-Abreu D, Gadgeel S,
Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ,
Powell SF, et al: Pembrolizumab plus chemotherapy in metastatic
non-small-cell lung cancer. N Engl J Med. 378:2078–2092. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cheng M and Hu S: Lung-resident γδ T cells
and their roles in lung diseases. Immunology. 51:375–384. 2017.
View Article : Google Scholar
|
|
71
|
Subhi-Issa N, Tovar Manzano D, Pereiro
Rodriguez A, Sanchez Ramon S, Perez Segura P and Ocaña A: γδ T
cells: Game changers in immune cell therapy for cancer. Cancers
(Basel). 17:10632025. View Article : Google Scholar
|
|
72
|
Lv J, Liu Z, Ren X, Song S, Zhang Y and
Wang Y: γδT cells, a key subset of T cell for cancer immunotherapy.
Front Immunol. 16:15621882025. View Article : Google Scholar
|
|
73
|
Jin C, Lagoudas GK, Zhao C, Bullman S,
Bhutkar A, Hu B, Ameh S, Sandel D, Liang XS, Mazzilli S, et al:
Commensal microbiota promote lung cancer development via γδ T
cells. Cell. 176:998–1013.e16. 2019. View Article : Google Scholar
|
|
74
|
Dhodapkar MV and Dhodapkar KM:
Tissue-resident memory-like T cells in tumor immunity: Clinical
implications. Semin Immunol. 49:1014152020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Hofmann M, Thimme R and Schamel WW: PD-1
and LAG-3: Synergistic fostering of T cell exhaustion. Signal
Transduct Target Ther. 9:2912024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ducimetière L, Vermeer M and Tugues S: The
interplay between innate lymphoid cells and the tumor
microenvironment. Front Immunol. 10:28952019. View Article : Google Scholar
|
|
77
|
Yuan X, Rasul F, Nashan B and Sun C:
Innate lymphoid cells and cancer: Role in tumor progression and
inhibition. Eur J Immunol. 51:2188–2205. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ebid N, Sharaky M, Elkhoely A, El Morsy EM
and Saad SY: Cross-talk signaling between non-small cell lung
cancer cell lines and fibroblasts attenuates the cytotoxic effect
of cisplatin. J Biochem Mol Toxicol. 39:e702012025. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang Q, Luo Y, Qian B, Cao X, Xu C, Guo
K, Wan R, Jiang Y, Wang T, Mei Z, et al: A systematic pan-cancer
analysis identifies LDHA as a novel predictor for immunological,
prognostic, and immunotherapy resistance. Aging (Albany NY).
16:8000–8018. 2024.PubMed/NCBI
|
|
80
|
Wang Z, Yan N, Sheng H, Xiao Y, Sun J and
Cao C: Single-cell transcriptomic analysis reveals an
immunosuppressive network between POSTN CAFs and ACKR1 ECs in
TKI-resistant lung cancer. Cancer Genomics Proteomics. 21:65–78.
2024. View Article : Google Scholar :
|
|
81
|
Wang Y, Meraz IM, Qudratullah M, Kotagiri
S, Han Y, Xi Y, Wang J and Lissanu Y: SMARCA4 mutation induces
tumor cell-intrinsic defects in enhancer landscape and resistance
to immunotherapy. bioRxiv. Jun 22–2024. View Article : Google Scholar
|
|
82
|
Huang H, Zhu X, Yu Y, Li Z, Yang Y, Xia L
and Lu S: EGFR mutations induce the suppression of CD8+ T cell and
anti-PD-1 resistance via ERK1/2-p90RSK-TGF-β axis in non-small cell
lung cancer. J Transl Med. 22:6532024. View Article : Google Scholar
|
|
83
|
Kobayashi N, Katakura S, Fukuda N,
Somekawa K, Kaneko A and Kaneko T: The impact of bevacizumab and
miR200c on EMT and EGFR-TKI resistance in EGFR-mutant lung cancer
organoids. Genes (Basel). 15:16242024. View Article : Google Scholar
|
|
84
|
Tan J, Zhu L, Shi J, Zhang J, Kuang J, Guo
Q, Zhu X, Chen Y, Zhou C and Gao X: Evaluation of drug resistance
for EGFR-TKIs in lung cancer via multicellular lung-on-a-chip. Eur
J Pharm Sci. 199:1068052024. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Pan X, Qian H, Sun Z, Yi Q, Liu Y, Lan G,
Chen J and Wang G: Investigating the role of disulfidptosis related
genes in radiotherapy resistance of lung adenocarcinoma. Front Med
(Lausanne). 11:14730802024. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Han R, Guo H, Shi J, Zhao S, Jia Y, Liu X,
Liu Y, Cheng L, Zhao C, Li X and Zhou C: Osimertinib in combination
with anti-angiogenesis therapy presents a promising option for
osimertinib-resistant non-small cell lung cancer. BMC Med.
22:1742024. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Shen A, Sun Y, Wang G, Meng X, Ren X, Wan
Q, Lv Q, Wang X, Ni J, Li M, et al: An adaptable nanoprobe
integrated with quantitative T1-Mapping MRI for accurate
differential diagnosis of Multidrug-resistant lung cancer. Adv
Healthc Mater. 12:e23006842023. View Article : Google Scholar
|
|
88
|
Lu J, Li J, Lin Z, Li H, Lou L, Ding W,
Ouyang S, Wu Y, Wen Y, Chen X, et al: Reprogramming of TAMs via the
STAT3/CD47-SIRPα axis promotes acquired resistance to EGFR-TKIs in
lung cancer. Cancer Lett. 564:2162052023. View Article : Google Scholar
|
|
89
|
Yuan Y: Spatial heterogeneity in the tumor
microenvironment. Cold Spring Harb Perspect Med. 6:a0265832016.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang Y, Liu B, Min Q, Yang X, Yan S, Ma Y,
Li S, Fan J, Wang Y, Dong B, et al: Spatial transcriptomics
delineates molecular features and cellular plasticity in lung
adenocarcinoma progression. Cell Discovery. 9:962023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang JT, Zhang J, Wang SR, Yan LX, Qin J,
Yin K, Chu XP, Wang MM, Hong HZ, Lv ZY, et al: Spatial
downregulation of CD74 signatures may drive invasive component
development in part-solid lung adenocarcinoma. iScience.
26:1076992023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang J, Song C, Tian Y and Yang X:
Single-cell RNA sequencing in lung cancer: Revealing phenotype
shaping of stromal cells in the microenvironment. Front Immunol.
12:8020802021. View Article : Google Scholar
|
|
93
|
Joo MS, Pyo KH, Chung JM and Cho BC:
Artificial intelligence-based non-small cell lung cancer
transcriptome RNA-sequence analysis technology selection guide.
Front Bioeng Biotechnol. 11:10819502023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Xie L, Xie D, Du Z, Xue S, Wang K, Yu X,
Liu X, Peng Q and Fang C: A novel therapeutic outlook:
Classification, applications and challenges of inhalable
micron/nanoparticle drug delivery systems in lung cancer (Review).
Int J Oncol. 64:382024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Hu M and Huang L: Strategies targeting
tumor immune and stromal microenvironment and their clinical
relevance. Adv Drug Deliv Rev. 183:1141372022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Qian W, Zhao M, Wang R and Li H:
Fibrinogen-like protein 1 (FGL1): The next immune checkpoint
target. J Hematol Oncol. 14:1472021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wang J, Sanmamed MF, Datar I, Su TT, Ji L,
Sun J, Chen L, Chen Y, Zhu G, Yin W, et al: Fibrinogen-like Protein
1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell.
176:334–347.e12. 2019. View Article : Google Scholar
|
|
98
|
Miao L, Wang Y, Lin CM, Xiong Y, Chen N,
Zhang L, Kim WY and Huang L: Nanoparticle modulation of the tumor
microenvironment enhances therapeutic efficacy of cisplatin. J
Control Release. 217:27–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Runa F, Hamalian S, Meade K, Shisgal P,
Gray PC and Kelber JA: Tumor microenvironment heterogeneity:
Challenges and opportunities. Curr Mol Biol Rep. 3:218–229. 2017.
View Article : Google Scholar
|
|
100
|
Guan XY, Guan XL and Jiao ZY: Improving
therapeutic resistance: Beginning with targeting the tumor
microenvironment. J Chemother. 34:492–516. 2022. View Article : Google Scholar
|
|
101
|
Lu J and Ramirez RA: The role of
checkpoint inhibition in Non-small cell lung cancer. Ochsner J.
17:379–387. 2017.PubMed/NCBI
|
|
102
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: Pembrolizumab versus chemotherapy for PD-L1-positive
Non-small-cell lung cancer. N Engl J Med. 375:1823–1833. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Hellmann MD, Paz-Ares L, Bernabe Caro R,
Zurawski B, Kim SW, Carcereny Costa E, Park K, Alexandru A,
Lupinacci L, de la Mora Jimenez E, et al: Nivolumab plus ipilimumab
in advanced Non-Small-Cell lung cancer. N Engl J Med.
381:2020–2031. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Forde PM, Chaft JE, Smith KN, Anagnostou
V, Cottrell TR, Hellmann MD, Zahurak M, Yang SC, Jones DR,
Broderick S, et al: Neoadjuvant PD-1 blockade in resectable lung
cancer. N Engl J Med. 378:1976–1986. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Niemeijer AN, Leung D, Huisman MC, Bahce
I, Hoekstra OS, van Dongen GAMS, Boellaard R, Du S, Hayes W, Smith
R, et al: Whole body PD-1 and PD-L1 positron emission tomography in
patients with non-small-cell lung cancer. Nat Commun. 9:46642018.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang Y, Zhou H and Zhang L: Which is the
optimal immunotherapy for advanced squamous non-small-cell lung
cancer in combination with chemotherapy: Anti-PD-1 or anti-PD-L1? J
Immunother Cancer. 6:1352018. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Bozorgmehr F, Hommertgen A, Krisam J,
Lasitschka F, Kuon J, Maenz M, Huber PE, König L, Kieser M, Debus
J, et al: Fostering efficacy of anti-PD-1-treatment: Nivolumab plus
radiotherapy in advanced non-small cell lung cancer-study protocol
of the FORCE trial. BMC Cancer. 19:10742019. View Article : Google Scholar
|
|
108
|
Zhao S, Ren S, Jiang T, Zhu B, Li X, Zhao
C, Jia Y, Shi J, Zhang L, Liu X, et al: Low-Dose apatinib optimizes
tumor microenvironment and potentiates antitumor effect of
PD-1/PD-L1 blockade in lung cancer. Cancer Immunol Res. 7:630–643.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Leighl NB, Redman MW, Rizvi N, Hirsch FR,
Mack PC, Schwartz LH, Wade JL, Irvin WJ, Reddy SC, Crawford J, et
al: Phase II study of durvalumab plus tremelimumab as therapy for
patients with previously treated anti-PD-1/PD-L1 resistant stage IV
squamous cell lung cancer (Lung-MAP substudy S1400F, NCT03373760).
J Immunother Cancer. 9:e0029732021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ott PA, Hu-Lieskovan S, Chmielowski B,
Govindan R, Naing A, Bhardwaj N, Margolin K, Awad MM, Hellmann MD,
Lin JJ, et al: A Phase Ib Trial of Personalized Neoantigen Therapy
Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-small Cell
Lung Cancer, or Bladder Cancer. Cell. 183:347–362.e24. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Awad MM, Govindan R, Balogh KN, Spigel DR,
Garon EB, Bushway ME, Poran A, Sheen JH, Kohler V, Esaulova E, et
al: Personalized neoantigen vaccine NEO-PV-01 with chemotherapy and
anti-PD-1 as first-line treatment for non-squamous non-small cell
lung cancer. Cancer Cell. 40:1010–1026.e11. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Li JH, Tian F, Qiu CS, Chen WJ, Xu DX,
Yang LQ and Li RJ: Relevant studies on effect of Fuzheng Sanjie
recipe in regulating immune microenvironment remodeling of TAMs in
Lewis lung cancer mice. Zhongguo Zhong Yao Za Zhi. 40:1161–1165.
2015.In Chinese. PubMed/NCBI
|
|
113
|
Gao J, Bi L, Jiang YC, Yang Y, Li BY and
Chen WP: Effect of water extract of ginseng on biological
bechaviors of lung cancer A549 cells and the expression of F-actin
in Co-culture system of TAMs and A549 cells. Zhongguo Zhong Xi Yi
Jie He Za Zhi. 37:345–350. 2017.In Chinese. PubMed/NCBI
|
|
114
|
Wen X, Wang Y, Su C, You Y, Jiang Z, Zhu D
and Fan Q: Integrating Multi-omics technologies with traditional
Chinese medicine to enhance cancer research and treatment. QJM. Apr
29–2025.Epub ahead of print. View Article : Google Scholar
|
|
115
|
Li Y, Cao F, Li M, Li P, Yu Y, Xiang L, Xu
T, Lei J, Tai YY, Zhu J, et al: Hydroxychloroquine induced lung
cancer suppression by enhancing chemo-sensitization and promoting
the transition of M2-TAMs to M1-like macrophages. J Exp Clin Cancer
Res. 37:2592018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Sarnaik AA, Hwu P, Mulé JJ and
Pilon-Thomas S: Tumor-infiltrating lymphocytes: A new hope. Cancer
Cell. 42:1315–1318. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Stachowiak M, Becker WJ, Olkhanud PB,
Moreno PA, Markowicz S, Berzofsky JA and Sarnowska E: Cancer cells
accelerate exhaustion of persistently activated mouse CD4+ T cells.
Oncoimmunology. 14:25213922025. View Article : Google Scholar :
|
|
118
|
Stachowiak M, Becker WJ, Olkhanud PB,
Moreno PA, Markowicz S, Berzofsky JA and Sarnowska E: Mechanisms
underlying immunosuppression by regulatory cells. Front Immunol.
15:13281932024. View Article : Google Scholar
|
|
119
|
Mastelic-Gavillet B, Navarro Rodrigo B,
Décombaz L, Wang H, Ercolano G, Ahmed R, Lozano LE, Ianaro A, Derré
L, Valerio M, et al: Adenosine mediates functional and metabolic
suppression of peripheral and tumor-infiltrating CD8+ T cells. J
Immunother Cancer. 7:2572019. View Article : Google Scholar :
|
|
120
|
Sumitomo R, Huang CL, Fujita M, Cho H and
Date H: Differential expression of PD-L1 and PD-L2 is associated
with the tumor microenvironment of TILs and M2 TAMs and tumor
differentiation in non-small cell lung cancer. Oncol Rep.
47:732022. View Article : Google Scholar :
|
|
121
|
Zhang Y, Zhang Z, Ding Y, Fang Y, Wang P,
Chu W, Jin Z, Yang X, Wang J, Lou J and Qian Q: Phase I clinical
trial of EGFR-specific CAR-T cells generated by the piggyBac
transposon system in advanced relapsed/refractory non-small cell
lung cancer patients. J Cancer Res Clin Oncol. 147:3725–3734. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Sterner RC and Sterner RM: CAR-T cell
therapy: Current limitations and potential strategies. Blood Cancer
J. 11:692021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Labanieh L and Mackall CL: CAR immune
cells: Design principles, resistance and the next generation.
Nature. 614:635–648. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kong Q, Zhu H, Gong W, Deng X, Liu B and
Dong J: Modified Bushen Yiqi formula enhances antitumor immunity by
reducing the chemotactic recruitment of M2-TAMs and PMN-MDSCs in
Lewis lung cancer-bearing mice. J Ethnopharmacol. 319:1171832024.
View Article : Google Scholar
|
|
125
|
Chen C, Hou J, Yu S, Li W, Wang X, Sun H,
Qin T, Claret FX, Guo H and Liu Z: Role of cancer-associated
fibroblasts in the resistance to antitumor therapy, and their
potential therapeutic mechanisms in non-small cell lung cancer.
Oncol Lett. 21:4132021. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Feng H, Cao B, Peng X and Wei Q:
Cancer-associated fibroblasts strengthen cell proliferation and
EGFR TKIs resistance through aryl hydrocarbon receptor dependent
signals in non-small cell lung cancer. BMC Cancer. 22:7642022.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Li F, Zhao S, Cui Y, Guo T, Qiang J, Xie
Q, Yu W, Guo W, Deng W, Gu C and Wu T: α1,6-Fucosyltransferase
(FUT8) regulates the cancer-promoting capacity of cancer-associated
fibroblasts (CAFs) by modifying EGFR core fucosylation (CF) in
non-small cell lung cancer (NSCLC). Am J Cancer Res. 10:816–837.
2020.
|
|
128
|
Yang F, Yan Y, Yang Y, Hong X, Wang M,
Yang Z, Liu B and Ye L: MiR-210 in exosomes derived from CAFs
promotes non-small cell lung cancer migration and invasion through
PTEN/PI3K/AKT pathway. Cell Signal. 73:1096752020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Chen Y, Zhu S, Yang L, Lu Y and Ye X:
Cancer-associated fibroblasts (CAFs) regulate lung cancer malignant
progression by transferring SERPINE2 (PN1) via exosomes. Curr Mol
Med. 25:1025–1037. 2025. View Article : Google Scholar
|
|
130
|
Sun Y, Ying K, Sun J, Wang Y, Qiu L, Ji M,
Sun L and Chen J: PRRX1-OLR1 axis supports CAFs-mediated lung
cancer progression and immune suppression. Cancer Cell Int.
24:2472024. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wang H, Wu C, Wan S, Zhang H, Zhou S and
Liu G: Shikonin attenuates lung cancer cell adhesion to
extracellular matrix and metastasis by inhibiting integrin β1
expression and the ERK1/2 signaling pathway. Toxicology.
308:104–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang
L, Zhang H, Chen X, Yang Y and Liu G: miRNA-29c suppresses lung
cancer cell adhesion to extracellular matrix and metastasis by
targeting integrin β1 and matrix metalloproteinase2 (MMP2). PLoS
One. 8:e701922013. View Article : Google Scholar
|
|
133
|
Bi HX, Shi HB, Zhang T and Cui G: PRDM14
promotes the migration of human non-small cell lung cancer through
extracellular matrix degradation in vitro. Chin Med J (Engl).
128:373–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Zhang T, Cui G, Yao YL, Guo Y, Wang QC, Li
XN and Feng WM: Inhibition of nonsmall cell lung cancer cell
migration by protein arginine methyltransferase 1-small hairpin RNA
through inhibiting Epithelial-mesenchymal transition, extracellular
matrix degradation, and src phosphorylation in vitro. Chin Med J
(Engl). 128:1202–1208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Wang Y, Zhang T, Guo L, Ren T and Yang Y:
Stromal extracellular matrix is a microenvironmental cue promoting
resistance to EGFR tyrosine kinase inhibitors in lung cancer cells.
Int J Biochem Cell Biol. 106:96–106. 2019. View Article : Google Scholar
|
|
136
|
Shie WY, Chu PH, Kuo MY, Chen HW, Lin MT,
Su XJ, Hong YL and Chou HE: Acidosis promotes the metastatic
colonization of lung cancer via remodeling of the extracellular
matrix and vasculogenic mimicry. Int J Oncol. 63:1362023.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Peláez R, Ochoa R, Pariente A,
Villanueva-Martínez Á, Pérez-Sala Á and Larráyoz IM: Sterculic acid
alters adhesion molecules expression and extracellular matrix
compounds to regulate migration of lung cancer cells. Cancers
(Basel). 13:43702021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Abdel-Hafez SM, Gallei M, Wagner S and
Schneider M: Inhalable nano-structured microparticles for
extracellular matrix modulation as a potential delivery system for
lung cancer. Eur J Pharm Biopharm. 204:1145122024. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Frezzetti D, Gallo M, Maiello MR,
D'Alessio A, Esposito C, Chicchinelli N, Normanno N and De Luca A:
VEGF as a potential target in lung cancer. Expert Opin Ther
Targets. 21:959–966. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Villaruz LC and Socinski MA: The role of
Anti-angiogenesis in non-small-cell lung cancer: An update. Curr
Oncol Rep. 17:262015. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Socinski MA, Nishio M, Jotte RM, Cappuzzo
F, Orlandi F, Stroyakovskiy D, Nogami N, Rodríguez-Abreu D,
Moro-Sibilot D, Thomas CA, et al: IMpower150 final overall survival
analyses for atezolizumab plus bevacizumab and chemotherapy in
First-line metastatic nonsquamous NSCLC. J Thorac Oncol.
16:1909–1924. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Qiang H, Wang Y, Zhang Y, Li J, Zhang L,
Du H, Ling X, Cao S, Zhou Y, Zhong R and Zhong H: Efficacy of
first-line chemotherapy combined with immunotherapy or
anti-angiogenic therapy in advanced KRAS-mutant non-small cell lung
cancer. Transl Oncol. 53:1023172025. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Cai Q, Hu K, Dong S, Li X, Hu S, Deng W
and Ou W: Tumor cavitation in patients with non-small-cell lung
cancer receiving anti-angiogenic therapy with apatinib. Transl Lung
Cancer Res. 13:1708–1717. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Zhang X, Sun Q, Chen R, Zhao M, Cai F, Cui
Z and Jiang H: Efficacy and safety of combining anti-angiogenic
therapy, radiotherapy, and PD-1 inhibitors in patients with driver
gene-negative non-small cell lung cancer brain metastases: A
retrospective study. BMC Cancer. 24:14922024. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Song JQ, Wang X, Zeng ZM, Liang PA, Zhong
CY and Liu AW: Efficacy of PD-1 Inhibitors combined with
anti-angiogenic therapy in driver gene mutation negative
non-small-cell lung cancer with brain metastases. Discov Med.
35:321–331. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Itatani Y, Kawada K, Yamamoto T and Sakai
Y: Resistance to Anti-angiogenic therapy in Cancer-alterations to
Anti-VEGF pathway. Int J Mol Sci. 19:12322018. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Bergers G and Hanahan D: Modes of
resistance to anti-angiogenic therapy. Nat Rev Cancer. 8:592–603.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wu Y, Yan Y, Guo Y, Niu M, Zhou B, Zhang
J, Zhou P, Chu Q, Mei Q, Yi M and Wu K: Anti-TGF-β/PD-L1 bispecific
antibody synergizes with radiotherapy to enhance antitumor immunity
and mitigate radiation-induced pulmonary fibrosis. J Hematol Oncol.
18:242025. View Article : Google Scholar
|
|
149
|
Huehls AM, Coupet TA and Sentman CL:
Bispecific T-cell engagers for cancer immunotherapy. Immunol Cell
Biol. 93:290–296. 2015. View Article : Google Scholar
|
|
150
|
Si Y, Pei X, Wang X, Han Q, Xu C and Zhang
B: An Anti-EGFR/anti-HER2 bispecific antibody with enhanced
antitumor activity against acquired Gefitinib-resistant NSCLC
cells. Protein Pept Lett. 28:1290–1297. 2021. View Article : Google Scholar
|
















