Prostate cancer (PCa) is a major public health
burden in men: In 2022, there were ~1.47 million new prostate
cancer cases globally, making it the second most common cancer in
men, and it accounted for ~4.1% of all male cancer mortalities
worldwide. In the United States alone, ~313,780 new cases and
35,770 mortalities from prostate cancer are projected in 2025
(1,2). Early and accurate diagnosis is
pivotal to improving clinical outcomes, yet the disease often
progresses silently in its initial stages. Conventional screening
approaches-such as serum prostate-specific antigen (PSA) testing
and digital rectal examination-have long served as cornerstones of
early detection (2). However,
their limited specificity and inability to distinguish indolent
from aggressive disease frequently result in overdiagnosis,
overtreatment and associated morbidity (3). These shortcomings highlight a need
for novel biomarkers that enable more precise disease
characterization, risk stratification and individualized treatment
planning (4).
Recent advances in genomics, transcriptomics and
metabolomics have revolutionized biomarker discovery, unveiling new
molecular signatures that bridge laboratory research and clinical
application (5). Genetic
alterations such as mutations in BRCA1/2, HOXB13 and
PTEN, as well as TMPRSS2-ERG gene fusions, have been
associated with PCa initiation, progression and therapeutic
resistance (6,7). These genetic insights not only
enhance the understanding of tumor biology but also aid in
identifying high-risk patients, predicting treatment response and
guiding precision interventions such as PARP inhibitor
therapy (8).
Parallel to these genomic developments, RNA-based
biomarkers-including prostate cancer antigen (PCA) 3, microRNAs
(miRNAs; such as miR-141 and miR-375) and long
non-coding RNAs (lncRNAs)-have emerged as promising tools for
non-invasive diagnosis (9).
Detection of these RNA molecules in urine, blood or exosomal (exo)
fractions notably improves diagnostic accuracy when used alongside
PSA testing, thereby minimizing unnecessary biopsies (10). The advent of liquid biopsy,
encompassing circulating tumor DNA (ctDNA) and exo RNA analysis,
further enhances real-time disease monitoring. This approach
enables dynamic assessment of tumor burden, therapeutic response
and the emergence of resistance-associated mutations in key genes
such as androgen receptor (AR) and BRCA1/2 (11,12). In addition to genomic and
transcriptomic markers, metabolic biomarkers provide valuable
insights into PCa pathophysiology. Metabolites such as citrate,
polyamines and sarcosine reflect the distinct metabolic
reprogramming that occurs during malignant transformation (13). Their quantification in
biofluids-including serum, urine and prostatic secretions-offers
complementary diagnostic information and helps differentiate benign
prostatic hyperplasia from malignant disease (14).
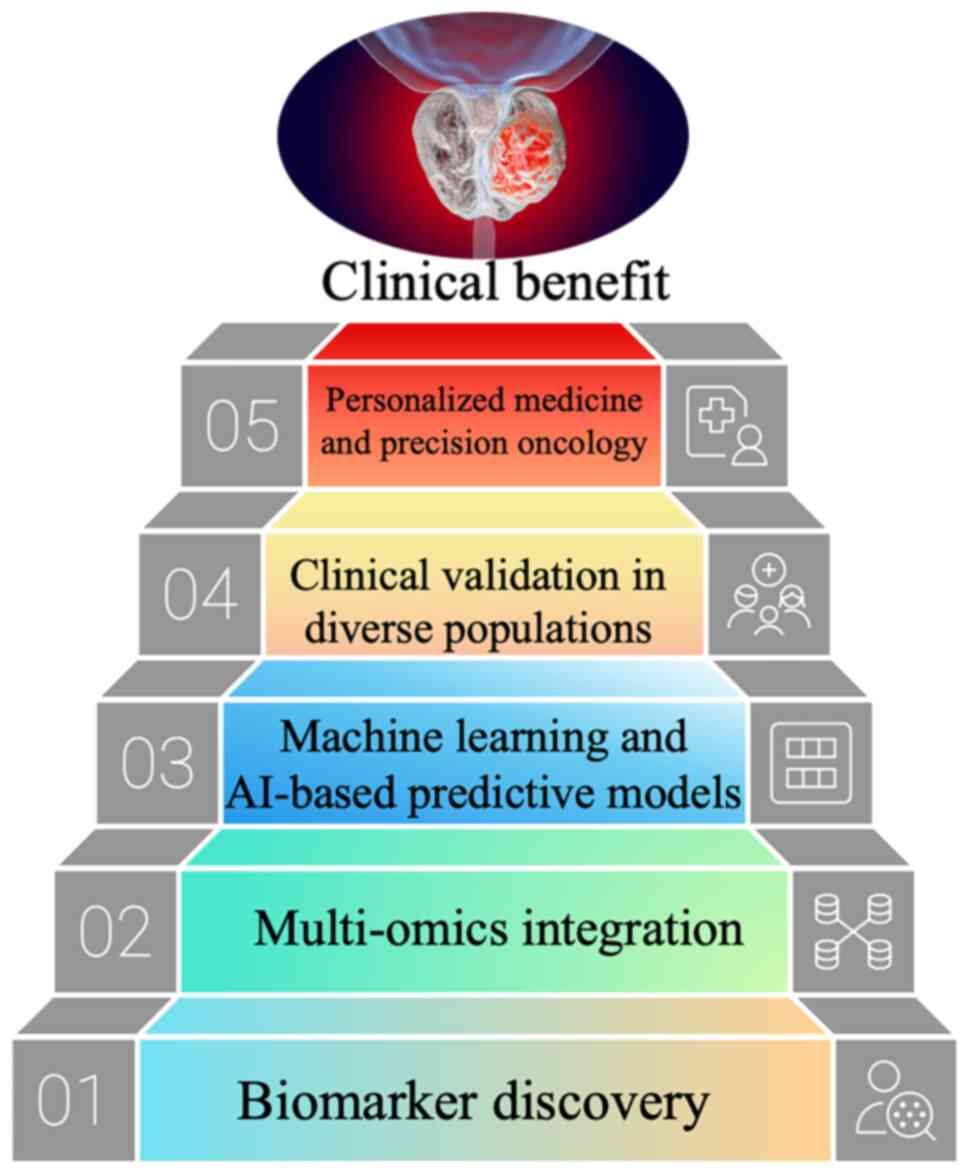
Collectively, these discoveries mark a paradigm
shift in PCa management. The integration of genetic, RNA and
metabolic biomarkers into diagnostic workflows has the potential to
transform routine screening, prognostic evaluation and therapeutic
decision-making. The present review synthesizes current evidence on
emerging biomarkers in PCa, emphasizing their translational
relevance, diagnostic performance and clinical feasibility. The
present review also discusses persistent challenges, such as
biomarker validation, inter-population variability and the
harmonization of multi-omics data-and explores future directions,
including artificial intelligence (AI)-assisted liquid biopsy
interpretation and multi-omics integration for advancing precision
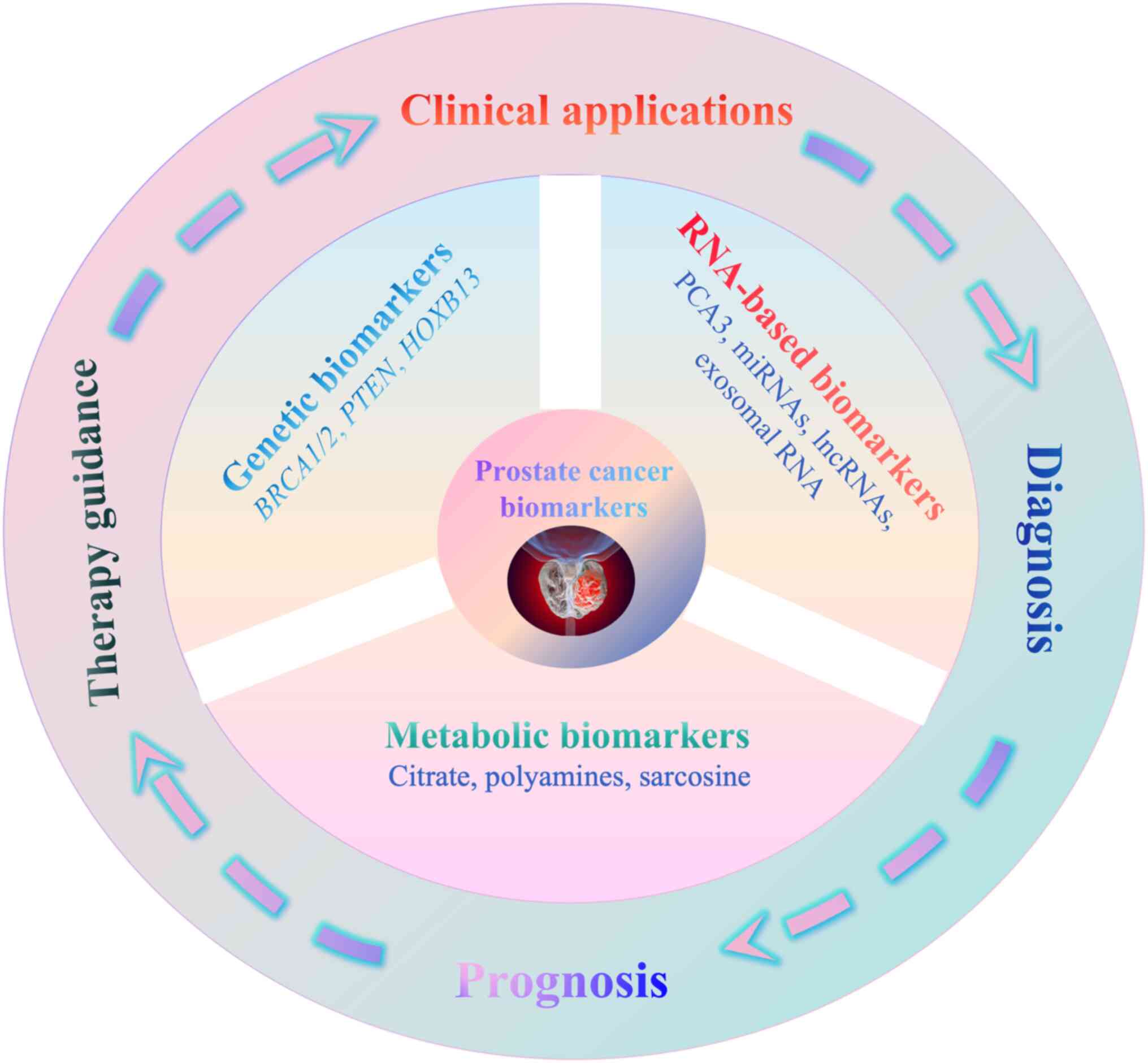
oncology (Fig. 1) (15).
An f/t PSA ratio <0.15 is associated with an
increased risk of malignancy and is a key factor in recommending
prostate biopsy, thereby improving diagnostic specificity (21,22). This metric is particularly
valuable within the diagnostic 'gray zone' of t-PSA (4-10 ng/ml)
(23). A ratio >0.25 typically
suggests a benign etiology, whereas a ratio <0.15 warrants
further invasive assessment (24,25). Notably, when t-PSA >10 ng/ml,
biopsy is generally indicated regardless of the f/t PSA ratio
(26). A large multicenter study
demonstrated that incorporating the f/t PSA ratio into clinical
decision-making reduced misclassification by ~43%, underscoring its
utility in enhancing diagnostic accuracy (27).
Emerging strategies seek to integrate the f/t PSA
ratio with other clinical parameters, including patient age, family
history and racial background, to develop multivariate predictive
models (28,29). These refined algorithms promise to
further personalize PCa risk stratification and support early
intervention strategies (30).
PSAD, defined as the ratio of serum PSA
concentration to prostate volume (PSAD=t-PSA/prostate volume),
refines diagnostic precision by accounting for gland size (31). Prostate volume is typically
quantified via transrectal ultrasound (TRUS) or, with increasing
frequency, MRI. A PSAD value >0.15 ng/ml/cm3 is
consistently associated with a heightened risk of PCa (32,33). This normalization mitigates the
confounding effect of benign gland enlargement, thereby reducing
false-positive results (34).
Longitudinal cohort studies have confirmed that an
elevated PSAD is a robust predictor of PCa incidence and
progression (35,36). Recent evidence indicates that
MRI-based PSAD calculations, benefiting from superior soft-tissue
resolution, outperform TRUS-derived values (37). Consequently, MRI-calculated PSAD
exhibits enhanced diagnostic sensitivity and specificity in
discriminating malignant from benign lesions, supporting its
integration into modern diagnostic pathways (38).
PSAV measures the dynamic change in PSA levels over
time, calculated as the absolute change in PSA per year
[PSAV=(PSA2-PSA1)/ΔTime in years] (39). A velocity >0.75 ng/ml per year
is considered clinically considerable and suspicious for underlying
malignancy (40). PSAV is
particularly useful for monitoring individuals with borderline PSA
levels and for assessing disease progression in patients'
post-treatment (41). A rapid
rise in PSA is often associated with aggressive tumor biology and a
worse prognosis (42).
PHI is an advanced blood test that combines three
distinct forms of PSA: f-PSA, t-PSA and a truncated form of
kallikrein (KLK)-related protein 4 (k-PSA, also known as hK4). It
is calculated using the following formula (46): PHI=(f-PSA)/(t-PSA) x t-PSA
√(t-PSA) x k-PSA.
The resulting score stratifies patients according to
their risk of harboring clinically considerable PCa, with higher
values indicating greater risk (47). The established clinical thresholds
guide management as follows: i) A PHI <15 suggests a low
likelihood of malignancy, ii) values between 15 and 35 fall into an
indeterminate range where adjunctive evaluation with
multiparametric MRI (mpMRI) is recommended, iii) a PHI >35
signifies high risk and typically warrants biopsy (48,49). PHI demonstrates superior
diagnostic accuracy compared with t-PSA or f/t PSA alone,
particularly within the t-PSA gray zone and has been shown to
reduce unnecessary biopsies by ~20% (50,51). Prospective validation studies
confirm that PHI effectively identifies aggressive PCa, improving
patient selection for invasive procedures (52,53).
The integration of PHI with mpMRI represents a
powerful diagnostic synergy. This combined model enhances detection
confidence for clinically considerable cancer while minimizing
overtreatment (54). Furthermore,
ongoing research into ethnicity-specific PHI thresholds highlights
the importance of personalized diagnostic standards to optimize
accuracy across diverse populations (55,56).
Transcriptomic analyses demonstrate distinct
expression patterns of multiple KLKs in prostate carcinoma compared
with benign tissue, underscoring their combined diagnostic and
prognostic potential (70,71).
KLK2, sharing ~78% sequence identity with KLK3, is
also AR-regulated and activates pro-PSA by cleaving its pro-peptide
(72). Due to its highly
restricted expression profile, KLK2 has emerged as an
attractive therapeutic target (73). Several innovative platforms are
currently in development, including: i) Radioligand therapies
delivering cytotoxic payloads selectively to KLK2-expressing cells
(74), ii) bispecific T-cell
engagers redirecting cytotoxic lymphocytes toward KLK2-positive
tumor cells (75); and iii)
chimeric antigen receptor T-cell therapies engineered to recognize
KLK2 on the cancer cell surface (76). Promising clinical candidates from
companies such as Johnson & Johnson and EpimAb Biotherapeutics
have advanced into Phase I trials, representing an encouraging step
toward precision immunotherapy in PCa (77,78).
Nucleic acid biomarkers, encompassing DNA- and
RNA-based alterations, have emerged as powerful tools for the
diagnosis, prognostic stratification and treatment selection of PCa
(79). Their analysis,
particularly through non-invasive liquid biopsies, offers a
real-time snapshot of tumor genetics and dynamics (80).
In PCa, intrachromosomal rearrangements such as
inversions or translocations within the 21q22 region can fuse the
TMPRSS2 promoter with the ERG coding sequence, creating the
TMPRSS2-ERG fusion gene (110).
This fusion places ERG expression under AR control, leading to
aberrant ERG overexpression in prostate epithelial cells and
promoting tumorigenesis (111).
The TMPRSS2-ERG fusion occurs in 40-60% of sporadic
prostate cancer types, representing the most frequent gene
rearrangement in PCa (112). Its
prevalence increases to 50-70% in familial and early-onset (≤55
years) cases (113).
Fusion-positive tumors are often associated with higher Gleason
scores (≥7), elevated PSA levels and increased risk of lymph node
and bone metastases, as well as castration-resistant progression
(114,115). However, the independent
prognostic value of TMPRSS2-ERG for overall survival remains
controversial, as it may depend on co-occurring genomic alterations
(116).
Mechanistically, ERG activation regulates multiple
downstream targets, including matrix metalloproteinases and
vascular endothelial growth factor, thereby enhancing cell
proliferation, invasion and angiogenesis (117). ERG can also repress DNA repair
genes such as BRCA1, leading to impaired repair capacity and
increased genomic instability (118). Notably, PTEN deletions
co-occur in 30-40% of TMPRSS2-ERG-positive tumors, acting
synergistically to drive cancer progression and enzalutamide
resistance (119). By contrast,
SPOP-mutated prostate cancer types exhibit a markedly lower
TMPRSS2-ERG fusion rate (<10%), indicating mutually exclusive
oncogenic pathways among PCa molecular subtypes (120).
Recent genomic studies have expanded understanding
of TMPRSS2-ERG-associated oncogenesis (121,122). Large-scale genome-wide
association studies have identified additional susceptibility loci
that may interact with known genes such as BRCA1/2,
broadening the scope of genetic risk assessment in PCa (123). Furthermore, emerging domestic
studies highlight that the TMPRSS2-ERG fusion can regulate PCa cell
behavior through modulation of non-coding RNAs, suggesting novel
avenues for targeted therapy and improved understanding of PCa
pathogenesis (124,125) (Table I).
Expression of ERG mRNA, a product of the TMPRSS2-ERG
fusion, and SPINK1 mRNA, often overexpressed in fusion-negative
types of cancer, provides complementary diagnostic and subtyping
information (132-134). miRNAs such as miR-141 and
miR-375 in blood or urine show promise as non-invasive biomarkers
(135,136). Panels combining multiple RNA
markers (such as PCA3, ERG and miRNAs) have demonstrated superior
diagnostic accuracy over single-marker tests (137,138). Furthermore, specific miRNA
expression patterns are implicated in modulating the tumor immune
microenvironment, suggesting potential for guiding immunotherapy
strategies (139,140).
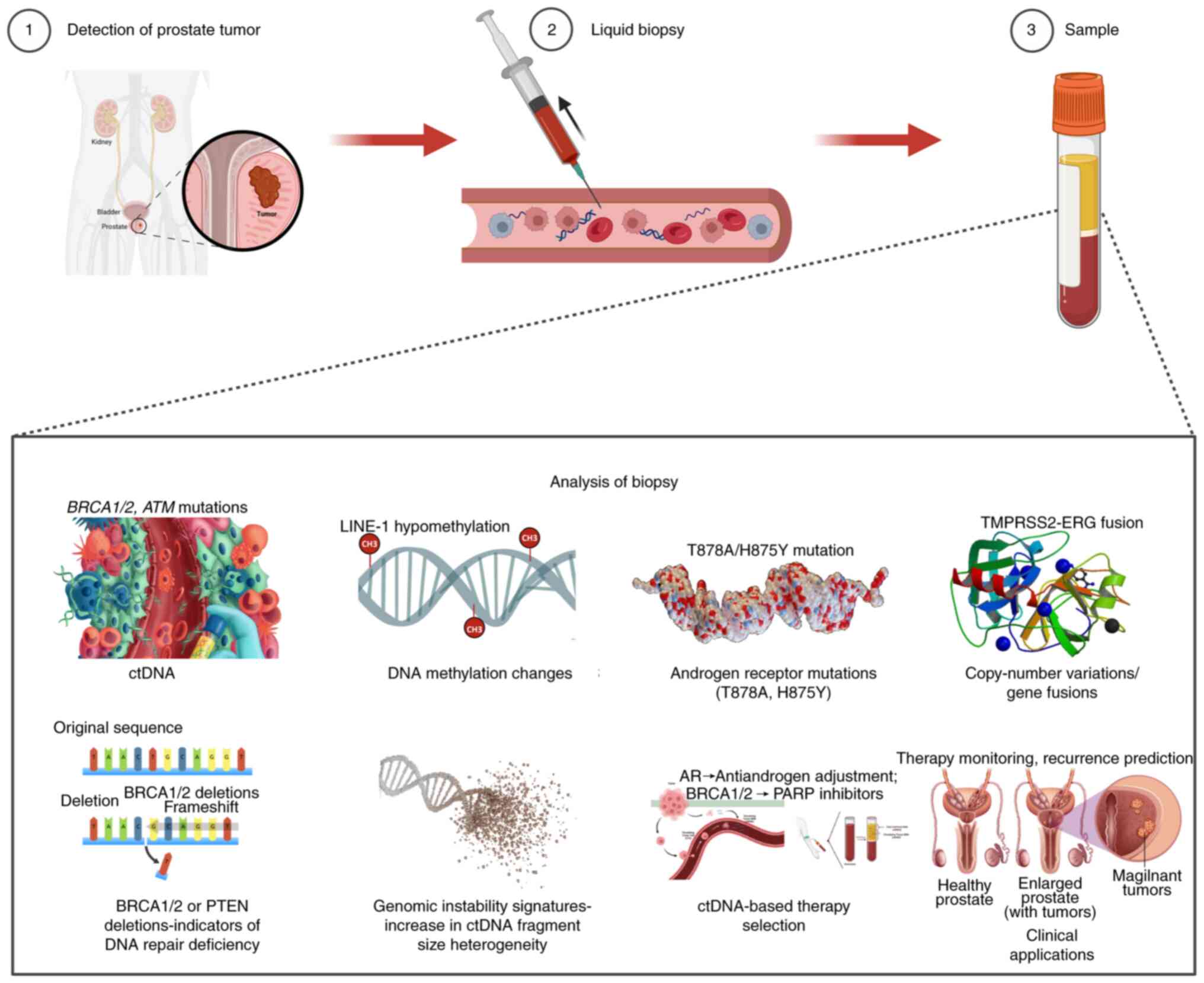
ctDNA, released into the bloodstream by tumor cells,
provides a non-invasive means to assess tumor genetics and
heterogeneity (141-143). Unlike prostate needle biopsies,
which are invasive and subject to sampling bias due to tumor
heterogeneity, ctDNA testing provides a non-invasive method to
detect genetic alterations throughout the body, offering a more
comprehensive outlook of tumor characteristics (144,145). This makes ctDNA testing
particularly valuable for monitoring patients with
castration-resistant prostate cancer or those with inaccessible
metastatic lesions (146,147).
In mCRPC, ctDNA analysis enables real-time
monitoring of AR mutations (such as T878A and H875Y) that confer
resistance to abiraterone and enzalutamide, allowing for dynamic
treatment adjustments (148). Of
all patients with mCRPC, ~20% harbor mutations in DNA repair genes
(such as BRCA1/2 and ATM) detectable in ctDNA, which
predict responsiveness to PARP inhibitors (149). ctDNA testing offers a
non-invasive method to detect these mutations, helping physicians
identify candidates for PARP inhibitor-targeted therapy
(150). Monitoring the abundance
of drug-resistant mutations in ctDNA, such as AR mutations, can
provide early indications of tumor progression, enabling timely
adjustments to treatment regimens (151). ctDNA dynamics also serve as a
sensitive marker for minimal residual disease and early recurrence
after radical prostatectomy (152,153). Emerging applications include
using ctDNA methylation profiles to predict responses to
immunotherapy (154,155), with ongoing clinical trials
validating its role in precision oncology (Fig. 2) (156).
Exosomes are extracellular vesicles (30-150 nm) that
carry nucleic acids, including RNAs, from their cell of origin,
reflecting the physiological and pathological state (157,158). In cancer, the RNA profiles in
exosomes undergo specific alterations that reflect tissue damage or
malignant transformation (159,160). For instance, miR-122 in exosomes
from liver cancer cells has been identified as a diagnostic marker
for liver cancer, while Aβ-related mRNA levels in exosomes from
cerebrospinal fluid can indicate Alzheimer's disease (161).
Beyond diagnostics, exosomes are being engineered as
therapeutic vehicles for the targeted delivery of siRNA or miRNA to
tumor cells, showing efficacy in preclinical models (165). For example exosomes loaded with
anti-miR-21 have been used to inhibit metastasis in breast cancer
and similar strategies are under investigation for PCa (166,167). Additionally, blocking the
release of metastasis-promoting miRNAs (such as miR-10b) in
exosomes from tumor cells has been shown to inhibit cancer cell
metastasis in lung cancer models, offering a potential strategy for
preventing PCa metastasis by interfering with exo RNA function
(Table II) (168).
Moreover, the regulation of exo lncRNA also presents
novel avenues for interfering with cancer proliferation and
invasion (169). In preclinical
studies, some researchers have successfully developed exosome
carriers specifically targeting PCa cells and loaded them with
anti-cancer RNA molecules (170-172). These carriers have demonstrated
promising effects in inhibiting tumor growth and metastasis in
animal models, providing new hope for targeted PCa therapies
(172). Similarly, some studies
have analyzed exo RNA profiles at different stages of PCa,
revealing that certain RNA characteristics can accurately determine
disease stage and prognosis, which can guide clinicians in
formulating more precise treatment plans (Fig. 3) (172,173).
Citric acid and polyamines have attracted increasing
attention as metabolic biomarkers for PCa diagnosis and management
(173). Citric acid, abundantly
present in prostatic fluid, blood and tissues, serves as an
indicator of prostate metabolism (174). Reduced citric acid levels are
typically associated with malignant transformation, offering
potential diagnostic utility in differentiating benign from
malignant lesions (175). The
use of magnetic resonance spectroscopy (MRS) enables non-invasive
quantification of citric acid, providing valuable metabolic
information for clinical decision-making (176). However, its diagnostic
sensitivity remains limited when used alone and combination with
other biomarkers such as PSA improves accuracy and specificity
(177).
Polyamines, such as putrescine, spermidine and
spermine-also show association with tumor aggressiveness and
metastatic potential (178,179). Elevated polyamine concentrations
in serum or urine may indicate enhanced tumor invasiveness and poor
prognosis (180). Monitoring
polyamine profiles could therefore assist in predicting disease
progression and evaluating treatment responses. Nevertheless,
benign conditions such as inflammation can modestly elevate
polyamine levels, limiting diagnostic specificity (181).
From a therapeutic perspective, abnormal metabolism
of citric acid and polyamines represents a promising target for
intervention (182). Strategies
aiming to restore mitochondrial metabolism or inhibit ornithine
decarboxylase, a key enzyme in polyamine synthesis, have
demonstrated potential in enhancing endocrine therapy and
overcoming drug resistance (182,183). Recent metabolomics-based studies
have identified urinary derivatives such as
N1-acetylputrescine, which, when analyzed together with
citric acid metabolites, notably improve early screening
sensitivity (184,185). Integration of MRS-derived citric
acid signals and serum polyamine levels with machine learning
algorithms has further enhanced diagnostic precision, particularly
for small lesions (186,187). Cross-ethnic studies have
revealed distinct metabolic profiles of citric acid and polyamines,
suggesting that population-specific diagnostic thresholds may
improve biomarker accuracy (188,189). Moreover, emerging evidence
indicates that gut microbial metabolites can modulate prostate
metabolism by influencing citric acid and polyamine pathways,
offering new insights into prostate carcinogenesis and potential
microbiota-targeted therapies (189).
Basal cell markers p63 and 34βE12 are indispensable
tools in PCa histopathology (190). In normal glands, basal cells
form a continuous layer expressing nuclear p63 and cytoplasmic
34βE12 (191-193). Their absence is a hallmark of
adenocarcinoma, reflecting the replacement of normal basal
epithelium by malignant cells (194,195).
Compared with p63, which is nuclear and
occasionally affected by tissue fixation, 34βE12 provides stable
cytoplasmic staining, allowing reliable detection even in
suboptimally preserved specimens (196,197). During prostate biopsy
evaluation, the concurrent loss of both markers indicates
carcinoma, with diagnostic sensitivity >95% and specificity
>90% (198-200). When basal cells appear partially
positive, additional markers such as PSA or α-methylacyl-CoA
racemase are used to confirm malignancy (201,202).
Recent advances combine p63 and 34βE12
immunostaining with AI-assisted pathology, markedly improving
interpretive accuracy and reducing observer variability (203,204). Single-cell sequencing and
organoid models have further elucidated the dynamic expression of
these markers across tumor grades, associating their loss with
malignancy progression, recurrence risk and treatment outcomes
(205-207).
DNA methylation represents one of the earliest and
most consistent molecular alterations in PCa. Among the numerous
genes identified, GSTP1 promoter methylation is the most
extensively validated (208).
GSTP1 encodes glutathione S-transferase, an enzyme involved
in detoxification and oxidative defense. Hypermethylation-induced
silencing of GSTP1 occurs in >90% of PCa cases but rarely
in benign tissues, making it a highly specific diagnostic indicator
(209). Combined detection of
GSTP1 methylation with urinary PCA3 enhances diagnostic
specificity >90% (210).
Genome-wide methylation profiling has recently
identified novel PCa-related epigenetic signatures with potential
diagnostic and prognostic applications (216). Combining methylation markers
with genetic and RNA-based assays, particularly in liquid biopsy
formats, offers a more comprehensive and non-invasive diagnostic
strategy (217). Studies also
indicate that non-coding RNAs may regulate methylation marker
expression, adding another layer of complexity to prostate
tumorigenesis (218,219). These findings highlight the
promise of methylation-based diagnostics and their integration into
precision oncology (Fig. 4)
(219).
The convergence of genetic, transcriptomic and
metabolic biomarkers has fundamentally transformed PCa research and
clinical management. Genetic alterations-such as BRCA1/2,
HOXB13 and TMPRSS2-ERG fusions-enable precise risk
stratification and therapeutic guidance (220). RNA-based biomarkers, including
PCA3, ERG mRNA and miRNA panels, have established robust,
non-invasive tools for early detection and disease monitoring
(221). Metabolic biomarkers
such as citric acid and polyamines complement these findings by
reflecting tumor aggressiveness and metabolic reprogramming
(222). Epigenetic and liquid
biopsy markers, particularly DNA methylation (such as GSTP1
and APC) and circulating nucleic acids (ctDNA, exo RNA), are
emerging as cornerstones of precision diagnostics (223). Their integration into clinical
workflows enables dynamic disease monitoring and individualized
therapy optimization (224).
Future research should focus on multi-omics
integration-combining genomic, transcriptomic, methylomic and
metabolomic data to construct comprehensive biomarker networks
(225). Incorporating AI and
machine learning will further enhance diagnostic accuracy and
predict therapeutic outcomes (226). Additionally, investigating
population-specific biomarker variations and tumor microenvironment
interactions will be vital for developing equitable, personalized
care strategies (227).
Ultimately, the continued refinement and clinical validation of
these emerging biomarkers will usher in a new era of PCa precision
medicine, enabling earlier detection, tailored therapy and improved
survival and quality of life for patients worldwide (Fig. 5) (228).
Not applicable.
YH, JM and XL contributed to the writing of the
original draft and the preparation of figures. YH contributed to
the data analysis. JM and XL provided writing review and editing,
as well as supervision. All authors read and approved the final
manuscript. Data authentication not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
The authors would like to express their sincere
gratitude to Mr. Zhenjun Xi and Mr. Li Wang from Chongqing Medical
and Pharmaceutical College, Chongqing, China for their invaluable
contributions to the manuscript; their meticulous efforts ensured
the accuracy and reliability of the research findings of the
present study.
The authors declare that financial support was received for the
research, authorship and publication of this article. This work was
funded by 2023 Chongqing Medical Scientific Research project (Joint
project of Chongqing Health Commission and Science and Technology
Bureau; grant no. 2023GGXM006), 2024 Scientific Research project of
Chongqing Medical and Pharmaceutical College (grant no.
ygzrc2024101), Chongqing Education Commission Natural Science
Foundation (grant no. KJQN202402821; grant no. KJQN202502819) and
2024 Chongqing Medical and Pharmaceutical College Innovation
Research Group Project (grant no. ygz2024401), Chongqing Science
and Health Joint Medical Research Project (grant no.
2024SQKWLHMS051), Natural Science Foundation of Chongqing (grant
no. CSTB2025NSCO-GPX1116) and Technological Innovation Project of
Shapingba District, Chongqing (grant no. 2025016, 2025017 and
2025031), Scientific Research Project of AnShun University, Guizhou
Province [grant no. asxybsjj (202307) and asxykypt (202402)] and
Natural Science Research Project of Guizhou Provincial Department
of Education [Qian Jiao He KY (2020) grant no. 063]
respectively.
|
1
|
Kratze TB, Mazzitelli N, Star J, Dahut WL,
Jemal A and Siegel RL: Prostate cancer statistics, 2025. CA Cancer
J Clin. 75:485–497. 2025.
|
|
2
|
Braga R, Morais S, Pacheco-Figueiredo L,
Araujo N and Lunet N: Prostate cancer screening: Knowledge,
attitudes, and practices by medical doctors in portugal. J Cancer
Educ. Jun 19–2025.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takashima Y, Yoshii K, Tanaka M and
Tashiro K: Ubiquitin-proteasome pathway-linked gene signatures as
prognostic indicators in prostate cancer. Anticancer Res.
45:1825–1841. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Elgehama AM, Yang Q, He Z, Ruegg L, You S
and Yang W: Identification of a RIPK2-regulated gene signature as a
candidate biomarker for RIPK2 activity and prognosis in prostate
cancer. bioRxiv [Preprint]. 2025.04.30.651490. 2025.PubMed/NCBI
|
|
5
|
Mellor J, Hunter E and Akoulitchev A:
Paradigm lost. Cancers (Basel). 17:21872025. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilkosz J, Sobieraj DW, Kaluzewski T,
Kaczmarek J, Szwalski J, Bednarek M, Morel A, Kasprzyk Z,
Kepczynski L, Salamunia J, et al: Personalized high-resolution
genetic diagnostics of prostate adenocarcinoma guided by
multiparametric magnetic resonance imaging: Results of a pilot
study. Int J Mol Sci. 26:56482025. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Waldman L, Silberman Y, Wang M,
Tackey C, Hanna L, Vesprini D, Emmenegger U, Eisen A and
Smoragiewicz M: Mainstream model of genetic testing for prostate
cancer at a large tertiary cancer centre. Clin Genitourin Cancer.
22:1020522024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gonzalez-Peramato P, Alvarez-Maestro M,
Heredia-Soto V, Mendiola Sabio M, Linares E, Serrano A,
Alvarez-Ossorio JL, Lopez Alcina E, Prieto L, Vazquez Alonso F, et
al: Comparing prostatype P-score and traditional risk models for
predicting prostate cancer outcomes in Spain. Actas Urol Esp (Engl
Ed). 49:5017882025.PubMed/NCBI
|
|
9
|
Stella M, Russo GI, Leonardi R, Carco D,
Gattuso G, Falzone L, Ferrara C, Caponnetto A, Battaglia R, Libra
M, et al: Extracellular RNAs from whole urine to distinguish
prostate cancer from benign prostatic hyperplasia. Int J Mol Sci.
25:100792024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pattou M, Neuzillet Y, Ghoneim T, Bosset
PO, Herve JM, Vanalderwerelt V, Bohin D, Lugagne PM, Soorojebally Y
and Lebret T: PSA density correlates to pathology T stage and ISUP
grade: Insights from a cohort of 3568 radical prostatectomy cases.
World J Urol. 43:4452025. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alahdal M, Perera RA, Moschovas MC, Patel
V and Perera RJ: Current advances of liquid biopsies in prostate
cancer: Molecular biomarkers. Mol Ther Oncolytics. 30:27–38. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Traphagen NA, Wheeler E, Li R, Lu F, Ahmed
B, Tewari AK, Balk SP, Nelson PS, Corey E, Long H, et al: Lack of
synergy between AR targeted therapies and PARP inhibitors in
homologous recombination-proficient prostate cancer. bioRxiv
[Preprint]. 2025.06.02.657429.2025.
|
|
13
|
Castellnou P, Gomez-Martinez M,
Gomez-Vallejo V, Baz Z, Lopez-Gallego F, Rondon-Lorefice I,
Zabala-Letona A, Poot AJ, Mendizabal I, Carracedo A, et al:
Unravelling the role of L- and D-alanine in prostate cancer: A
positron emission tomography study in a genetic mouse model. Nucl
Med Biol. 148-149:1090482025. View Article : Google Scholar
|
|
14
|
Coradduzza D, Sanna A, Di Lorenzo B,
Congiargiu A, Marra S, Cossu M, Tedde A, De Miglio MR, Zinellu A,
Mangoni AA, et al: Associations between plasma and urinary heavy
metal concentrations and the risk of prostate cancer. Sci Rep.
15:142742025. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mitura P, Paja W, Mlynarczyk G, Kowalski
R, Bar K and Depciuch J: Urine-based Raman markers for prostate
cancer diagnosis: A machine learning approach using fingerprint and
lipid spectral region. Spectrochim Acta A Mol Biomol Spectrosc.
344(Pt 1): 1266612026. View Article : Google Scholar
|
|
16
|
Sharma D, Mitra D, Bansal VK and Singh KV:
Study of the correlation between multi-parametric MRI (MP-MRI)
prostate findings and transrectal ultrasound (TRUS)-guided prostate
biopsy results in patients with raised serum PSA. Cureus.
17:e862162025.PubMed/NCBI
|
|
17
|
Huang S, Hart JC, Smith JF, Bench S, Yepes
LR, Griscom B and Clark-Langone KM: Tissue of origin
characterization of cell free DNA in seminal plasma: Implications
for new liquid biopsies. PLoS One. 20:e03177122025. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mo H, Hu C, Li P, Miao Z, Zhang H, Shen G
and Sun J: PSA proportion of index lesion: a novel index for
predicting prostate pathology outcomes in biopsy-naive patients
with PSA < 10 ng/ml and PI-RADS 3 lesions. World J Urol.
43:4442025. View Article : Google Scholar
|
|
19
|
Wang X, He Z, He X, Zeng R, He H, Zhang K
and Xu Y: High-efficiency detection of Total-PSA and Free-PSA in
whole blood by microfluidic chip integrated with electromagnetic
co-preprocessing. ACS Sens. 10:3878–3887. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arcay Ozturk A, Erkilic M, Bural GG, Aydin
F and Boz A: Physiological biodistribution on Ga68-PSMA PET/CT and
the factors effecting biodistribution. Ann Nucl Med. 38:894–903.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schulze A, Christoph F, Sachs M, Schroeder
J, Stephan C, Schostak M and Koenig F: Use of the prostate health
index and density in 3 outpatient centers to avoid unnecessary
prostate biopsies. Urol Int. 104:181–186. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Chang Y, Song L, Ren J, Yin L, Cao
W, Xu W and Ren S: Robotic prostatectomy for large-volume prostates
in prostate cancer: A retrospective analysis of 50 cases (>100
ml). BMC Cancer. 25:7012025. View Article : Google Scholar
|
|
23
|
Zheng B, Mo F, Shi X, Li W, Shen Q, Zhang
L, Liao Z, Fan C, Liu Y, Zhong J, et al: An automatic
deep-radiomics framework for prostate cancer diagnosis and
stratification in patients with serum prostate-specific antigen of
4.0-10.0 ng/mL: A multicenter retrospective study. Acad Radiol.
32:2709–2722. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qiu L, Tian T, Ma Z, Sun J, Zhao Y, Yang
L, Ji Y, Yi L, Yan L, Xu C and Li D: Predictive value of the
pretreatment serum sialic acid/total protein ratio for bone
metastases in newly diagnosed prostate cancer patients: development
of a nomogram model. Gland Surg. 14:1066–1078. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rastogi S, Sato N, Lee S, Lee MJ, McKinney
Y, Lindenberg L, Mena E, Thomas A, Shrestha RL and Choyke PL:
Circulating tumor cells are detectable and independent of PSA and
PSMA-PET metrics in localized high-risk and biochemically recurrent
prostate cancer. medRxiv [Preprint]. 2025.07.09.25331014.
2025.PubMed/NCBI
|
|
26
|
Fan YP, Zhang YT, Zhang G, Ma L, Lv Y, Li
J, Luan Y, Zhang YX, Chen YT, Ren HY, et al: Proteomic profiling of
urinary large extracellular vesicles for the diagnosis of prostate
cancer. Anal Chem. 97:17368–17379. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen D, Ma Y, Yang A, Hu L, Zhou H, Xu J,
Chen S, Nie D, Feng W, Cai H, et al: Dual-Enhanced SERS satellite
immuno-nanocomplex for multiple PSA-Mediated PHI assay toward
clinical prostate cancer screening. Adv Sci (Weinh).
12:e24117472025. View Article : Google Scholar
|
|
28
|
Yao W, Wu J, Kong Y, Xu F, Zhou Y, Sun Q,
Gao Q, Cai Z, Yang C and Huang Y: Associations of systemic
immune-inflammation index with high risk for prostate cancer in
middle-aged and older US males: A population-based study. Immun
Inflamm Dis. 12:e13272024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hochma E, Firer MA and Minnes R:
Near-infrared and sono-enhanced photodynamic therapy of prostate
cancer cells using phyto-second harmonic generation nanoconjugates.
Polymers (Basel). 17:18312025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Luo B, Zhou J, Zhan X, Ying B, Lan F and
Wu Y: Smartphone-based free-to-total prostate specific antigen
ratio detection system using a colorimetric reaction integrated
with proximity-induced bio-barcode and CRISPR/Cas12a assay. Small.
20:e23102122024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Serkan Y, Caner E, Serkan A, Bulent K,
Yasin V, Adem A and Omer Y: Significance of inflammation markers to
predict curative treatment for prostate cancer patients on active
surveillance. J Clin Lab Anal. 39:e700592025. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stojadinovic M, Jurisevic N, Stojadinovic
M and Jankovic S: Predictive clinical characteristics for adverse
pathological outcomes in intermediate- and high-risk prostate
cancer during biopsy. Int Urol Nephrol. 57:4077–4086. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yorioka MAW, Murta CB, Leite KRM, Cardili
L, de Mello ES, de Carvalho Fazoli AJ, Cordeiro MD, Coelho RF,
Viana PCC, Kohama CSN, et al: ERG and PTEN role on active
surveillance for low-risk prostate cancer in the multiparametric
MRI Era. Prostate. 85:364–373. 2025. View Article : Google Scholar
|
|
34
|
Weng Y and Ji B: Serum levels of prostate
specific antigen, free PSA, (-2)proPSA, fPSA/tPSA ratio, prostate
health index, and glycosylation patterns of free PSA in patients
with benign prostatic hyperplasia pharmacotherapy: Limitations and
future directions. Prostate. 85:1248–1249. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Encarnacion Navarro JA, Morillo Macias V,
Borras Calbo M, De la Fuente Munoz I, Lozano Martinez A, Garcia
Martinez V, Fernandez Fornos L, Guijarro Roche M, Amr Rey O and
Garcia Gomez R: Multicenter real-world study: 432 patients with
apalutamide in metastatic hormone-sensitive prostate cancer. Curr
Oncol. 32:1192025. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gjelsvik YM, Myklebust TA, Fosså SD, Haug
ES, Kvåle R, Ursin G and Johannesen TB: A nationwide, longitudinal
collection of patient-reported outcomes from prostate cancer
patients and controls. Qual Life Res. 34:2689–2700. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zeng Y, Liao H, Jiang G, Wu G and Zhong J:
Diagnostic value of 18F-prostate-specific membrane antigen-1007
PET/MRI versus 18F-prostate-specific membrane antigen-1007
PET/computed tomography for biochemical recurrence of prostate
cancer after radical prostatectomy. Nucl Med Commun. 46:1052–1060.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Williams N, Remmers S, Jay A, Santoro K
and O'Callaghan M: External validation of MRI-based prostate cancer
risk calculators in an Australian cohort. ANZ J Surg. 95:1906–1911.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Van der Eecken H, De Cock D, Roussel E,
Giesen A, Vansevenant B, Goeman L, Quackels T and Joniau S:
Nutritional supplement with fermented soy in patients under active
surveillance for low-risk or intermediate-risk prostate cancer:
Results from the PRAEMUNE trial. Cancers (Basel). 16:36342024.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo P, A G, Chen T, Guo Y, Tang Y, Pan J,
Wang B, Gong R, Chen G and Huang S: A hematological and
inflammatory marker-based model for prostate carcinoma diagnosis.
Am J Cancer Res. 15:2551–2563. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cirulli GO, Davis M, Stephens A, Chiarelli
G, Finati M, Chase M, Tinsley S, Arora S, Sood A, Lughezzani G, et
al: Midlife baseline prostate-specific antigen, velocity, and
doubling time association with lethal prostate cancer and
mortality. Cancer. 131:e355632025. View Article : Google Scholar
|
|
42
|
Kobayashi M, Kijima T, Yashi M and Kamai
T: Prostate-specific antigen kinetics contributes to decision
making for biopsy referral: the predictive implication for PSA
retest in patients with elevated PSA levels. Prostate Int.
11:27–33. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Javaeed A, Ghauri SK, Ibrahim A and Doheim
MF: Prostate-specific antigen velocity in diagnosis and prognosis
of prostate cancer - a systematic review. Oncol Rev. 14:4492020.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ono K, Hiyoshi Y, Ono A, Ouchi M, Kosumi
K, Eto K, Ida S, Iwatsuki M, Baba Y, Miyamoto Y, et al: Locally
advanced rectal cancer in a young adult affected with dyskeratosis
congenita (Zinsser-Cole-Engman syndrome): A case report. Surg Case
Rep. 10:2062024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dai F, He Y, Duan J, Lin K, Lv Q, Zhao Z,
Zou Y, Jiang J, Zheng Z and Qiu X: Global trends in the use of
artificial intelligence for urological tumor histopathology: A
20-year bibliometric analysis. Digit Health.
11:205520762513488342025. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Aktas S, Yucetas U, Yucetas E, Ates HA,
Gene C and Erkan E: The efficacy of prostate health index (PHI) in
predicting pathological progression in low-risk localized prostate
cancer cases under active surveillance protocol. World J Urol.
43:4312025. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Assam KD, Pierce JM, Galloway LA and
Tosoian JJ: Blood- and urine-based biomarkers for the detection of
clinically significant prostate cancer: A contemporary review. Curr
Opin Urol. 35:590–596. 2025. View Article : Google Scholar
|
|
48
|
Gupta VK, Cortese BD and Talwar R:
Cost-effectiveness of serum, urine, and tissue-based prostate
cancer biomarkers. Curr Opin Urol. 35:412–417. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jeon SS, Song W, Kang M, Sung HH, Jeon HG,
Jeong BC, Seo SI and Chung JH: Diagnostic value of prostate health
index in patients with no index lesion on mpMRI or negative
previous combined biopsy. Investig Clin Urol. 66:124–129. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jirasko M, Vitak R, Pecen L, Pinkeova A,
Tkac J, Bertok T, Bergman N and Kucera R: Serum levels of prostate
specific antigen, free PSA, (-2)proPSA, fPSA/tPSA ratio, prostate
health index, and glycosylation patterns of free PSA in patients
with benign prostatic hyperplasia pharmacotherapy. Prostate.
85:65–72. 2025. View Article : Google Scholar
|
|
51
|
Vakili S, Beheshti I, Barzegar Behrooz A,
Los MJ, Vitorino R and Ghavami S: Transforming prostate cancer
care: Innovations in diagnosis, treatment, and future directions.
Int J Mol Sci. 26:53862025. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Britton CJ, Andrews JR, Arafa A, Kim Y,
Latuche LR, Schulte PJ, Joshi VB, Ahmed ME, Jeffrey Karnes R and
Lucien F: Prostate extracellular vesicles and prognostic biomarkers
of clinically significant prostate cancer: A prospective
single-institution pilot study. Prostate. 85:594–602. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lee IT, Hou CM, Vo TTT, Hong JH, Huang CY,
Wang YL, Chen YL and Chiang CH: Optimizing prostate cancer care:
Clinical utility of the prostate health index. Prostate.
85:1357–1368. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chung Y and Hong SK: Evaluating prostate
cancer diagnostic methods: The role and relevance of digital rectal
examination in modern era. Investig Clin Urol. 66:181–187. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fasulo V, Chiarelli G, Garofano G,
Ripamonti CB, Barile M, Bianchi P, Morenghi E, Benetti A, Aljoulani
M and Finocchiaro A: Impact of prostate cancer screening in
European ancestry un-affected men with germline DNA repair
pathogenic variants. BJUI Compass. 6:e4242025. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chang CH, Yu PH, Hsieh PF, Hong JH, Chiang
CH, Cheng HM, Wu HC, Huang CY and Lin TP: Prostate health index
density aids the diagnosis of prostate cancer detected using
magnetic resonance imaging targeted prostate biopsy in Taiwanese
multicenter study. J Chin Med Assoc. 87:678–685. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Stuardo-Parada A, Lopez-Munoz R,
Villarroel-Espindola F, Figueroa CD and Ehrenfeld P: Minireview:
Functional roles of tissue kallikrein, kinins, and
kallikrein-related peptidases in lung cancer. Med Oncol.
40:2242023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kelam N, Ogorevc M, Gotovac I, Kuzmic
Prusac I, Vukojevic K, Saraga-Babic M and Mardesic S: Analysis of
kallikrein 6, Acetyl-α-tubulin, and aquaporin 1 and 2 expression
patterns during normal human nephrogenesis and in congenital
anomalies of the kidney and urinary tract (CAKUT). Genes (Basel).
16:4992025. View Article : Google Scholar
|
|
59
|
Sasiadek L, Bielecka E, Falkowski K,
Kulczycka M, Bereta G, Maksylewicz A, Zubrzycka N, Dobosz E, Koziel
J, Drukala J, et al: Human tissue kallikrein 14 induces the
expression of IL-6, IL-8, and CXCL1 in skin fibroblasts through
protease-activated receptor 1 signaling. FEBS J. 292(21):
5659–5675. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Giuffre M, Lista AD, Paulson N and Masuoka
L: A phase 1C, open label, single ascending dose study to evaluate
the safety, tolerability, and pharmacokinetics of DM199
administered intravenously with a polyvinyl chloride bag in adult
healthy subjects and adults recently taking angiotensin-converting
enzyme inhibitors. Clin Pharmacol Drug Dev. 14:452–460. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Singh SP, Dhanasekara CS, Melkus MW, Bose
C, Khan SY, Sardela de Miranda F, Mahecha MF, Gukhool PJ, Tonk SS,
Jun SR, et al: Relevance of cellular homeostasis-related gene
expression signatures in distinct molecular subtypes of breast
cancer. Biomedicines. 13:10582025. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kumar N, Kole L and Singh AK: Precision
assay for detection and quantification of anti-prostate specific
antigen antibodies using indirect ELISA. Infect Disord Drug
Targets. May 15–2025.Epub ahead of print. View Article : Google Scholar
|
|
63
|
Boumali R, David E, Chaaya N, Lucas M, Ait
Amiri S, Lefort V, Nina-Diogo A, Salmain M, Petropoulos I, Corce V,
et al: Deferasirox derivatives as inhibitors of kallikrein-related
peptidases associated to neurodegenerative diseases. ChemMedChem.
20:e2025001872025. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Koistinen H, Kovanen RM, Hollenberg MD,
Dufour A, Radisky ES, Stenman UH, Batra J, Clements J, Hooper JD,
Diamandis E, et al: The roles of proteases in prostate cancer.
IUBMB Life. 75:493–513. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Angel MR, Loeffler D, Charifa A, Sinit RB,
Amery T, Cengiz B, Beer TM and Thomas GV: TMPRSS2 expression in
lung tissue of prostatic adenocarcinoma patients: A pathologic
perspective on androgen deprivation therapy. medRxiv [Preprint].
2025.05.03.25326931. 2025.PubMed/NCBI
|
|
66
|
Srinivasan S, Kryza T, Batra J and
Clements J: Remodelling of the tumour microenvironment by the
kallikrein-related peptidases. Nat Rev Cancer. 22:223–238. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Stein MN, Vinceneux A, Robbrecht D, Doger
B, Autio KA, Schweizer MT, Calvo E, Medina L, Van Dongen M, Deville
JL, et al: Real-world effectiveness of darolutamide in metastatic
castration-resistant prostate cancer. J Clin Oncol. 43:2515–2526.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhao J, Sun Y, Ren L, Huang S and Zhang J:
Antagonism of androgen receptor signaling by aloe-emodin. Food Chem
Toxicol. 181:1140922023. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Boyukozer FB, Tanoglu EG, Ozen M, Ittmann
M and Aslan ES: Kallikrein gene family as biomarkers for recurrent
prostate cancer. Croat Med J. 61:450–456. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wenta T, Nastaly P, Lipinska B and
Manninen A: Remodeling of the extracellular matrix by serine
proteases as a prerequisite for cancer initiation and progression.
Matrix Biol. 134:197–219. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Matin F, Jeet V, Srinivasan S, Cristino
AS, Panchadsaram J, Clements JA and Batra J; Australian Prostate
Cancer BioResource: MicroRNA-3162-5p-mediated crosstalk between
kallikrein family members including prostate-specific antigen in
prostate cancer. Clin Chem. 65:771–780. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Nakayama H, Sekine Y, Oka D, Miyazawa Y,
Arai S, Koike H, Matsui H, Shibata Y and Suzuki K: Combination
therapy with novel androgen receptor antagonists and statin for
castration-resistant prostate cancer. Prostate. 82:314–322. 2022.
View Article : Google Scholar
|
|
73
|
Karan D, Dubey S, Gunewardena S, Iczkowski
KA, Singh M, Liu P, Poletti A, Choo YM, Chen HZ and Hamann MT:
Manzamine A reduces androgen receptor transcription and synthesis
by blocking E2F8-DNA interactions and effectively inhibits prostate
tumor growth in mice. Mol Oncol. 18:1966–1979. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Nordstrand A, Bovinder Ylitalo E, Thysell
E, Jernberg E, Crnalic S, Widmark A, Bergh A, Lerner UH and
Wikstrom P: Bone cell activity in clinical prostate cancer bone
metastasis and its inverse relation to tumor cell androgen receptor
activity. Int J Mol Sci. 19:12232018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang J and Chadha JS: Developmental
therapeutics in metastatic prostate cancer: New targets and new
strategies. Cancers (Basel). 16:30982024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Isaacs JT, Brennen WN, Christensen SB and
Denmeade SR: Mipsagargin: The beginning-not the end-of thapsigargin
prodrug-based cancer therapeutics. Molecules. 26:74692021.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ma Z, Shi B, Wei Z, Guo W, Hu L, Li Y, Wu
X, Gong S, Wu D and Wu C: Abstract 3520: EM1031, a novel KLK2 x CD3
bispecific T-cell engager with highly effective efficacy in
preclinical models. Cancer Res. 85(8_Supplement_l): S35202025.
View Article : Google Scholar
|
|
78
|
Shen F, Smith R, McDevitt T, Menard K,
Tian S, Chu G, Chaudhary R, McCann J, Oyer H, Wang SC, et al: Human
kallikrein 2: A novel lineage-specific surface target in prostate
cancer. Clin Cancer Res. 31:4543–4556. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Vietri MT, Della Pepa C, Caliendo G,
Mignano A, Albanese L, Zitiello M, Stilo M and Molinari AM:
Expanding the genomic landscape of HBOC and cancer risk among
mutation carriers. Int J Mol Sci. 26:59282025. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nyberg T, Frost D, Barrowdale D, Evans DG,
Bancroft E, Adlard J, Ahmed M, Barwell J, Brady AF, Brewer C, et
al: Prostate cancer risks for male BRCA1 and BRCA2 mutation
carriers: A prospective cohort study. Eur Urol. 77:24–35. 2020.
View Article : Google Scholar :
|
|
81
|
Yatrano S, Pepe P, Pepe L, Vella N, Alario
C, Chiaranda A, Taranto C, Scillieri R, Mauceri C and Fraggetta F:
BRCA mutations and prostate cancer: should urologist improve daily
clinical practice? Arch Ital Urol Androl. 97:136352025.
|
|
82
|
Fizazi K, Azad AA, Matsubara N, Carles J,
Fay AP, De Giorgi U, Young Joung J, Fong PCC, Voog E, Jones RJ, et
al: Talazoparib plus enzalutamide in men with HRR-deficient
metastatic castration-resistant prostate cancer: Final overall
survival results from the randomised, placebo-controlled, phase 3
TALAPRO-2 trial. Lancet. 406:461–474. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Amini AE, Hunter AE, Almashad A, Feng AJ,
Patel ND, O'Dea MR, McCormick SR, Rodgers LH and Salari K: Magnetic
resonance imaging-based prostate cancer screening in carriers of
pathogenic germline mutations: Interim results from the initial
screening round of the prostate cancer genetic risk evaluation and
screening study. Eur Urol Oncol. 7:1358–1366. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li X and Mao J: Research progress on the
role of lipoxygenase and its inhibitors in prostate cancer. Future
Oncol. 20:3549–3568. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Santos-Pereira M, Pereira SC, Matos B,
Fardilha M, Oliveira PF and Alves MG: Increased susceptibility to
prostate cancer biomarkers in the offspring of male mouse
progenitors with lifelong or early life exposure to high-fat diet.
Eur J Nutr. 64:2122025. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Biyikoglu M, Tanriverdi R, Bozlu M, Send
S, Fidanci SB, Tamer L and Akbay E: Evaluation of homeobox protein
B13 (HOXB13) gene G84E mutation in patients with prostate cancer.
World J Urol. 42:4762024. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Sulaiman KM and Hama Salih RM: Study of
HOXB13 gene variants in prostate cancer patients. Cureus.
16:e725132024.PubMed/NCBI
|
|
88
|
Barashi NS, Li T, Angappulige DH, Zhang B,
O'Gorman H, Nottingham CU, Shetty AS, Ippolito JE, Andriole GL,
Mahajan NP, et al: Symptomatic benign prostatic hyperplasia with
suppressed epigenetic regulator HOXB13 shows a lower incidence of
prostate cancer development. Cancers (Basel). 16:2132024.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
de Angelis M, Siech C, Jannello LMI, Di
Bello F, Penaranda NR, Scilipoti P, Goyal JA, Tian Z, Longo N, de
Cobelli O, et al: Does race/ethnicity affect secondary bladder and
rectal cancer rates after prostate cancer radiation? Results from a
population-based study. J Racial Ethn Health Disparities. Jul
11–2025.Epub ahead of print. View Article : Google Scholar
|
|
90
|
Wei J, Shi Z, Na R, Wang CH, Resurreccion
WK, Zheng SL, Hulick PJ, Cooney KA, Helfand BT, Isaacs WB and Xu J:
Germline HOXB13 G84E mutation carriers and risk to twenty common
types of cancer: Results from the UK Biobank. Br J Cancer.
123:1356–1359. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chartier J, Chansavang A, Jouinot A,
Hamzaoui N, Srikaran A, Moliere D, Huillard O, Thibault C, Tiako M,
Sibony M, et al: Predisposition to prostate cancer and clinical
implications in a real-life cohort. Eur J Hum Genet. 33:1163–1172.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Fraser M, Livingstone J, Wrana JL, Finelli
A, He HH, van der Kwast T, Zlotta AR, Bristow RG and Boutros PC:
Somatic driver mutation prevalence in 1844 prostate cancers
identifies ZNRF3 loss as a predictor of metastatic relapse. Nat
Commun. 12:62482021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Poon DM, Yuan J, Wong OL, Yang B, Chiu ST,
Cheung KY, Chiu G and Yu SK: Stereotactic total ablative
radiotherapy with MR-LINAC for synchronous oligometastatic prostate
cancer. Front Oncol. 15:16076102025. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Garcia de Herreros M, Jimenez N, Padrosa
J, Aversa C, Ferrer-Mileo L, Garcia-Esteve S, Rodriguez-Carunchio
L, Trias I, Fernandez-Manas L, Marin-Aguilera M, et al: Clinical
and transcriptomic characterization of metastatic hormone-sensitive
prostate cancer patients with low PTEN expression. Int J Mol Sci.
26:62442025. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Einspieler H, Ofner H, Ozenil M,
Spielvogel CP, Langrate IK, Hassler MR, Nics L, Bamminger K,
Baltzer PAT, Shariat SF, et al: Does PARP1 up-regulation correlate
with PSMA expression in patients with metastatic
castration-resistant prostate cancer studied with ((18)F)PARPi and
((68)Ga) PSMA PET/CT? Eur J Nucl Med Mol Imaging. July 19–2025.Epub
ahead of print. View Article : Google Scholar
|
|
96
|
Handa N, Li EV, Michael J, Proudfoot JA,
Weiner AB, Alam R, Alshalalfa M, Hao Y, Hakansson A, Zhao X, et al:
Prevalence of potential candidates for targeted therapies according
to treatment-related transcriptomic signatures among 140 548
patients with nonmetastatic prostate cancer. Eur Urol Oncol.
8:1050–1058. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Yin L, Yang F, Wang W, Zhang L, Cao Z, Shi
H, Pan K, Wu L, Xiao H and Xing N: PSMA-Targeted nanoparticles with
PI3K/mTOR dual inhibitor downregulate P-Glycoprotein and inactivate
myeloid-derived suppressor cells for enhanced chemotherapy and
immunotherapy in prostate cancer. Adv Mater. 37:e24153222025.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Miyachi S, Sasaki T, Kato M, Higashi S,
Masui S, Nishikawa K, Fujiwara T, Kitajima T, Hashizume Y, Uchida
K, et al: Comprehensive genomic profiling tests for Japanese
patients with metastatic castration-resistant prostate cancer: A
single-institution experience. Hinyokika Kiyo. 71:171–179. 2025.In
Japanese. PubMed/NCBI
|
|
99
|
Giantini-Larsen A, Ramos AD, Martin D,
Panageas KS, Kostrzewa CE, Abou-Mrad Z, Schmitt A, Bromberg JF,
Safonov A, Rudin CM, et al: Integration of next generation
sequencing data to inform survival prediction of patients with
spine metastasis. Cancers (Basel). 17:22182025. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Kldiashvili E, Abiatari I, Kekelia E,
Iordanishvili S, Metreveli T and Dumbadze E: SOX2, PIWI proteins,
and MALAT1 - plasma-based emerging biomarkers for cancer detection
and monitoring. PLoS One. 20:e03285572025. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Yazaki S, Pei X, Powell S, Khan A, Setton
J and Riaz N: AlphaMissense for identifying pathogenic missense
mutations in DNA damage repair genes in cancer. JCO Precis Oncol.
9:e24009082025. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Hommerding M, Hommerding O, Bernhardt M,
Kreft T, Sanders C, Tischler V, Basitta P, Pelusi N, Wulf AL,
Ohlmann CH, et al: Real-world data on the prevalence of BRCA 1/2
and HRR gene mutations in patients with primary and metastatic
castration resistant prostate cancer. World J Urol. 42:4912024.
View Article : Google Scholar
|
|
103
|
Kwon A and Joung JY: Precision targeting
in metastatic prostate cancer: molecular insights to therapeutic
frontiers. Biomolecules. 15:6252025. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Naisam S, Mohan A, Sivakumar GS,
Gopalakrishnan S and Sreekumar N: Discovering potential therapeutic
agents for lupus nephritis: Insights from in silico research.
Medinformatics. 2:226–240. 2025.
|
|
105
|
Norgaard M, Rusan M, Kondrup K, Sorensen
EMG, Weiss S, Bjerre MT, Fredsoe J, Vang S, Jensen JB, De Laere B,
et al: Deep targeted sequencing of circulating tumor DNA to inform
treatment in patients with metastatic castration-resistant prostate
cancer. J Exp Clin Cancer Res. 44:1202025. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Dhabhai B, Dhakar R, Ranga V, Surolia P,
Menon AM, Sharma NK, Dakal TC and Lohar D: Integrated
bioinformatics approach showed linagliptin as potential drug for
prevention of cardiac arrest and cancer. Medinformatics. 1:131–141.
2023. View Article : Google Scholar
|
|
107
|
Yoon CE, Kang S, Rhew SA, Kwon HJ, Shin D,
Moon HW, Kim MY and Lee JY: Genetic alterations of prostate cancer:
In localized and metastatic prostate cancer. BMC Urol. 25:1662025.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wierbilowicz K, Yang CS, Almaghasilah A,
Wesolowski PA, Pracht P, Dworak NM, Masur J, Wijngaarden S,
Filippov DV, Wales DJ, et al: Parp7 generates an ADP-ribosyl degron
that controls negative feedback of androgen signaling. EMBO J.
44:4720–4744. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Fragkoulis C, Glykas I, Tzelves L,
Stamatakos PV, Papadopoulos G, Stathouros G, Dellis A, Ntoumas K,
Kostopoulou A, Deliveliotis C and Papatsoris A: Clinical impact of
ERG and PTEN status in prostate cancer patients underwent radical
prostatectomy. Arch Ital Urol Androl. 94:390–395. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Lorenzoni M, De Felice D, Beccaceci G, Di
Donato G, Foletto V, Genovesi S, Bertossi A, Cambuli F, Lorenzin F,
Savino A, et al: ETS-related gene (ERG) undermines genome stability
in mouse prostate progenitors via Gsk3β dependent Nkx3.1
degradation. Cancer Lett. 534:2156122022. View Article : Google Scholar
|
|
111
|
Ionescu CA, Cozaru GC, Aschie M, Leopa N,
Cimpineanu B, Voinea F, Matei E, Mitroi A, Deacu M, Iorga I and
Pundiche M: Toward personalized surgery in advanced prostate
cancer: Stratification by PTEN, AR-V7, TP53, TMPRSS2-ERG, and ERBB2
genetic alterations. Chirurgia (Bucur). 120:265–274. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Check MH, Ernst SE and Sfanos KS: Use of
droplet digital PCR for consistent detection of TMPRSS2:ERG gene
fusion transcripts initiated in vitro. Prostate. 85:1282–1289.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Raspin K, O'Malley DE, Marthick JR,
Donovan S, Malley RC, Banks A, Redwig F, Skala M, Dickinson JL and
FitzGerald LM: Analysis of a large prostate cancer family
identifies novel and recurrent gene fusion events providing
evidence for inherited predisposition. Prostate. 82:540–550. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Xie J, Xiang J, Shen Y and Shao S:
Mechanistic insights into the tools for intracellular protein
delivery. Chem Bio Eng. 2:132–155. 2024. View Article : Google Scholar
|
|
115
|
Dematteis A, Miszczyk M, Cormio A,
Matsukawa A, Gontero P and Shariat SF: The role of radiotherapy in
pelvic nodal recurrence following definitive treatment for prostate
cancer. Curr Opin Urol. 35:574–582. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Feng EM, Vo-Phamhi J, Subramanian AN, Dias
M, Foye A, Vinson J, Hong JC, Freedland SJ, Alumkal JJ, Beltran H,
et al: Racial variation in the advanced prostate cancer genome.
Prostate Cancer Prostatic Dis. 28:902–907. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Hadjimichael AC, Foukas AF, Savvidou OD,
Mavrogenis AF, Psyrri AK and Papagelopoulos PJ: The anti-neoplastic
effect of doxycycline in osteosarcoma as a metalloproteinase (MMP)
inhibitor: A systematic review. Clin Sarcoma Res. 10:72020.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Saad F, Armstrong AJ, Shore N, George DJ,
Oya M, Sugimoto M, McKay RR, Hussain M and Clarke NW: Olaparib
monotherapy or in combination with abiraterone for the treatment of
patients with metastatic castration-resistant prostate cancer
(mCRPC) and a BRCA mutation. Target Oncol. 20:445–466. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
McGrath CB, Shreves AH, Shanahan MR, Guard
HE, Nhliziyo MV, Pernar CH, Penney KL, Lotan TL, Fiorentino M,
Mucci LA and Stopsack KH: Etiology of prostate cancer with the
TMPRSS2:ERG fusion: A systematic review of risk factors. Int J
Cancer. 156:1898–1908. 2025. View Article : Google Scholar :
|
|
120
|
Zhang H, Kong L, Li J, Liu Z, Zhao Y, Lv
X, Wu L, Chai L, You H, Jin J, et al: SPOP mutations increase PARP
inhibitor sensitivity via CK2/PIAS1/SPOP axis in prostate cancer.
JCI Insight. 10:e1868712025. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Ding T, Li X, Zhang L, Wei Z, Xiong C,
Wang H, Hao X and Zeng X: Comparison of androgen receptor mutation
detection between plasma extracellular vesicle DNA and cell-free
DNA and its relationship to prostate cancer prognosis. Ann Med.
56:24267702024. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Bugoye FC, Torrorey-Sawe R, Biegon R,
Dharsee N, Mafumiko F, Kibona H, Aboud S, Patel K and Mining S:
Exploring therapeutic applications of PTEN, TMPRSS2:ERG fusion, and
tumour molecular subtypes in prostate cancer management. Front
Oncol. 15:15212042025. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Urabe F and Takemura K: Evaluating the
genetic landscape of prostate cancer: New insights from BRCA1/2,
ATM and CDK12 mutations. BMJ Oncol. 4:e0007172025. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Sekar A, Chatterjee R, Selvadurai BR, Pal
LR, Verma A, Nataraj NB, Garcia DD, Giri S, Genna A, Karatekin F,
et al: TMPRSS2-ERG confers resistance to antiandrogens: Mechanism
and therapeutic implications. bioRxiv [Preprint].
2025.01.17.633582. 2025.
|
|
125
|
Qi L, Li X, Liu Z, Zhang P and Liu L: A
depth-wise separable residual neural network for PCDH8 status
prediction in thyroid cancer pathological images. Intell Oncol.
1:290–298. 2025. View Article : Google Scholar
|
|
126
|
Cheng B, Luo T, Wu Y, Hu J, Yang C, Wu J,
Luo Y, Shangguan W, Li W, Yang L, et al: Urinary exosomal
FAM153C-RPL19 chimeric RNA as a diagnostic and prognostic biomarker
for prostate cancer in Chinese patients. Cancer Lett.
631:2179382025. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Mugoni V, Ciani Y, Nardella C and
Demichelis F: Circulating RNAs in prostate cancer patients. Cancer
Lett. 524:57–69. 2022. View Article : Google Scholar
|
|
128
|
Zvirble M, Vaicekauskaite I, Survila Z,
Bosas P, Dobrovolskiene N, Mlynska A, Sabaliauskaite R and
Pasukoniene V: Liquid-based diagnostic panels for prostate cancer:
The synergistic role of soluble PD-L1, PD-1, and mRNA biomarkers.
Int J Mol Sci. 26:7042025. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Crocetto F, Musone M, Chianese S, Conforti
P, Digitale Selvaggio G, Caputo VF, Falabella R, Del Giudice F,
Giulioni C, Cafarelli A, et al: Blood and urine-based biomarkers in
prostate cancer: Current advances, clinical applications, and
future directions. J Liq Biopsy. 9:1003052025. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Cengiz T, Yurtbay A, Muslu O, Aydin Simsek
S, Ozbalci AB, Coskun HS, Baris YS and Dabak N: Evaluation of the
diagnostic accuracy of percutaneous core needle biopsy in bone and
soft tissue tumors. Acta Chir Orthop Traumatol Cech. 91:376–384.
2024. View Article : Google Scholar
|
|
131
|
Aferin U, Bahtiyar N, Onaran I and Ozkara
H: Are elevated mitochondrial DNA fragments in prostatic
inflammation a potential biomarker for prostate cancer? Arch Ital
Urol Androl. 95:116102023.PubMed/NCBI
|
|
132
|
Valentini V, Santi R, Silvestri V, Saieva
C, Roviello G, Amorosi A, Comperat E, Ottini L and Nesi G: CD44
methylation levels in androgen-deprived prostate cancer: A putative
epigenetic modulator of tumor progression. Int J Mol Sci.
26:25162025. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Su XA, Stopsack KH, Schmidt DR, Ma D, Li
Z, Scheet PA, Penney KL, Lotan TL, Abida W, DeArment EG, et al:
RAD21 promotes oncogenesis and lethal progression of prostate
cancer. Proc Natl Acad Sci USA. 121:e24055431212024. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Montoya Perez I, Jambor I, Pahikkala T,
Airola A, Merisaari H, Saunavaara J, Alinezhad S, Vaananen RM,
Tallgren T, Verho J, et al: Prostate cancer risk stratification in
men with a clinical suspicion of prostate cancer using a unique
biparametric mri and expression of 11 genes in apparently benign
tissue: Evaluation using machine-learning techniques. J Magn Reson
Imaging. 51:1540–1553. 2020. View Article : Google Scholar
|
|
135
|
Gan L, Li W, Chen Q, Cheng L, Zhang F,
Zhong H, Lu Y, Zheng L and Qian B: MicroRNA-145 in urologic tumors:
Biological roles, regulatory networks, and clinical translation.
Front Pharmacol. 16:16096462025. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Rafiq A and Kanavarioti A: The potential
and limitations of the MinION/Yenos Platform for miRNA-Enabled
early cancer detection. Int J Mol Sci. 26:38222025. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Davey M, Benzina S, Savoie M, Breault G,
Ghosh A and Ouellette RG: Affinity vesicles captured urinary
extracellular provide mRNA and miRNA biomarkers for improved
accuracy of prostate cancer detection: A pilot study. Int J Mol
Sci. 21:83302020. View Article : Google Scholar
|
|
138
|
Fuso GA, Bianciardi G, Mei R, Brusa I,
Emiliani S, Fortunati E and Nanni C: Radiotracing the future:
Non-FDG radiotracers nuclear medicine. Semin Nucl Med. 55:648–663.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Cicatiello AG, Musone M, Imperatore S,
Giulioni C, La Rocca R, Cafarelli A, Del Giudice F, Dentice M and
Crocetto F: Circulating miRNAs in genitourinary cancer: Pioneering
advances in early detection and diagnosis. J Liq Biopsy.
8:1002962025. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Tong G, Jing X, Yang Y, Wang X, Lu J, Hu
J, Wang Y, Jumuddin FA, Zhang W and Lv Y: Screening and validation
of key genes involved in castration-resistant prostate cancer based
on transcriptomics sequencing. Sci Rep. 15:256482025. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Muniz M, Childs DS, Andrews J, Mahmoud AM,
Park S, Sartor O, Kase AM, Riaz IB, Stish BJ, Chaudhuri AA, et al:
Deferral of systemic therapy in patients with oligorecurrent
prostate cancer treated with metastasis-directed radiotherapy. Ann
Transl Med. 13:292025. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Kwan EM, Ng SWS, Tolmeijer SH, Emmett L,
Sandhu S, Buteau JP, Iravani A, Joshua AM, Francis RJ, Subhash V,
et al: Lutetium-177-PSMA-617 or cabazitaxel in metastatic prostate
cancer: Circulating tumor DNA analysis of the randomized phase 2
TheraP trial. Nat Med. 31:2722–2736. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Shiota M, Matsubara N, Kato T, Eto M,
Osawa T, Abe T, Shinohara N, Nishimoto K, Yasumizu Y, Tanaka N, et
al: Prediction of undetectable circulating tumor DNA by
comprehensive genomic profiling assay in metastatic prostate
cancer: The SCRUM-Japan MONSTAR SCREEN project. World J Urol.
42:5262024. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Peng N, Chen D, Yang M, Zeng C and Li W:
PARP inhibitors for HRR-deficient metastatic castration-resistant
prostate cancer: Mechanisms and clinical strategies. Eur J Pharm
Sci. 212:1071932025. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Liang A, Gulati S, Huang Q, Dowst H, Lim
A, Zarrin-Khameh N, Godoy G, Noor AB, Castro P, Scheurer ME, et al:
Real-world effectiveness of darolutamide in metastatic
castration-resistant prostate cancer. Endocr Relat Cancer.
32:e2401882025. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Sweeney CJ, Petry R, Xu C, Childress M, He
J, Fabrizio D, Gjoerup O, Morley S, Catlett T, Assaf ZJ, et al:
Circulating Tumor DNA assessment for treatment monitoring adds
value to PSA in metastatic castration-resistant prostate cancer.
Clin Cancer Res. 30:4115–4122. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Dariane C and Timsit MO: DNA-Damage-Repair
gene alterations in genitourinary malignancies. Eur Surg Res.
63:155–164. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Okuda Y, Kato T, Ishizuya Y, Hayashi T,
Yamamoto Y, Hatano K, Kawashima A, Murai J and Nonomura N: PARP
Inhibitors in Genitourinary Cancer: A New Paradigm Beyond Prostate
Cancer. Int J Urol. 32:1091–1101. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Virga A, Urbini M, Polano M, Petracci E,
Tedaldi G, Gurioli G, Marisi G, Angeli D, Ambrosini-Spaltro A, De
Luca G, et al: Mutational and low-coverage whole genome sequencing
identifies actionable DNA repair alterations in prostate cancer
plasma DNA. Sci Rep. 15:212962025. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Slootbeek PHJ, Tolmeijer SH, Mehra N and
Schalken JA: Therapeutic biomarkers in metastatic
castration-resistant prostate cancer: does the state matter? Crit
Rev Clin Lab Sci. 61:178–204. 2024. View Article : Google Scholar
|
|
151
|
Zhang X, Zhou F, Lu T, Zhang S, Wei X, Qiu
X, Xu L, Guo H and Zhuang J: Neoadjuvant darolutamide plus androgen
deprivation therapy for high-risk and locally advanced prostate
cancer: A multicenter, open-label, single-arm, phase II trial.
World J Urol. 43:582025. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Wong J, Tian Y, Patel MS, Avasthi K,
Hanson C, Larsen M, Ampaw E, Fadlullah MZH, Finklestein J, Tan AC,
et al: Plasma cell-free DNA methylation-based prognosis in
metastatic castrate-resistant prostate cancer. Res Sq [Preprint]
rs.3.rs-6331572. 2025.
|
|
153
|
Crumbaker M, Goldstein LD, Murray DH, Tao
J, Pathmanandavel S, Boulter N, Ratnayake L, Joshua AM, Kummerfeld
S and Emmett L: Circulating tumour DNA biomarkers associated with
outcomes in metastatic prostate cancer treated with
Lutetium-177-PSMA-617. Eur Urol Open Sci. 57:30–36. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Fei X, Du X, Gong Y, Liu J, Fan L, Wang J,
Wang Y, Zhu Y, Pan J, Dong B and Xue W: Early plasma circulating
tumor DNA as a potential biomarker of disease recurrence in
non-metastatic prostate cancer. Cancer Res Treat. 55:969–977. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Earland N, Chen K, Semenkovich NP, Chauhan
PS, Zevallos JP and Chaudhuri AA: Emerging roles of circulating
tumor DNA for increased precision and personalization in radiation
oncology. Semin Radiat Oncol. 33:262–278. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Lianidou E: Detection and relevance of
epigenetic markers on ctDNA: Recent advances and future outlook.
Mol Oncol. 15:1683–1700. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Huang YX, Wang HY, Zhang YH, Lin YF, Qiao
XQ and Hu LH: Bibliometric analysis of exosomes in the biomarker
research field. Se Pu. 43:498–507. 2025.PubMed/NCBI
|
|
158
|
Aseervatham J: Dynamic role of exosome
microRNAs in cancer cell signaling and their emerging role as
noninvasive biomarkers. Biology (Basel). 12:7102023.PubMed/NCBI
|
|
159
|
Trujillo B, Wu A, Wetterskog D and Attard
G: Blood-based liquid biopsies for prostate cancer: clinical
opportunities and challenges. Br J Cancer. 127:1394–1402. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Pudova E, Kobelyatskaya A, Katunina I,
Snezhkina A, Nyushko K, Fedorova M, Pavlov V, Bulavkina E, Dalina
A, Tkachev S, et al: Docetaxel resistance in castration-resistant
prostate cancer: Transcriptomic determinants and the effect of
inhibiting Wnt/β-catenin signaling by XAV939. Int J Mol Sci.
23:128372022. View Article : Google Scholar
|
|
161
|
Chen Z, Fu S, Shan Y, He Z, Gu J, Wu H,
Lin J, Huang Y, Wang H, Lu Y and Ding M: Circ_0001047 inhibits
prostate cancer progression and enhances abiraterone sensitivity
via miR-122-5p/FKBP5/PHLPPl/AKT axis in vitro. Discov Oncol.
15:5692024. View Article : Google Scholar
|
|
162
|
Song X, Li T, Zhou W, Feng C, Zhou Z, Chen
Y, Li D, Chen L, Zhao J, Zhang Y and Han B: CAF-derived exosomal
miR-196b-5p after androgen deprivation therapy promotes
epithelial-mesenchymal transition in prostate cancer cells through
HOXC8/NF-KB signaling pathway. Biol Direct. 20:802025. View Article : Google Scholar
|
|
163
|
Pecoraro G, Leone I, Nuzzo S, Negueruela
S, Smaldone G and Buono L: Co-modulation of a circular form of
PCDH11Y during neuroendocrine differentiation of prostate cancer.
Front Oncol. 15:15024052025. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Julio MK: Elevating prostate cancer
diagnostics through extracellular vesicle miRNAs. Gene. 1496622025.
View Article : Google Scholar
|
|
165
|
Qin X, Niu R, Tan Y, Huang Y, Ren W, Zhou
W, Wu H, Zhang J, Xu M, Zhou X, et al: Exosomal PSM-E inhibits
macrophage M2 polarization to suppress prostate cancer metastasis
through the RACK1 signaling axis. Biomark Res. 12:1382024.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Zhang J, Chen B, Gan C, Sun H, Zhang J and
Feng L: A comprehensive review of small interfering RNAs (siRNAs):
Mechanism, therapeutic targets, and delivery strategies for cancer
therapy. Int J Nanomedicine. 18:7605–7635. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Kren BT, Henzler CM, Ahmed K and Trembley
JH: Impact of protein kinase CK2 downregulation and inhibition on
oncomir clusters 17 ~ 92 and 106b ~ 25 in prostate, breast, and
head and neck cancers. Mol Med. 30:1752024. View Article : Google Scholar
|
|
168
|
Shukla KK, Choudhary GR, Sankanagoudar S,
Misra S, Vishnoi JR, Pareek P, Pilla KK, Pandey SN and Sharma P:
Deregulation of miR-10b and miR-21 correlate with cancer stem cells
expansion through the apoptotic pathway in prostate cancer. Asian
Pac J Cancer Prev. 24:2105–2119. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Qiu J, Wu J, Zhou N, Lai C, Huang X, Liu
C, Yuan X and Xu K: An EcDNA gene-based risk model and functional
verification of a key ec-lncRNA AC016394.2 for prostate cancer.
Cancer Cell Int. 25:2402025. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Gupta P, Dutta S, Kumar P, Kaushik M,
Ashique S and Bhowmick M: Targeted management: Unlocking the
crucial role of PROTACs in cancer treatment. Curr Drug Discov
Technol. Feb 26–2025.Epub ahead of print. View Article : Google Scholar
|
|
171
|
Miao G, Shang Z and Wang X, Zhang J, Xu M,
He P, Zhong Q, Zhao X, Tan G and Wang X: Concurrent inhibition of
tumor growth and metastasis by a lipidated nanophotosensitizer
tracing and disabling tumor extracellular vesicles. Nat Cancer.
6:1438–1457. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Wang C, Feng Y, Rong X, Yan J, Lv B, Jiang
H, Duan L and Jiang J: Mesenchymal stromal cell exosomes for drug
delivery of prostate cancer treatments: A review. Stem Cell Res
Ther. 16:182025. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Bellomo G, Marcocci F, Bianchini D,
Mezzenga E, D'Errico V, Menghi E, Zannoli R and Sarnelli A: MR
spectroscopy in prostate cancer: New algorithms to optimize
metabolite quantification. PLoS One. 11:e01657302016. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Gungoren M, Koyuncu I, Kilinc E and Baysal
Z: Docetaxel-Loaded, Dipeptide-Functionalized
Y-Fe(2)O(3) magnetic nanoparticles: Synthesis,
characterization, and targeting to prostate cancer. Chem Biodivers.
22:e2024032452025. View Article : Google Scholar
|
|
175
|
Galey L, Olanrewaju A, Nabi H, Paquette
JS, Pouliot F and Audet-Walsh E: PSA, an outdated biomarker for
prostate cancer: In search of a more specific biomarker, citrate
takes the spotlight. J Steroid Biochem Mol Biol. 243:1065882024.
View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Jupin M, van Heijster FHA and Heerschap A:
Metabolite interactions in prostatic fluid mimics assessed by (1)H
NMR. MAGMA. 35:683–694. 2022. View Article : Google Scholar
|
|
177
|
Ma T, Xie W, Xu Z, Gao W, Zhou J, Wang Y
and Chan FL: Estrogen-related receptor alpha (ERRalpha) controls
the sternness and cellular energetics of prostate cancer cells via
its direct regulation of citrate metabolism and zinc
transportation. Cell Death Dis. 16:1542025. View Article : Google Scholar
|
|
178
|
Feng D, Zhang J, Niu H, Zheng X, Jia M, Lu
Q, Wang J, Guo W, Sun Q, Yuan H and Lou H: Spermidine inactivates
proteasome activity and enhances ferroptosis in prostate cancer.
Acta Pharm Sin B. 15:2095–2113. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Li QZ, Zuo ZW, Zhou ZR and Ji Y: Poly
amine homeostasis-based strategies for cancer: The role of
combination regimens. Eur J Pharmacol. 910:1744562021. View Article : Google Scholar
|
|
180
|
Tamura R, Chen J, De Jaeger M, Morris JF,
Scott DA, Vangheluwe P and Looger LL: Genetically encoded
fluorescent sensors for visualizing polyamine levels uptake, and
distribution. bioRxiv [Preprint]. 2024.08.21.609037. 2024.
|
|
181
|
Hunt RL, Oh J, Jain A, Kuo TH, Berardi D,
Jian W, Song D, Wu Q, Goodman AL, Palm NW, et al: Discovery of a
widespread polyamine-low-molecular-weight thiol hybrid pathway in
clostridioides difficile. ACS Infect Dis. 11:2246–2264. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Zong Y, Li H, Liao P, Chen L, Pan Y, Zheng
Y, Zhang C, Liu D, Zheng M and Gao J: Mitochondrial dysfunction:
Mechanisms and advances in therapy. Signal Transduct Target Ther.
9:1242024. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Kumar R, Jonnatan S, Sanin DE, Vakkala V,
Kadam A, Kumar S, Dalrymple SL, Zhao L, Foley J, Holbert CE, et al:
Androgen receptor drives polyamine synthesis creating a
vulnerability for prostate cancer. medRxiv [Preprint].
2024.12.12.24318845. 2024.PubMed/NCBI
|
|
184
|
Danilova EY, Maslova AO, Stavrianidi AN,
Nosyrev AE, Maltseva LD and Morozova OL: CKD urine metabolomics:
Modern concepts and approaches. Pathophysiology. 30:443–466. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Zhu D, Dai J, Jia J, Kanagaraj T,
Rajalakshmi K, Muthusamy S, Geng L and Yuan G: Biogenic synthesis
of N-doped carbon dots from S. cumini seeds for prostate cancer
biomarker citrate detection, its live cancer cell imaging.
Spectrochim Acta A Mol Biomol Spectrosc. 329:1255682025. View Article : Google Scholar
|
|
186
|
Jimenez Gutierrez GE, Borbolla Jimenez FV,
Munoz LG, Tapia Guerrero YS, Murillo Melo NM, Cristobal-Luna JM,
Leyva Garcia N, Cordero-Martinez J and Magana JJ: The molecular
role of polyamines in age-related diseases: An update. Int J Mol
Sci. 24:164692023. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Lista S, Gonzalez-Dominguez R, Lopez-Ortiz
S, Gonzalez-Dominguez A, Menendez H, Martin-Hernandez J, Lucia A,
Emanuele E, Centonze D, Imbimbo BP, et al: Integrative metabolomics
science in Alzheimer's disease: Relevance and future perspectives.
Ageing Res Rev. 89:1019872023. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Sena LA: Polyamine metabolism in prostate
cancer. Curr Opin Oncol. 37:223–232. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Hu P, Yuan M, Guo B, Lin J, Yan S, Huang
H, Chen JL, Wang S and Ma Y: Citric acid promotes immune function
by modulating the intestinal barrier. Int J Mol Sci. 25:12392024.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Wu WF, Song XY, Warner M, Imamov O,
Antonson P and Gustafsson JA: The role of estrogen receptor beta in
maintaining basal cells and modulating the immune environment in
the prostate. Proc Natl Acad Sci USA. 122:e25057971222025.
View Article : Google Scholar
|
|
191
|
Ramatlho P, Rugemalila MM, Tawe L, Pain D,
Choga OT, Ndlovu AK, Koobotse MO, Lal P, Rebbeck TR, Paganotti GM,
et al: The role of immunohistochemistry for AMACR/p504s and p63 in
distinguishing prostate cancer from benign prostate tissue samples
in botswana. Res Rep Urol. 17:1–10. 2025.PubMed/NCBI
|
|
192
|
Spieker AJ, Gordetsky JB, Maris AS, Dehan
LM, Denney JE, Arnold Egloff SA, Scarpato K, Barocas DA and
Giannico GA: PTEN expression and morphological patterns in
prostatic adenocarcinoma. Histopathology. 79:1061–1071. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Zhang HZ, Wang SY, Guo YN and Zhao M:
Isolated intraductal carcinoma of the prostate: A
clinicopathological and molecular analysis. Zhonghua Bing Li Xue Za
Zhi. 53:803–808. 2024.In Chinese. PubMed/NCBI
|
|
194
|
Martini C, Logan JM, Sorvina A,
Prabhakaran S, Ung BSY, Johnson IRD, Hickey SM, Brooks RD; kConFab
Consortium; Caruso MC, et al: Distinct patterns of biomarker
expression for atypical intraductal proliferations in prostate
cancer. Virchows Arch. 485:723–728. 2024. View Article : Google Scholar :
|
|
195
|
Akhoundova D, Fischer S, Triscott J,
Lehner M, Thienger P, Maletti S, Jacquet M, Lubis DSH, Bubendorf L,
Jochum W and Rubin MA: Rare histologic transformation of a CTNNB1
(β-catenin) mutated prostate cancer with aggressive clinical
course. Diagn Pathol. 19:832024. View Article : Google Scholar
|
|
196
|
Ayala Soriano C, Benitez Barzaga M, Chhina
A, Jain M and Nava VE: Hepatoid prostatic carcinoma with adrenal
metastasis and novel genetic alterations. Diagn Cytopathol.
50:E310–E314. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Ruschoff JH, Stratton S, Roberts E, Clark
S, Sebastiao N, Fankhauser CD, Eberli D, Moch H, Wild PJ and Rupp
NJ: A novel 5x multiplex immunohistochemical staining reveals PSMA
as a helpful marker in prostate cancer with low p504s expression.
Pathol Res Pract. 228:1536672021. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Zhou L, Xu J, Wang S, Yang X, Li C, Zhou
J, Zhang P, Xu H and Wang C: Papillary renal neoplasm with reverse
polarity: A clinicopathologic study of 7 cases. Int J Surg Pathol.
28:728–734. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Nolan E, Lindeman GJ and Visvader JE:
Deciphering breast cancer: From biology to the clinic. Cell.
186:1708–1728. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Chenlo M, Aliyev E, Rodrigues JS,
Vieiro-Balo P, Blanco Freire MN, Cameselle-Teijeiro JM and Alvarez
CV: Sequential Colocalization of ERa, PR, and AR hormone receptors
using confocal microscopy enables new insights into normal breast
and prostate tissue and cancers. Cancers (Basel). 12:35912020.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Comperat E, Renard-Penna R, Ondet V,
Cancel-Tassin G and Cussenot O: Diagnosis of prostate cancer in one
day: The benefits of cytology in tumour detection. Cytopathology.
32:211–216. 2021. View Article : Google Scholar
|
|
202
|
Bee J, Zhou X, Tipirneni Y, Chen S, Qi J,
Iczkowski KA, Dall'Era M and Marcu L: Rapid discrimination of
high-grade prostate cancer using label-free fluorescence lifetime
measurements. medRxiv [Preprint]. 2025.05.23.25328257. 2025.
|
|
203
|
Riddi VL, Harahap EU, Sitinjak D and
Nirmawati R: Ductal adenocarcinoma of the prostate presenting with
bladder invasion and bone metastasis: A case report. Cureus.
17:e860002025.PubMed/NCBI
|
|
204
|
Salvi M, Manini C, Lopez JI, Fenoglio D
and Molinari F: Deep learning approach for accurate prostate cancer
identification and stratification using combined immunostaining of
cytokeratin, p63, and racemase. Comput Med Imaging Graph.
109:1022882023. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Bolis M, Bossi D, Vallerga A, Ceserani V,
Cavalli M, Impellizzieri D, Di Rito L, Zoni E, Mosole S, Elia AR,
et al: Dynamic prostate cancer transcriptome analysis delineates
the trajectory to disease progression. Nat Commun. 12:70332021.
View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Conte MI, Fuentes-Trillo A and Dominguez
Conde C: Opportunities and tradeoffs in single-cell transcriptomic
technologies. Trends Genet. 40:83–93. 2024. View Article : Google Scholar
|
|
207
|
Zhou L, Zhang C, Zhang Y and Shi C:
Application of organoid models in prostate cancer research. Front
Oncol. 11:7364312021. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Mharrach I, Aqerrout M, Tadlaoui KA,
Laraqui A, Ghazzaly AE and Ennaji MM: Methylation status of GSTP1
and APC promoter genes as potential biomarkers for early detection
and tumor aggressiveness in prostate cancer: Insights from a
Moroccan cohort. Mol Biol Rep. 52:7112025. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Aloke C, Onisuru OO and Achilonu I:
Glutathione S-transferase: A versatile and dynamic enzyme. Biochem
Biophys Res Commun. 734:1507742024. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Gupta H, Inoue H, Nakai Y, Nakayama M,
Jones T, Hicks JL, Kumar B, Gurel M, Nelson WG, De Marzo AM and
Yegnasubramanian S: Progressive spreading of DNA methylation in the
GSTP1 promoter CpG island across transitions from precursors to
invasive prostate cancer. Cancer Prev Res (Phila). 16:449–460.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Ji R, Zhang L, Xing Y, Zhu Y, Mao K, Wang
M, Xuan K, Yang Y, Lo R, Luo B and Lu Z: Clinical significance of
urine-based automated detection of GSTP1 methylation for the
diagnosis of suspected prostate cancer patients. Transl Androl
Urol. 14:1204–1213. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Cussenot O, Timms KM, Perrot E, Blanchet
P, Brureau L, Solimeno C, Fromont G, Comperat E and Cancel-Tassin
G: Tumour-based mutational profiles predict visceral metastasis
outcome and early death in prostate cancer patients. Eur Urol
Oncol. 7:597–604. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Turan G, Turan M, Basbinar Y and Ellidokuz
H: Prostate cancer in Turkiye: Trend analysis of incidence and
mortality rates. Semin Oncol. 52:1523912025. View Article : Google Scholar
|
|
214
|
Lam D, Clark S, Stirzaker C and Pidsley R:
Advances in prognostic methylation biomarkers for prostate cancer.
Cancers (Basel). 12:29932020. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Eismann L, von Walter P, Jung A, Chaloupka
M, Rodler S, Westhofen T, Buchner A, Stief CG, Stadler T and
Schlenker B: Methylation status of various gene loci in localized
prostate cancer: Novel biomarkers for diagnostics and biochemical
recurrence. Urol Oncol. 41:325 el–325 e8. 2023. View Article : Google Scholar
|
|
216
|
Deng Y, Cai B and Wang X: A fully
automated quantitative analysis method based on deep learning
algorithms for immunohistochemical staining expression intensities.
Intell Oncol. 1:256–264. 2025. View Article : Google Scholar
|
|
217
|
Friedemann M, Horn F, Gutewort K, Tautz L,
Jandeck C, Bechmann N, Sukocheva O, Wirth MP, Fuessel S and
Menschikowski M: Increased sensitivity of detection of RASSF1A and
GSTP1 DNA fragments in serum of prostate cancer patients:
Optimisation of diagnostics using OBBPA-ddPCR. Cancers (Basel).
13:44592021. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Tufail M: PTEN-mediated resistance in
cancer: From foundation to future therapies. Toxicol Rep.
14:1019872025. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Bucker L and Lehmann U: CDH1 (E-cadherin)
gene methylation in human breast cancer: Critical appraisal of a
long and twisted story. Cancers (Basel). 14:43772022. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Corso G, Magnoni F, Dal Molin M, Marino E,
Nicosia L, Pesapane F, Noonan DM and Albini A: Second-hit CDH1 gene
mechanisms in hereditary diffuse gastric and lobular breast cancer
syndrome: Frequency and impact on tumorigenesis. Hum Mol Genet.
34:1345–1352. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Zhang W, Zhang K, Ma Y, Song Y, Qi T,
Xiong G, Zhang Y, Kan C, Zhang J, Han F and Sun X: Secreted
frizzled-related proteins: A promising therapeutic target for
cancer therapy through Wnt signaling inhibition. Biomed
Pharmacother. 166:1153442023. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Fiu C, Tang H, Hu N and Fi T: Methylomics
and cancer: The current state of methylation profiling and marker
development for clinical care. Cancer Cell Int. 23:2422023.
View Article : Google Scholar
|
|
223
|
Zhou Y, Tao L, Qiu J, Xu J, Yang X, Zhang
Y, Tian X, Guan X, Cen X and Zhao Y: Tumor biomarkers for
diagnosis, prognosis and targeted therapy. Signal Transduct Target
Ther. 9:1322024. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Ibrahim J, Peeters M, Van Camp G and Op de
Beeck K: Methylation biomarkers for early cancer detection and
diagnosis: Current and future perspectives. Eur J Cancer.
178:91–113. 2023. View Article : Google Scholar
|
|
225
|
Bao Y, Wang X, Zeng B, Shi Y, Huang Y,
Huang Y, Shang S, Shan F and Ma F: Research progress of liquid
biopsy based on DNA methylation in tumor diagnosis and Treatment.
Biomolecules. 14:16342024. View Article : Google Scholar
|
|
226
|
Wan JCM, Sasieni P and Rosenfeld N:
Promises and pitfalls of multi-cancer early detection using liquid
biopsy tests. Nat Rev Clin Oncol. 22:566–580. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Fi X and Fi X, Wang F, Hou Y, Fiu Y, Mao
J, Zhang F and Fi X: Advancing Traditional Chinese medicine
research through network pharmacology: Strategies for target
identification, mechanism elucidation and innovative therapeutic
applications. Am J Chin Med. 53:2021–2042. 2025. View Article : Google Scholar
|
|
228
|
Maliye R, Babu S, Harris AE, Patke R,
Ntekim AI, Rutland CS, James V, Mongan NP, Madhusudan S, Jeyapalan
JN and Semenova MM: Evaluation of human papillomavirus as a risk
factor in prostate cancer pathogenesis. Discov Oncol. 16:13752025.
View Article : Google Scholar : PubMed/NCBI
|