Introduction
Circular RNAs (circRNAs) are single-stranded,
covalently closed RNA molecules that are produced from pre-mRNAs
through a process termed backsplicing and were initially proposed
to be splicing-associated noise (1). However, it has since been shown that
circRNAs possess a range of biological functions (2). Previous reports on malignant
diseases have demonstrated that circRNAs are differentially
expressed in different histologic types and stage-specific
variations in their expression have also been observed (3,4).
circRNAs modulate a range of cellular functions, including invasive
potential, stemness, chemosensitivity and proliferative capacity
(5-7). A unique biological function of
circRNAs is their sponge-like adsorption of miRNAs to regulate
translation, transmit intercellular information and regulate
protein-bound functions (8,9),
highlighting them as a unique class of functional RNAs that possess
significant potential as gene regulators in humans (10). In this context, several reports
indicate the potential roles of circRNAs in altering the
chemosensitivity of various malignancies by modulating miRNAs
(7,11). Peng et al (7) reported that circCUL2 regulated
cisplatin sensitivity through miR-142-3p/ROCK2-mediated autophagy
activation in gastric cancer. Yu et al (12) revealed that circ_0092367 inhibited
epithelial-mesenchymal transition and improved gemcitabine
sensitivity in pancreatic cancer (PC) by regulating the
miR-1206/ESRP1 axis. Additionally, a single circRNA can harbor
multiple miRNA binding sites (13,14). Yamada et al (13) reported an analysis of the
regulatory network of circ_0004365, which bound to 33 miRNAs to
regulate the expression of 187 mRNAs. Another characteristic
biological feature of circRNAs is their enrichment and stability in
serum exosomes owing to their circular structure (15). Given this unique feature, circRNAs
have been reported as potential liquid biomarkers for early
diagnosis and prognostic and chemosensitivity prediction in various
malignant diseases (16-18).
Gemcitabine is a key drug widely used in PC in
various clinical situations, including as palliative systemic
therapy for unresectable disease and as perioperative chemotherapy
for resectable/borderline resectable disease (19,20). Although gemcitabine can markedly
improve outcomes in patients with PC, its efficacy is modest, with
a response rate of only 24% (21). To overcome these obstacles to the
clinical application of gemcitabine, improving gemcitabine
sensitivity and establishing an efficient method for predicting
sensitivity prior to use are crucial for developing
improved-tailored treatment strategies for patients with PC to
improve treatment outcomes. Given the biological characteristics of
circRNAs (including regulation of a range of cellular functions and
their stability in serum), they are promising targets for
investigation to address the aforementioned clinical issues in PC
treatment. Indeed, several circRNAs have been reported to be
associated with gemcitabine sensitivity in patients with PC
(12,22,23). However, only limited information
is available regarding the biological mechanisms underlying their
roles in PC, particularly in relation to gemcitabine sensitivity
and no reports have investigated their potential as a liquid
biomarker in the treatment of PC. Therefore, the aim of the present
study was to identify the circRNAs associated with gemcitabine
sensitivity in PC, elucidate their mechanisms and further
investigate whether candidate circRNAs may be useful as a liquid
biomarker in predicting gemcitabine sensitivity. Specifically,
beyond elucidating the molecular mechanisms of circRNAs in
gemcitabine sensitivity in vitro, these findings were
translated into a clinical context. The potential of these in
vitro-identified circRNAs was evaluated as liquid biomarkers to
predict the therapeutic response to gemcitabine-based neoadjuvant
therapy using a cohort of patients who had undergone
gemcitabine-based treatment.
Materials and methods
Cell culture
In the present study, three human PC cell lines:
MIAPaCa-2 (RRID: CVCL_0428) purchased from the Japan Cancer
Research Resources Bank, Panc-1 (RRID: CVCL_0480) purchased from
the American Type Culture Collection and PSN-1 (RRID: CVCL_1644)
obtained from the European Collection of Authenticated Cell
Culture. All cell lines were authenticated using STR profiling
within the last 3 years and routinely tested for mycoplasma
contamination. The cells were cultured at 37°C in 5% CO2
atmosphere with at least 95% humidity in DMEM supplemented with 10%
FBS and 100 U/ml penicillin and streptomycin each (Thermo Fisher
Scientific, Inc.).
Establishment of gemcitabine-resistant
cell lines
Gemcitabine-resistant (GR) cell lines were generated
by exposure to gradually increasing concentrations of the drug for
2 months as previously described (24). Parental MIAPaCa-2 cells were
exposed to gemcitabine at an initial concentration of 1 ng/ml. When
the cells adapted to the drug, the gemcitabine concentration was
increased incrementally, with a final concentration of 20 ng/ml for
MIAPaCa-2. Through this process, GR cells were established
(25). A total of three stable GR
cell lines were established from MIAPaCa-2, termed GR3, GR8 and
GR10. The GR cell lines showed ~100% cell viability even at
gemcitabine concentrations of 200 ng/ml (Fig. S1). Gemcitabine was purchased from
Eli Lilly (Eli Lilly and Company).
Reverse transcription-quantitative (RT-q)
PCR
RT-qPCR was performed as previously described
(26). Briefly, total RNA from
cultured cells was extracted using QIAzol® reagent and a
miRNeasy mini kit (Qiagen GmbH), while total RNA from serum was
extracted using a miRNeasy Serum/Plasma Advanced kit (Qiagen GmbH)
and reverse transcribed into cDNA using a Reverse Transcription
System kit, according to the manufacturer's protocol (Promega
Corporation). qPCR was performed using THUNDERBIRD SYBR qPCR Mix
(Toyobo Co., Ltd.) on a QuantiStudio7 amplifier (Thermo Fisher
Scientific, Inc.). PCR cycling conditions were as follows: An
initial holding stage at 94°C for 60 sec, followed by 40 cycles of
denaturation at 95°C for 15 sec and annealing/extension at 62°C for
60 sec. The 2−ΔΔCq method was used to analyze the
relative levels of target genes (27). All primer sequences are listed in
Table SI. All experiments were
performed in triplicate.
circ72309 overexpression
For circ72309 overexpression, the identified 580-bp
sequence was synthesized as an artificial oligonucleotide and
subcloned into pRP[Exp]-EGFP Laccase2 MCS Exon Vector
(VectorBuilder Inc.), flanked by a GT-AG splice-site. Constructs
were transfected into cells using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Confirmation of circ72309 transfection was
assessed using RT-qPCR 48 h after transfection. The GR cell lines
overexpressing circ72309 were termed GR_circ72309 overexpressing
(OE). Cells transfected with pRP[Exp]-EGFP/Neo-CMV>ORF_Stuffer
(vector ID: VB900145-9829wty) served as the negative control and
were termed GR_NC.
RNase R digestion
For RNase R digestion assay, total RNA was treated
with or without 5 U/µg RNase R (AR Brown Co., Ltd.) and
incubated at 37°C for 2 h. After purifying RNA using an RNA
Clean-up XS kit (Takara Bio, Inc.), RT-qPCR was performed.
circRNA sequencing and bioinformatics
analysis
circRNA sequencing was performed as previously
described (13). Briefly, raw
sequencing reads underwent quality assessment using FastQC (version
0.11.9; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/,
RRID:SCR_014583), followed by adaptor trimming and removal of
low-quality reads using fastp (version 0.19.5; https://github.com/OpenGene/fastp). Cleaned reads
were aligned to the human reference genome (GRCh38; https://www.gencodegenes.org/human/) with STAR
(version 2.7.10a; https://github.com/alexdobin/STAR, RRID:SCR_004463),
and circRNAs were identified using CIRCexplorer2 (version 2.4.0;
https://circexplorer2.readthedocs.io/) and
circRNA_finder (https://github.com/orzechoj/circRNA_finder).
Expression quantification and normalization were performed using
edgeR (version 3.42.4; https://bioconductor.org/packages/edgeR/,
RRID:SCR_012802) prior to differential expression analysis. To
compare differences in the circRNA expression profiles between
MIAPaCa-2 and GR3 cells, the fold change was calculated between the
groups for each circRNA. Student's t-test was used to determine
statistical significance and a significant difference in circRNA
levels was defined as a false discovery rate (FDR)<0.05.
mRNA sequencing and bioinformatics
analysis
To compare differences in the mRNA expression
profiles between GR3_ NC and GR3_circ72309 OE cells, mRNA
sequencing was performed 48 h after transfection. All sequencing
libraries were then sequenced at single-end 100 bp on an Illumina
NovaSeq 6000 (Illumina, Inc.) at the Research Institute for
Microbial Diseases of the University of Osaka (Osaka, Japan). Raw
gene expression was quantified across all gene exons using the
top-expressed isoform as a proxy for gene expression and
differential gene expression analyses were performed using edgeR
(version 3.42.4; http://bioconductor.org/packages/edgeR/, RRID:
SCR_012802) using replicates to compute within-group dispersion.
Undetectable mRNAs were excluded from the analysis. Differentially
expressed genes (DEGs) were defined as having an FDR<0.05 and a
log2 fold change >1.0. Gene Set Enrichment Analysis
(GSEA) (RRID: SCR_003199) was performed using clusterProfiler
(version 4.8.3; https://bioconductor.org/packages/clusterProfiler/,
RRID: SCR_016884). The sequencing coverage and quality statistics
for each sample are summarized in Table SII.
MTT assay
To assess the drug response of PC cell lines, 3,000
treated cells were seeded per well in 96-well plates. The following
day, fresh medium containing gemcitabine at concentrations of 0,
1.5625, 3.125, 6.25, 12.5, 25, 50, 100 or 200 ng/ml was added to
the cells and incubated for 72 h. The 12 mM MTT (~10 µl)
solution was added to each well and incubated at 37°C for 4 h.
Subsequently, 100 µl SDS-HCl solution was added to each well
and agitated for 1 h at room temperature on a shaker; subsequently,
the absorbance at 570 nm was measured using a Model 680 Microplate
Reader (Bio-Rad Laboratories, Inc.) and the IC50 was
calculated. Dose-response curves were analyzed using non-linear
regression using a four-parameter logistic (4PL) sigmoidal model
[log(concentration) vs. normalized cell viability]. IC50
values were derived from the fitted curves and goodness of fit was
assessed by the coefficient of determination (R2)
(28). The fitted curves are
presented as solid lines. For resistant clones that did not display
a typical sigmoidal decline, median values are shown using dashed
lines.
Apoptosis assay
The Annexin V-FITC Apoptosis Detection Kit
(BioVision, Inc.; Abcam) was used to assess apoptosis according to
the manufacturer's protocol. The cells were digested with trypsin
and washed with PBS twice. The cell count was adjusted to
1×105; subsequently, the cells were stained with
propidium iodide and Annexin V-FITC according to the manufacturer's
protocol. Analysis was performed using a flow cytometer (BD
Biosciences) and the data were measured using FlowJo version 10
(RRID: SCR_008520; BD Biosciences). For each sample, at least
10,000 events were acquired within the live cell gate. All
experiments were independently repeated three times (29).
Western blotting
Total protein was extracted from short interfering
(si)RNA knockdown and overexpression cells using RIPA lysis buffer
containing protease inhibitors (MilliporeSigma). Total protein
concentration was determined using a Bradford protein assay kit
(Bio-Rad Laboratories, Inc.). The samples were heated at 95°C for 5
min in loading buffer, loaded into wells of 15% acrylamide gels
(Bio-Rad Laboratories, Inc.) along with a protein ladder and
resolved by SDS-PAGE at 200V. Resolved proteins were subsequently
transferred onto PVDF membranes (Bio-Rad Laboratories, Inc.) for 1
h and blocked using Blocking One (Nacalai Tesque, Inc.) at room
temperature for 30 min. The blots were treated with an
anti-equilibrative nucleoside transporter-1 (ENT1) polyclonal
rabbit antibody (ProteinTech Group, Inc.; 1:600; cat. no.
29862-1-AP; RRID: AB_2935484) and an anti-cytidine deaminase (CDA)
polyclonal rabbit antibody (Invitrogen; Thermo Fisher Scientific,
Inc.; 1:5,000; cat. no. PA5-84630; RRID: AB_2791781) in Can Get
Signal® solution 1 (Toyobo Co., Ltd.) overnight at 4°C
on a shaker, while an anti-βactin polyclonal rabbit antibody
(Abcam; 1:1,000; cat. no. 1784-1; RRID: AB_598136) was used as the
loading control. After washing three times, horseradish
peroxidase-conjugated anti-rabbit IgG antibody (Cytiva; cat. no.
NA934, RRID: AB_772206) and HRP-conjugated anti-human sheep IgG
whole antibody (Cytiva cat. no. NA933; RRID: AB_772208) were used
as secondary antibodies. Signals were visualized using an ECL Prime
Western Blotting Detection kit (Cytiva) and membranes were scanned
using a ChemiDoc™ Touch MP (Bio-Rad Laboratories, Inc.) (30). Band intensities were quantified
using ImageJ (version 1.54g; https://imagej.nih.gov/ij/, National Institutes of
Health) using a lane-based densitometric method (31). The values were normalized to the
corresponding β-actin loading control. Quantitative results were
presented as the mean ± SE from at least three independent
experiments.
Reactive oxygen species (ROS)
quantification
A ROS Assay Kit-Highly Sensitive
(2′,7′-Dichlorodihydrofluorescein diacetate) containing DCFH-DA
(Dojindo Molecular Technologies, Inc.) was used to calculate the
ROS activity according to the manufacturer's protocol. Briefly,
~1×105 cells were treated with DCFH-DA as indicated,
incubated for 30 min in PBS, trypsinized and analyzed by flow
cytometry. For each sample, at least 15,000 events were acquired
within the live cell gate and the mean fluorescence intensity (MFI)
of DCF was calculated using FlowJo (version 10; BD Biosciences).
Each experiment was performed in triplicate (n=3 independent
biological replicates). N-acetylcysteine (NAC; 5 mmol/l) was used
as an ROS scavenger. To inhibit ROS, PC cell lines were treated for
36 h with NAC. PBS-treated cells served as the control.
Knockdown of circ72309
circ72309 siRNA targeting the sequence of the
backsplicing junction of circ72309 was synthesized. Silencer™;
Select Negative Control #1 siRNA (Invitrogen; Thermo Fisher
Scientific, Inc.; cat. no. 4390843) was used as the negative
control. This reagent is proprietary and the manufacturer does not
disclose its nucleotide sequence; only product identification and
catalog information are publicly available. MIAPaCa-2, PANC1 and
PSN-1 were transfected with circ72309 siRNAs using
Lipofectamine® RNAiMAX transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. A total of 2×105 cells/well were seeded in a
6-well plate and 50 nM siRNA was added to each well with the
transfection reagent and cultured for 24 h at 37°C in 5%
CO2. The sequence of the circ72309 siRNA is listed in
Table SI. Successful knockdown
of circ72309 by siRNA transfection was confirmed (Fig. S2).
Identification of target miRNAs for
circ72309
Circular RNA Interactome (RRID: SCR_016304)
(32) was used to identify target
miRNAs for circ72309 (Table
SIII) and among these miRNAs, TargetScan (RRID: SCR_010845,
targetscan. org/vert_80/) was used to explore the candidate mRNAs
regulating gemcitabine metabolism targeted by relevant miRNAs
(Table SIV). Expression of these
target miRNAs was compared using RT-qPCR between GR_NC cells and
GR_circ72309 OE cells in all three GR cell lines to confirm the
effect of circ72309 on the expression of these miRNAs.
Clinical effect of circ72309 expression
in patients with PC
Data were retrospectively collected from patients
with PC aged ≥20, excluding those with intrapapillary mucinous
carcinoma, between January 2016 and December 2023. A total of 31
patients with resectable status according to the definition of the
National Comprehensive Cancer Network Guidelines (Pancreatic
Adenocarcinoma, version 1.2023) (33), who received gemcitabine plus S-1
for neoadjuvant chemotherapy (excluding chemoradiotherapy), were
recruited. The median age was 73 (50-84) years, and the sex ratio
was 16 males (53%) and 14 females (47%). Borderline resectable and
unresectable cases were excluded as they frequently received
heterogeneous treatment regimens, including multiple lines of
chemotherapy and/or radiotherapy, which could confound the accurate
assessment of chemosensitivity to gemcitabine. By contrast,
patients with resectable status were treated under a standardized
protocol with gemcitabine plus S-1. This homogeneity provided a
consistent clinical background to appropriately evaluate the
predictive value of serum circ72309. The levels of circ72309 in the
pre-treatment serum were measured using RT-qPCR. Serum samples were
stored at -80°C without repeated freeze-thaw cycles. The cases were
divided into two groups according to the median circ72309 levels
and the relationship between the levels of circ72309 and the
therapeutic effect of neoadjuvant chemotherapy was examined. The
Response Evaluation Criteria in Solid Tumors (RECIST)
classification (34) and
progression-free survival (PFS) were used to assess the therapeutic
effect. Briefly, in the RECIST classification, target lesions were
defined as primary tumors and lymph nodes (with short axes of ≥15
mm) and the response criteria was defined as follows: i) Complete
response (CR), the disappearance of all target lesions; ii) partial
response (PR), ≥30% decrease in the sum of diameters of target
lesions compared to the respective baseline; and iii) progressive
disease (PD), ≥20% increase in the sum of diameters of target
lesions compared to the respective baseline (including the baseline
sum if that was the smallest on study); in addition to the relative
increase of 20%, the sum should also have demonstrated an absolute
increase of ≥5 mm; and the appearance of one or more new lesions
was also considered progression; and iv) stable disease (SD),
neither sufficient shrinkage to qualify for PR nor sufficient
increase to qualify for PD, taking the smallest sum diameters while
on study as the baseline. PFS was calculated from the day of
neoadjuvant chemotherapy initiation to the day of mortality from
any cause or to the day of tumor progression and was censored on
the last day that the patient was documented to be alive without
tumor progression. Post-operative recurrence was monitored using
regular imaging tests, including enhanced abdominal computed
tomography (CT) and tracking changes in the levels of tumor
markers. Post-operative adjuvant treatment was routinely performed
unless clinical contraindications, such as gemcitabine or S-1, were
present for 6 months (35,36).
Local recurrence was defined as a recurrence near the resection
site, including the lymph node dissection site and the area where
the tumor was dissected. Distant recurrence was defined as
metastasis to other organs.
Statistical analysis
Categorical characteristics are presented as
frequencies and percentages. Numerical values are presented as the
median and range. Normality was tested using a Shapiro-Wilk test
and equality of variances was assessed using a Levene's test.
Specifically, a Student's t-test was used for RT-qPCR and western
blotting data after confirming normality and homogeneity of
variance. Wilcoxon tests were used for non-normally distributed
continuous variables in clinical datasets. Categorical variables
were analyzed using a χ2 or Fisher's exact test, as
appropriate. Kaplan-Meier analysis with a log-rank test was used
for survival comparisons and Cox proportional hazards models were
applied for univariate and multivariate survival analyses. For all
analyses involving multiple comparisons, appropriate post hoc tests
with Bonferroni corrections were applied if differences were
considered significant by ANOVA. JMP® Pro version 14
(SAS Institute Inc.; RRID: SCR_022199) and R Project for
Statistical Computing (version 4.3.2; https://www.r-project.org/, RRID: SCR_001905) were
used for statistical analyses. P<0.05 was considered to indicate
a statistically significant difference.
Results
circ72309 expression is upregulated in
parent PC cell lines and culture supernatant
circRNA sequencing was performed on parent PC cell
lines (MIAPaCa-2, gemcitabine IC50=12.5 ng/ml; n=3) and
a gemcitabine-resistant cell line (GR3; gemcitabine IC50
not reached; n=3) to extract candidate circRNAs associated with
gemcitabine sensitivity (GSE297252, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE297252).
A total of 29 differentially expressed circRNAs (20 upregulated and
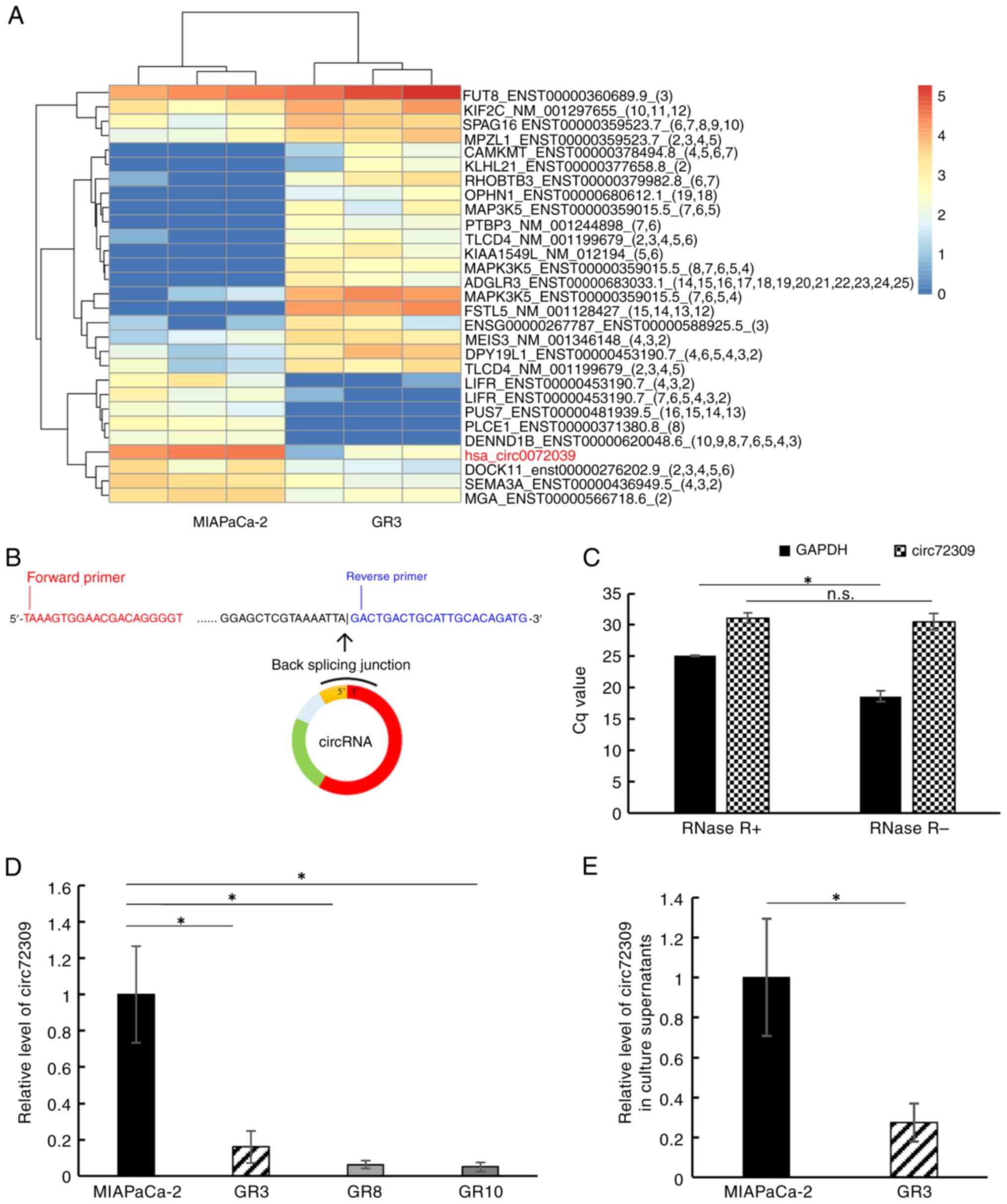
9 downregulated) were identified by circRNA sequencing (Fig. 1A). Based on a literature search,
circ72309 was identified as potentially relevant; it has a full
length of 7,248 bp, a spliced length of 580 bp and is located on
human chromosome 5 (chr5:38523520-38530768). To confirm the levels
of circ72309 by RT-qPCR, the primers specific for circ72309 were
designed to amplify the backsplicing junction sequence (Fig. 1B and Table SV) and the primers were subjected
to RNase R treatment to ascertain whether they were capable of
recognizing circular RNA. The Cq value of GAPDH, a linear
RNA, was increased by RNase R treatment; however, the Cq value of
circ72309 was unchanged by RNase R treatment, confirming that the
primers recognized the backsplicing junction of circ72309 (Fig. 1C). Subsequently, the levels of
circ72309 in three gemcitabine-resistant cell lines (GR3, GR8 and
GR10) were confirmed by RT-qPCR; circ72309 was markedly lower in GR
cell lines than in MIAPaCa-2 (Fig.
1D). Furthermore, the culture supernatant levels of circ72309
in GR3 was observed to be markedly lower than that in MIAPaCa-2
cells, as well as in cells when compared between MIPaCa-2 and GR3
(Fig. 1E).
circ72309 induces cell apoptosis and
improves gemcitabine sensitivity in gemcitabine-resistant PC cell
lines
The identified 580-bp sequence was synthesized as an
artificial oligonucleotide, subcloned into the pRP[Exp]-EGFP
Laccase2 MCS Exon Vector (vector ID: VB230525-1184qve), flanked by
a GT-AG splice site (Fig. 2A) and
transfected into GR3, GR8 and GR10 cells. The circ72309 levels in
the transfected cell lines were markedly increased following
transfection (Fig. S2). Each
overexpressed gemcitabine-resistant cell line (GR3_circ72309 OE,
GR8_circ72309 OE and GR10_circ72309 OE) was assessed using MTT and
apoptosis assays. The pRP[Exp]-EGFP/Neo-CMV>ORF_Stuffer (vector
ID: VB900145-9829wty) was used as a control (GR_NC) for comparison.
In the MTT assay, a concentration-dependent decrease in cell
viability was evident in GR_circ72309 OE, indicating enhanced
sensitivity to gemcitabine. The fitted curves followed a typical
sigmoidal profile, with IC50 values of 13.3 ng/ml
(R2=0.974) for GR3_circ72309 OE, 130 ng/ml
(R2=0.951) for GR8_circ72309 OE and 16.9 ng/ml
(R2=0.984) for GR10_circ72309 OE (Fig. 2B). In resistant clones, cell
viability remained relatively high even at the maximal drug
concentrations tested and the curves did not follow a typical
sigmoidal profile, consistent with a resistant phenotype. The
effect of si_circ72309 on gemcitabine sensitivity was evaluated
using MIAPaCa-2 (IC50, 23.3 ng/ml), Panc-1
(IC50, 24.7 ng/ml) and PSN-1 (IC50, 6.0
ng/ml) cells. Next, untreated parent cells, scramble control (Scr)
transfected cells and GR3, GR8 and GR10 si_circ72309 transfected
cells were assessed. In MIAPaCa-2, no difference in gemcitabine
sensitivity was observed between MIAPaCa-2_Parent and MIAPaCa-2_Scr
(IC50, MIAPaCa-2_ Parent vs. MIAPaCa-2_Scr, 23.3 ng/ml
vs. 23 ng/ml). However, MIAPaCa-2_si_circ72309 demonstrated
increased resistance to gemcitabine, with an IC50 of 180
ng/ml. Similarly, gemcitabine resistance was markedly augmented in
Panc-1_si_circ72309 (IC50, not reached) relative to
Panc-1_Parent (IC50, 24.7 ng/ml) and Panc-1_Scr
(IC50, 21.3 ng/ml). No effect on gemcitabine sensitivity
was observed in PSN-1 (Fig. 2C).
The apoptosis rate after 20 ng/ml gemcitabine treatment was
examined using the annexin V assay, which showed a significant
increase in early and late apoptotic cells (Q3 and Q2) in
GR_circ72309 OE [rate of apoptosis cells (Q2+Q3): GR3_NC vs.
GR3_circ72309 OE, 18.0 vs. 38.3%, P=0.002; GR8_NC vs. GR8_circ72309
OE, 18.7% vs. 43.9%, P=0.009; GR10_NC vs. GR10_circ72309 OE, 12.4%
vs. 32.6%, P=0.045, Fig. 2D].
This showed that overexpression of circ72309 in
gemcitabine-resistant cell lines induced apoptosis in the presence
of gemcitabine, thereby enhancing gemcitabine sensitivity.
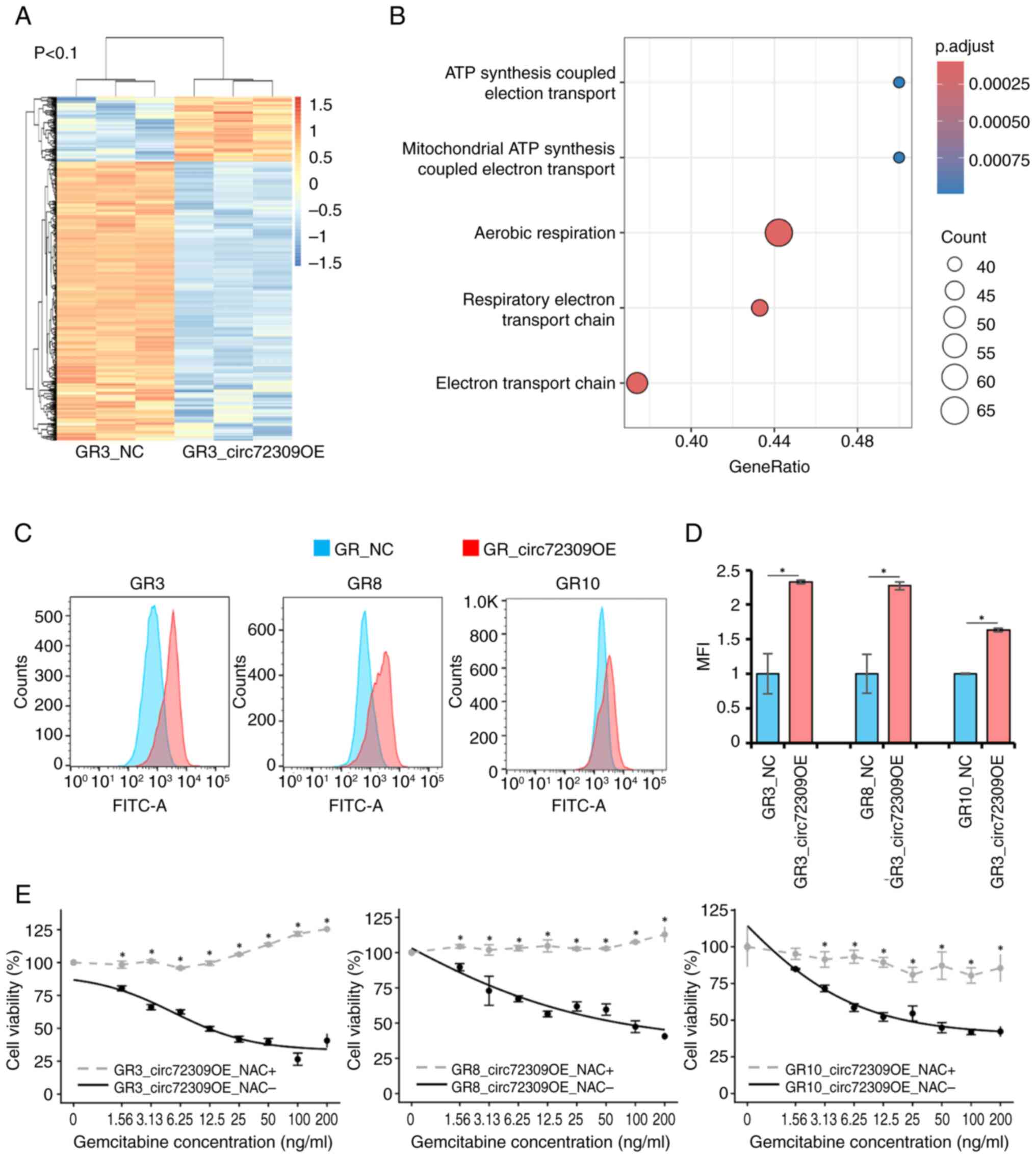
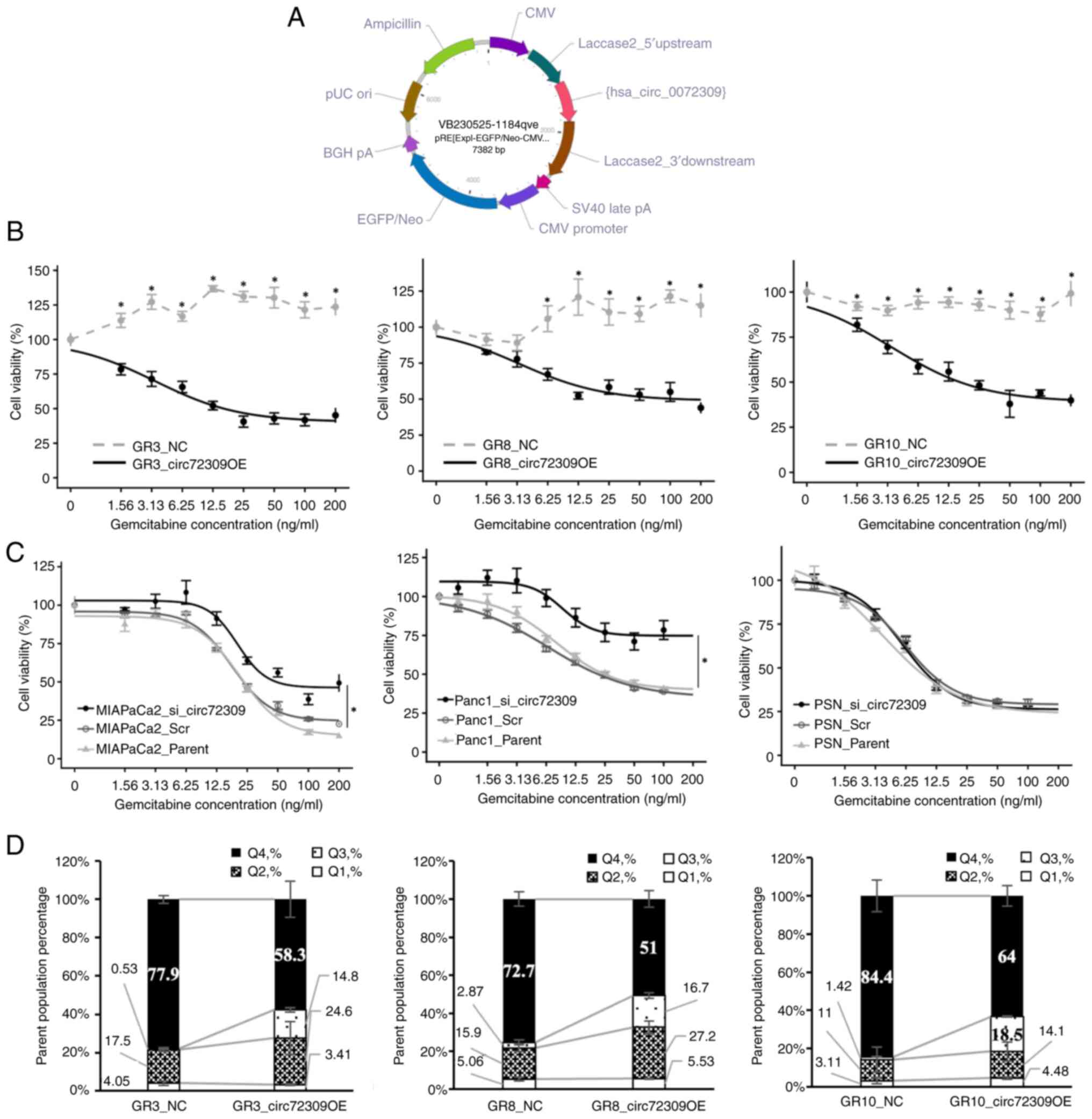 | Figure 2Effects of circ72309 overexpression
on cellular function in gemcitabine-resistant cell lines. (A)
Vector schema for overexpression of circ72309. (B) Dose-response
curves of gemcitabine in GR_NC and GR_circ72309OE clones.
GR_circ72309OE clones showed a concentration-dependent decrease in
viability with a typical sigmoidal fitting (IC50 values:
GR3, 13.3 ng/ml, R2=0.974; GR8, 130 ng/ml,
R2=0.951; GR10, 16.9 ng/ml, R2=0.984),
whereas GR_NC cells retained high viability even at maximal
gemcitabine concentrations, consistent with the resistant
phenotype. Fitted curves are shown as solid lines, while resistant
clone values without a typical sigmoidal decline are represented as
dashed lines. (C) MTT assays were used to assess the effects of
si-circ72309 in PC cell lines (MIAPaCa-2, PSN-1 and Panc-1).
Dose-response curves were analyzed using non-linear regression with
a four-parameter logistic model; all fitted curves showed excellent
agreement with the experimental data (R2>0.95).
Fitted curves are shown as solid lines. (D) Apoptosis assays
following treatment with 20 ng/ml gemcitabine.
*P<0.05. circRNA, circular RNA; PC, pancreatic
cancer; GR, gemcitabine resistant; NC, negative control; siRNA,
small interfering RNA. |
Overexpression of circ72309 increases
gemcitabine sensitivity by increasing ROS activity and expression
of the gemcitabine transporter
To investigate the mechanism by which overexpression
of circ72309 improved gemcitabine sensitivity, GR3_NC (n=3) and
GR3_circ72309 OE (n=3) were submitted for mRNA sequencing (Fig. 3A). The differentially expressed
mRNAs (545 in total; 103 upregulated and 442 downregulated) were
identified (GSE297082, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE297082).
GSEA showed that the oxidative phosphorylation and ATP production
pathway in mitochondria were enhanced in GR3_circ72309 OE (Fig. 3B), suggesting that ROS activity
may be involved in the increase in gemcitabine sensitivity. The ROS
activity in GR_circ72309 OE showed an increase compared to GR_NC
(Fig. 3C and D). A previous
report revealed that the effect of ROS reduces CDA, which
metabolizes gemcitabine to the inactive form
2′,2′-Difluoro-2′-deoxyuridine (dFdU) (37). Subsequently, GR_circ72309 OE were
treated with a ROS scavenger, NAC, to suppress ROS activity and the
effect on gemcitabine sensitivity was then assessed. As a control,
the same volume of PBS was added to GR_circ72309 OE_NAC-. In the
MTT assay, GR_circ72309 OE_NAC+ promoted gemcitabine resistance,
indicating that NAC counteracted the gemcitabine sensitivity
observed following overexpression of circ72309 (IC50:
GR3_circ72309 OE_NAC-vs. GR3_circ72309 OE_NAC+, 12.0 ng/ml vs. not
reached; GR8_circ72309 OE_NAC-vs. GR8_circ72309 OE_NAC+, 69.6 ng/ml
vs. not reached; GR10_circ72309 OE_NAC-vs. GR10_circ72309 OE_NAC+,
20.8 ng/ml vs. not reached; Fig.
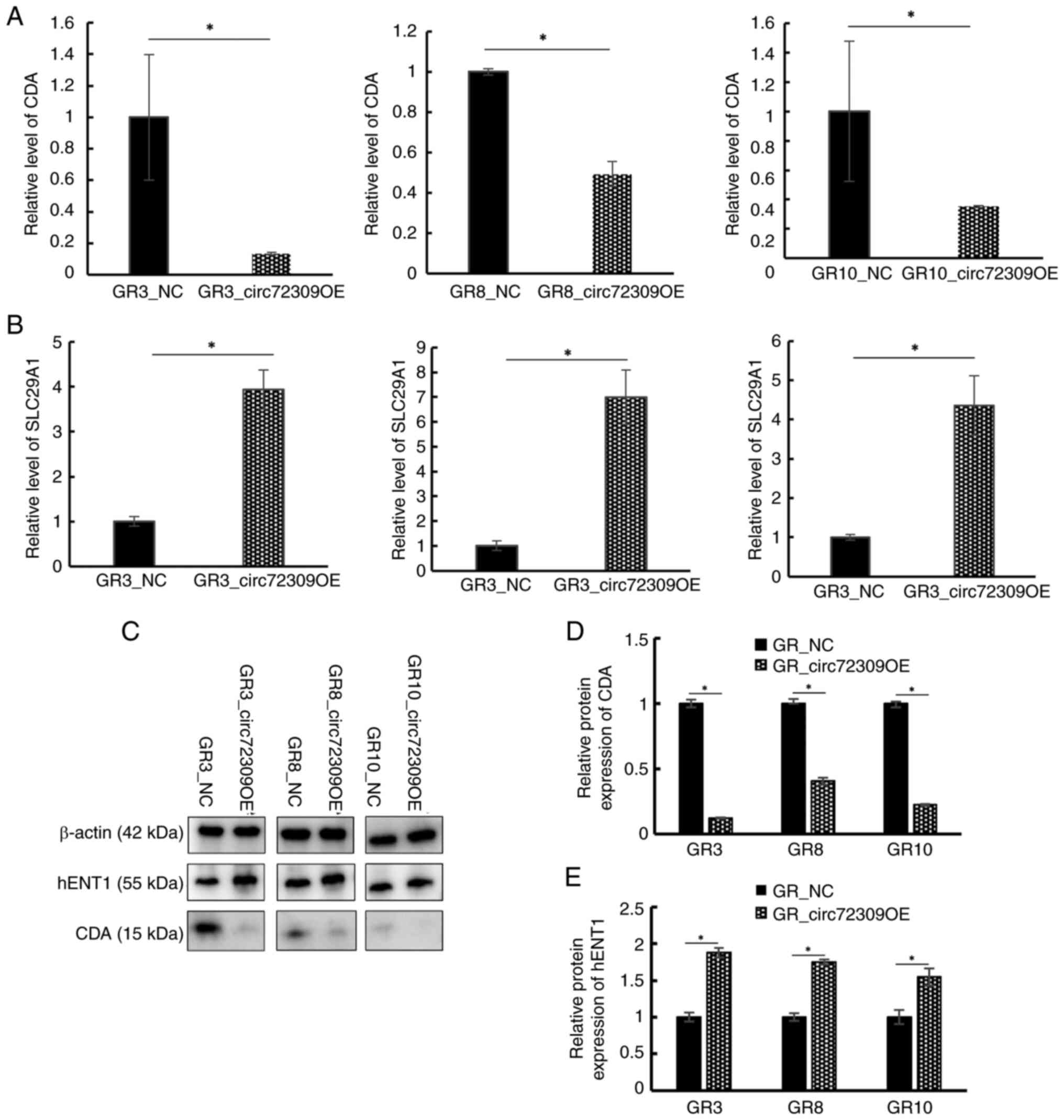
3E). When CDA expression was confirmed by RT-qPCR and western
blotting, its expression was markedly reduced in GR_circ72309 OE
(Fig. 4A, C and D). mRNA
sequencing results confirmed the changes in genes related to
gemcitabine metabolism and showed a significant increase in
SLC29A1 expression (log fold change, 1.16; P<0.001). The
gene encodes human equilibrative nucleoside transporter 1 (hENT1),
a transmembrane glycoprotein that localizes to the plasma and
mitochondrial membranes and mediates the cellular uptake of
nucleosides from the surrounding medium. When the expression of
SLC29A1 was confirmed by RT-qPCR and western blotting, the
expression was markedly increased in GR_circ72309 OE cells
(Fig. 4B, C and E).
Target miRNAs for circ72309 regulating
the expression of SLC29A1
Based on the CircInteractome database, circ72309 has
adsorption sites for 26 miRNAs (Table SIII). Among these miRNAs,
miR-1225-3p and miR-1234 were identified by TargetScan as having
binding sites with SLC29A1 (Table SIV). Further, it was confirmed
that the expression of miR-1225-3p and miR-1234 in GR cell lines
was markedly decreased by circ72309 overexpression (Fig. S3).
High circ72309 levels in the
pre-treatment serum of patients with PC are associated with
improved gemcitabine-based neoadjuvant treatment efficacy and
recurrence-free survival
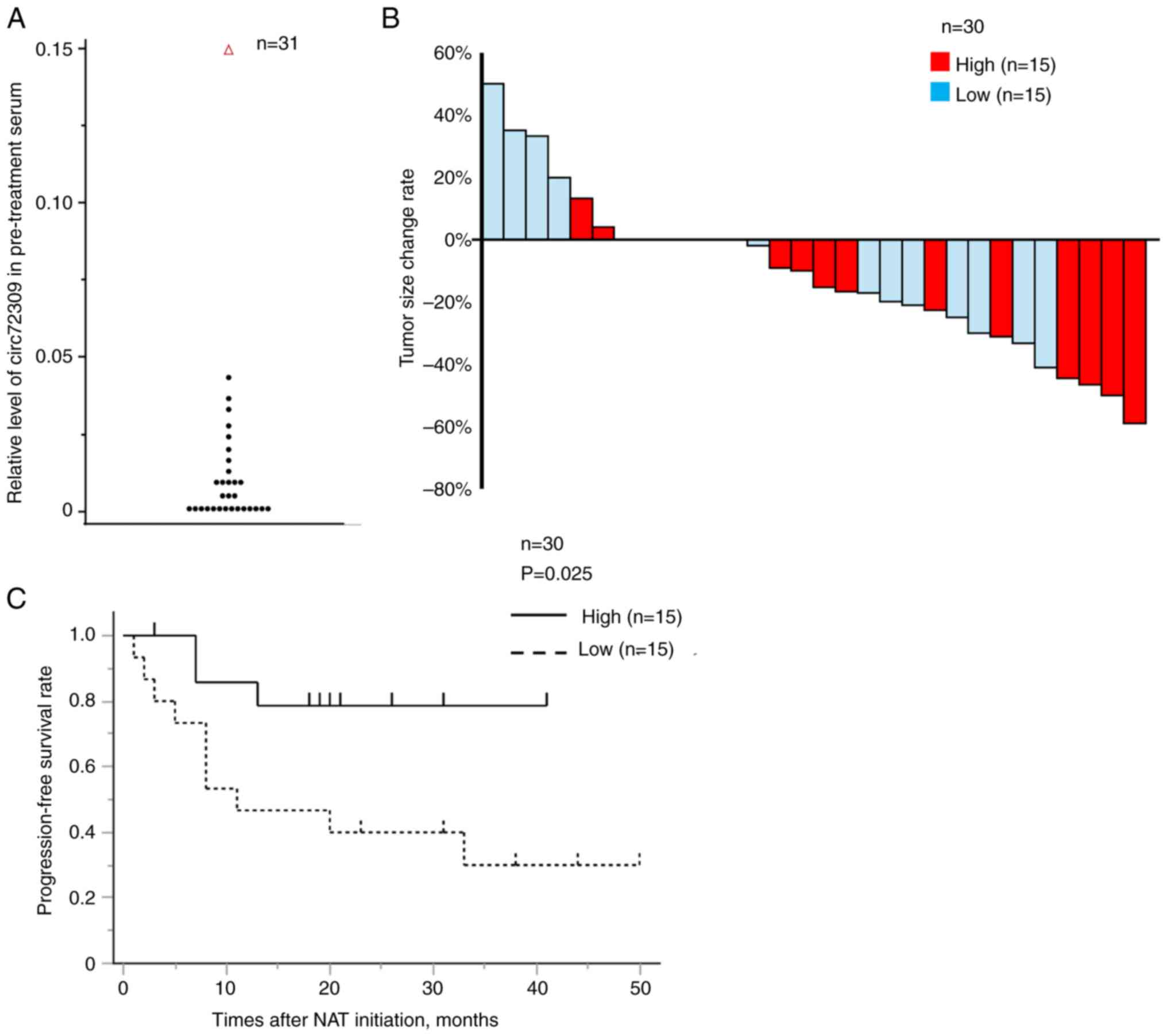
A total of 31 patients with PC who received
gemcitabine plus S-1 for neoadjuvant chemotherapy were included.
The levels of circ72309 are shown in Fig. 5A; one case showing an outlier
value was excluded (shown as Δ in Fig. 5A). The patients' pre-treatment
characteristics (n=30) are shown in Table I. A total of 3 patients could not
undergo surgery owing to tumor progression during neoadjuvant
chemotherapy and surgery was performed on 27 patients (90%). In 13
patients who underwent resection and for whom post-operative serum
samples were available, serum circ72309 levels were measured after
tumor resection and were found to be markedly reduced (Fig. S4). The median pre-treatment tumor
size was 20 (0-50) mm and the median pre-treatment CA19-9 levels
were 53.3 (0.3-2,729) U/ml. The 30 cases were stratified into high
and low groups based on the median circ72309 level; Table I shows the pre-treatment
characteristics of the two groups. The pre-treatment CA19-9 levels
were markedly lower in the high circ72309 group. The pre-treatment
tumor size did not differ markedly between the two groups. Fig. 5B shows a waterfall plot of the
rate of change in tumor size. ~18 patients (60%) had some degree of
tumor shrinkage on CT imaging. The RECIST classification
demonstrated a markedly improved response in the high circ72309
group, whereas PD cases were observed only in the low circ72309
group (Table I; PD/SD/PR, high
vs. low, 5/10/0 vs. 2/8/5, P=0.016). PFS was significantly higher
in the high circ72309 group (Fig.
5C; median PFS rates, high vs. low, not reached vs. 9.5 months,
P=0.035). The results of univariate and multivariate analyses of
prognostic factors for PFS are shown in Table II. Multivariate analysis revealed
that pre-treatment serum circ72309 levels were an independent
prognostic factor for PFS (P=0.042).
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Parameter | All cases,
n=30 | High circ72309
group, n=15 | Low circ72309
group, n=15 | P-value |
|---|
| Age, yearsc | 73 (50-84) | 73 (50-82) | 72 (55-84) | 0.781 |
| Sex, n (%) | | | | 0.026a |
| Male | 16 (53%) | 5 (33%) | 11 (73%) | |
| Female | 14 (47%) | 10 (67%) | 4 (27%) | |
| Tumor location, n
(%) | | | | 0.063 |
| Ph | 13 (43%) | 4 (27%) | 9 (60%) | |
| Pb-t | 17 (57%) | 11 (73%) | 6 (40%) | |
| Pre-treatment
CA19-9 level, U/mlc | 53.3
(0.3-2,729) | 19.3 (0.3-693) | 204
(15.6-2,729) | 0.007b |
| Elevated CA19-9
before treatment, n (%) | 18 (60%) | 6 (40%) | 12 (80%) | 0.023a |
| Preoperative CA19-9
level, U/mlc | 21.6 (0.3-767) | 11.5 (0.3-357) | 63.5 (9.3-767) | 0.002b |
| Pre-treatment
DUPAN2, U/mlc | 31 (25-680) | 26 (25-680) | 61 (25-670) | 0.534 |
| Preoperative
DUPAN2, U/mlc | 56 (25-430) | 55 (25-270) | 61 (25-430) | 0.715 |
| Pre-treatment tumor
size, mmc | 20 (0-51) | 16 (0-30) | 20 (10-51) | 0.084 |
| Preoperative tumor
size, mmc | 15 (0-50) | 11 (0-26) | 20 (10-50) | 0.006b |
| Resected cases,
% | 27 (90%) | 15 (100%) | 12 (80%) | 0.039a |
| RECIST
classification, PD/SD/PR, n | 5/18/7 | 0/10/5 | 5/8/2 | 0.016a |
 | Table IIPrognostic factors associated with
progression-free survival. |
Table II
Prognostic factors associated with
progression-free survival.
| Factor | n | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years | | | | | |
| >65 | 23 | 1.83
(0.40-8.3) | 0.403 | 3.96
(0.44-35.8) | 0.220 |
| ≤65 | 7 | Reference | | | |
| Sex | | | | | |
| Male | 16 | 2.1 (0.64-6.8) | 0.203 | 0.24
(0.03-2.15) | 0.202 |
| Female | 14 | Reference | | | |
| Tumor location | | | | | |
| Ph | 13 | 1.97
(0.66-5.9) | 0.225 | 1.27
(0.29-5.5) | 0.753 |
| Pb-t | 17 | Reference | | | |
| Pre-treatment
CA19-9 level, U/ml | | | | | |
| >37 | 12 | 2.27
(0.76-10.1) | 0.098 | 4.23 (0.26-68) | 0.308 |
| ≤37 | 18 | Reference | | | |
| Pre-treatment
DUPAN2, U/ml | | | | | |
| >25 | 15 | 1.55
(0.40-6.0) | 0.519 | 0.80
(0.12-5.2) | 0.810 |
| ≤25 | 15 | Reference | | | |
| Pre-treatment tumor
size, mm | | | | | |
| >20 | 11 | 1.30
(0.43-3.9) | 0.638 | 1.27
(0.29-5.5) | 0.753 |
| ≤20 | 19 | Reference | | | |
| Pre-treatment
circ72309 level | | | | | |
| Low | 15 | 3.64 (1.0-13) | 0.0340 | 5.50 (1.2-31) | 0.042a |
| High | 15 | Reference | | | |
Discussion
The roles of noncoding RNAs, including miRNAs,
circRNAs and other molecules, in epigenetic modifications of
biological functions have attracted attention for improving
chemosensitivity in patients with PC (38,39). However, research findings on the
effect of miRNAs on improving gemcitabine sensitivity have been
generally modest and unsatisfactory (40-42). Iwagami et al (43) reported that miR320c inhibits the
tumor suppressor gene, SMARCC1 and Hasegawa et al (44) reported that miR-1246 induces
cancer stem cell-like properties in PC cells. Silencing this miRNA
improved gemcitabine sensitivity; however, the effect was limited
to a <20% reduction in cell viability (43,44). A potential reason why the
influence of miRNAs in improving gemcitabine sensitivity is limited
is that a single miRNA affects only a limited number of mRNAs,
although the mechanism by which gemcitabine exerts its antitumor
effect is multifactorial. Indeed, intracellular translocation of
gemcitabine is primarily mediated by the hENT1-mediated mechanism
and multiple steps of phosphorylation are required for
intracellular conversion of gemcitabine to its active form,
gemcitabine triphosphate (45).
Additionally, there is an intracellular gemcitabine inactivation
mechanism mediated by CDA (45).
The present study demonstrated that circ72309 regulated multiple
RNAs involved in different points of the gemcitabine metabolic
pathway (including CDA and SLC29A1) and that the
overexpression of circ72309 resulted in a significant increase in
gemcitabine sensitivity in gemcitabine-resistant PC cell lines,
resulting in a concentration-dependent reduction in cell viability
by >50%.
In the present study, CDA was identified as involved
in the intracellular gemcitabine metabolic pathway regulated by
circ72309 and overexpression of circ72309 markedly decreased
intracellular CDA expression in gemcitabine-resistant PC cell
lines. CDA is a nucleoside enzyme that metabolizes gemcitabine to
its inactive form, dFdU (37).
The results of the present study indicated that overexpression of
circ72309 increased aerobic respiration, ATP production and
electron transport pathways in the mitochondria and subsequently
increased ROS activity. Previous studies have shown that increases
in intracellular ROS levels render PC cells susceptible to
gemcitabine chemotherapy (46).
Additionally, increased intracellular ROS levels were shown to
enhance gemcitabine sensitivity by downregulating CDA expression
(37). In the present study, NAC
(a ROS scavenger) suppressed the decrease in CDA levels induced by
circ72309 overexpression (Fig.
S5). Therefore, it is hypothesized that the overexpression of
circ72309 decreases CDA expression by increasing ROS activity.
Another mRNA involved in the gemcitabine metabolic
pathway regulated by circ72309, as revealed in the present study,
was SLC29A1, which encodes the gemcitabine intracellular
transporter, hENT1 (47).
Gemcitabine is hydrophilic and, therefore, unable to pass through
the cell membrane via passive diffusion. It is imported via a
nucleoside transporter, hENT1 (47). Previous reports demonstrate that
high hENT1 expression is associated with a favorable response to
gemcitabine in patients with PC (48) and indeed, an improved prognosis
has been reported in patients with PC with high hENT1 expression
who have been treated with gemcitabine (49,50). The results of the present study
indicated that circ72309 increased SLC29A1/hENT1 expression,
which may partly explain the increased gemcitabine sensitivity in
gemcitabine-resistant PC cell lines following circ72309
overexpression. It was also shown that circ72309 decreased the
expression of miR-1225-3p and miR-1234, which possess binding sites
for SLC29A1, thereby potentially increasing
SLC29A1/hENT1 expression. These preliminary results
suggested that one of the possible biological mechanisms regarding
the improved gemcitabine sensitivity induced by circ72309 is that
the overexpression of circ72309 decreased miR-1225-3p and miR-1234
by adsorption to circ72309 and consequently increased the
expression of SLC29A1/hENT1 by suppressing the translational
regulation of these miRNAs.
As aforementioned, two possible mechanisms of how
circ72309 improves gemcitabine sensitivity in gemcitabine-resistant
PC cell lines have been indicated in the present study. NAC
markedly reversed the improvement in gemcitabine sensitivity
induced by the overexpression of circ72309. These observations
indicated that the increase in gemcitabine sensitivity via
circ72309 overexpression may be primarily mediated by its function
of regulating the intracellular ROS levels and the subsequent
decrease in CDA levels. Previous reports indicated that high hENT1
expression in resected PC specimens was associated with a favorable
response to gemcitabine-based adjuvant therapy and improved
survival only in patients with low mRNA levels of CDA,
indicating that the effect of hENT1 may be at least partly
dependent on the level of CDA (51).
As aforementioned, circRNAs in serum are stable due
to their circular structure (15)
and this has resulted in increased attention for their potential as
liquid biomarkers (16-18). Wang et al (17) reported that higher levels of serum
circSETDB1 had a shorter PFS in patients with serous ovarian cancer
who were treated with platinum-taxane-combined chemotherapy. When
assessing the potential of serum circRNAs as liquid biomarkers, the
origin of relevant circRNAs should be taken into consideration as
the expression of circRNAs in normal tissues has not yet been fully
examined. As circ72309 expression in the cell culture supernatant
of gemcitabine-resistant cell lines (such as cells with low
circ72309 expression) was lower than that of parental cells and a
significant decrease in circ72309 levels was observed after
resection of the primary tumor from patients with PC in the present
study, it could be speculated that the primary tumor was the
predominant source of serum circ72309. Therefore, serum circ72309
levels may serve as a liquid biomarker representing a phenotype of
primary tumor regarding gemcitabine sensitivity modulated by
circ72309 levels in patients with PC. The present study revealed
that among patients with PC treated with a gemcitabine-based
neoadjuvant chemotherapy, those with high serum circ72309 levels
experienced improved therapeutic efficacy than those with low
circ72309 levels and that high serum circ72309 levels were an
independent predictor of a favorable PFS. These observations
indicated that serum circ72309 levels may be used to predict the
therapeutic efficacy of gemcitabine-based neoadjuvant chemotherapy
in PC and may be useful for therapeutic selection strategies. The
results of the in vitro experiments further supported this
concept, revealing the detailed molecular mechanisms of circ72309
in modulating gemcitabine sensitivity in PC cell lines. Beyond the
mechanistic insights, the therapeutic translation of circ72309 may
benefit from emerging delivery technologies. Recent work has
emphasized the role of virus-like particles (VLPs) as protective
and programmable scaffolds for circRNA delivery, which enhance
stability, cellular entry and tissue-targeting potential (52). Considering that circ72309
overexpression improved gemcitabine sensitivity by modulating both
CDA and hENT1 in resistant cell lines, it is conceivable that
VLP-mediated administration of circ72309 may serve as a novel
approach for overcoming gemcitabine resistance in PC. As
chemoresistance represents a major clinical challenge across
multiple tumor types, the findings of the present study may also
have broader implications. A recent review highlighted that several
molecular determinants of chemoresistance are shared across
different cancer types (53).
While the present study focused only on PC, it would be of interest
to explore whether circ72309 similarly contributes to
chemoresistance in other types of cancer. Given that the mechanisms
identified here involve key regulators of gemcitabine metabolism,
including CDA and hENT1, which are also implicated in the response
to nucleoside analogs across various types of cancer, circ72309 may
also play a role in overcoming gemcitabine resistance in other
tumor types treated with gemcitabine. Such investigations may
further expand the translational relevance of the findings of the
present study and inform the development of novel therapeutic
strategies.
The present study had several limitations. First,
the in vitro analyses were performed only in
gemcitabine-resistant cell lines derived from MIAPaCa-2 cells and
validation in other PC models is required. Second, the clinical
analysis included a relatively small number of patients, which may
limit the generalizability of the results. Third, although serum
circ72309 levels were associated with therapeutic response and
prognosis, a clinically applicable cut-off value in this cohort was
not determined. Further investigation is required to ascertain the
feasibility of using serum circ72309 levels as a liquid biomarker
for the selection of a potentially effective treatment plan based
on the pre-treatment serum levels.
In conclusion, circ72309 influences multiple steps
in the intracellular gemcitabine metabolic pathway and induces
significant increases in gemcitabine sensitivity in
gemcitabine-resistant PC cell lines, indicating that circRNAs are a
promising epigenetic target in modulating cellular biology
(Fig. S6). Furthermore, serum
circ72309 levels may potentially serve as a liquid biomarker for
predicting gemcitabine sensitivity in PC and may serve as a
valuable tool for selecting optimal therapeutic agents and
facilitating personalized strategy in neoadjuvant treatment for
patients with PC.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
NS, HT, SK, KS, SH, YI, DY, YT, HA, TA, TN, JS, KT,
RC, YD and HE conceived and designed the study. NS performed the
experiments. NS, HT and KT collected the data and confirm the
authenticity of all the raw data. NS, HT, SK, KS, SH, YI, DY, YT,
HA, TA, TN, JS, KT, RC, YD and HE analyzed the data. NS, HT, KS,
YI, DY, YT, TN and SK drafted the manuscript. YD and HE revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the ethical standards described in the 1975 Declaration of Helsinki
and its later revisions and approved by the Ethics Committees of
the University of Osaka Hospital (approval no. 24536). Written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang Y, Luo D, Shao Y, Shan Z, Liu Q, Weng
J, He W, Zhang R, Li Q, Wang Z and Li X: circCAPRIN1 interacts with
STAT2 to promote tumor progression and lipid synthesis via
upregulating ACC1 expression in colorectal cancer. Cancer Commun
(Lond). 43:100–122. 2023. View Article : Google Scholar
|
|
4
|
Wu H, Wang B, Wang L and Xue Y: circular
RNAs 0000515 and 0011385 as potential biomarkers for disease
monitoring and determining prognosis in pancreatic ductal
adenocarcinoma. Oncol Lett. 23:562022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ling S, He Y, Li X, Hu M, Ma Y, Li Y, Lu
Z, Shen S, Kong B, Zou X, et al: CircRHOT1 mediated cell
proliferation, apoptosis and invasion of pancreatic cancer cells by
sponging miR-125a-3p. J Cell Mol Med. 24:9881–9889. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li C, Zhang J, Yang X, Hu C, Chu T, Zhong
R, Shen Y, Hu F, Pan F, Xu J, et al: hsa_circ_0003222 accelerates
stemness and progression of non-small cell lung cancer by sponging
miR-527. Cell Death Dis. 12:8072021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X,
Li X, Dang Y and Zhang G: circCUL2 regulates gastric cancer
malignant transformation and cisplatin resistance by modulating
autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 19:1562020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng D, Wang J, Dong Z and Li X:
Cancer-related circular RNA: Diverse biological functions. Cancer
Cell Int. 21:112021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang M, Yu F, Li P and Wang K: Emerging
function and clinical significance of exosomal circRNAs in cancer.
Mol Ther Nucleic Acids. 21:367–383. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu H, Zhao X, Wang J, Jiang X, Cheng Y, He
Y, Sun L and Zhang G: Circular RNA CDR1as alleviates
Cisplatin-based chemoresistance by suppressing MiR-1299 in ovarian
cancer. Front Genet. 12:8154482021. View Article : Google Scholar
|
|
12
|
Yu S, Wang M, Zhang H, Guo X and Qin R:
Circ_0092367 inhibits EMT and gemcitabine resistance in pancreatic
cancer via regulating the miR-1206/ESRP1 Axis. Genes (Basel).
12:17012021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamada M, Tanaka K, Yamamoto K, Matsumoto
H, Yamasaki M, Yamashita K, Makino T, Saito T, Yamamoto K,
Takahashi T, et al: Association between circ_0004365 and cisplatin
resistance in esophageal squamous cell carcinoma. Oncol Lett.
26:4672023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mao S, Cheng Y, Huang Y, Xiong H and Gong
C: Comprehensive analysis of the exosomal circRNA-miRNA-mRNA
network in breast cancer. J Gene Med. 25:e35002023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang XJ, Wang Y, Wang HT, Liang ZF, Ji C,
Li XX, Zhang LL, Ji RB, Xu WR, Jin JH and Qian H: Exosomal
hsa_circ_000200 as a potential biomarker and metastasis enhancer of
gastric cancer via miR-4659a/b-3p/HBEGF axis. Cancer Cell Int.
23:1512023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang W, Wang J, Zhang X and Liu G: Serum
circSETDB1 is a promising biomarker for predicting response to
platinum-taxane-combined chemotherapy and relapse in high-grade
serous ovarian cancer. Onco Targets Ther. 12:7451–7457. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu C, Jun E, Okugawa Y, Toiyama Y,
Borazanci E, Bolton J, Taketomi A, Kim SC, Shang D, Von Hoff D, et
al: A circulating panel of circRNA biomarkers for the noninvasive
and early detection of pancreatic ductal adenocarcinoma.
Gastroenterology. 166:178–190.e16. 2024. View Article : Google Scholar
|
|
19
|
Takahashi H, Ohigashi H, Gotoh K,
Marubashi S, Yamada T, Murata M, Ioka T, Uehara H, Yano M and
Ishikawa O: Preoperative gemcitabine-based chemoradiation therapy
for resectable and borderline resectable pancreatic cancer. Ann
Surg. 258:1040–1050. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Von Hoff DD, Ervin T, Arena FP, Chiorean
EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, et
al: Increased survival in pancreatic cancer with nab-paclitaxel
plus gemcitabine. N Engl J Med. 369:1691–1703. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Burris H and Storniolo AM: Assessing
clinical benefit in the treatment of pancreas cancer: Gemcitabine
compared to 5-fluorouracil. Eur J Cancer. 33(Suppl 1): S18–S22.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen M, Liu X, Lu J, Teng H, Yu C, Liu Y
and Zheng Y: Dysregulation of the circ_0087502/miR-1179/TGFBR2
pathway supports gemcitabine resistance in pancreatic cancer.
Cancer Biol Ther. 24:22585662023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu Y, Zhou S, Cheng G, Ruan Y, Tian Y, Lv
K, Han S and Zhou X: CircLMTK2 silencing attenuates gemcitabine
resistance in pancreatic cancer by sponging miR-485-5p and to
target PAK1. J Oncol. 2022:19115922022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goan YG, Zhou B, Hu E, Mi S and Yen Y:
Overexpression of ribonucleotide reductase as a mechanism of
resistance to 2,2-difluorodeoxycytidine in the human KB cancer cell
line. Cancer Res. 59:4204–4207. 1999.PubMed/NCBI
|
|
25
|
Nakahira S, Nakamori S, Tsujie M,
Takahashi Y, Okami J, Yoshioka S, Yamasaki M, Marubashi S, Takemasa
I, Miyamoto A, et al: Involvement of ribonucleotide reductase M1
subunit overexpression in gemcitabine resistance of human
pancreatic cancer. Int J Cancer. 120:1355–1363. 2007. View Article : Google Scholar
|
|
26
|
Oyama K, Iwagami Y, Kobayashi S, Sasaki K,
Yamada D, Tomimaru Y, Noda T, Asaoka T, Takahashi H, Tanemura M, et
al: Removal of gemcitabine-induced senescent cancer cells by
targeting glutaminase1 improves the therapeutic effect in
pancreatic ductal adenocarcinoma. Int J Cancer. 154:912–925. 2024.
View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Scherbakov AM, Vorontsova SK, Khamidullina
AI, Mrdjanovic J, Andreeva OE, Bogdanov FB, Salnikova DI, Jurisic
V, Zavarzin IV and Shirinian VZ: Novel pentacyclic derivatives and
benzylidenes of the progesterone series cause anti-estrogenic and
antiproliferative effects and induce apoptosis in breast cancer
cells. Invest New Drugs. 41:142–152. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Radenković N, Milutinović M, Nikodijević
D, Jovankić J and Jurišić V: Sample preparation of adherent cell
lines for flow cytometry: Protocol Optimization-our experience with
SW-480 colorectal cancer cell line. Indian J Clin Biochem.
40:74–79. 2025. View Article : Google Scholar
|
|
30
|
Ueda H, Takahashi H, Kobayashi S, Kubo M,
Sasaki K, Iwagami Y, Yamada D, Tomimaru Y, Asaoka T, Noda T, et al:
miR-6855-5p enhances radioresistance and promotes migration of
pancreatic cancer by inducing Epithelial-mesenchymal transition via
suppressing FOXA1: Potential of plasma exosomal miR-6855-5p as an
indicator of radiosensitivity in patients with pancreatic cancer.
Ann Surg Oncol. 32:720–735. 2025. View Article : Google Scholar
|
|
31
|
Taylor SC, Berkelman T, Yadav G and
Hammond M: A defined methodology for reliable quantification of
Western blot data. Mol Biotechnol. 55:217–226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar :
|
|
33
|
National Comprehensive Cancer Network
(NCCN) guidelines. (Pancreatic Adenocarcinoma (version 1.2023)).
Available at: https://www.nccn.org/guidelines/category_1. Accessed
June 12, 2023
|
|
34
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar
|
|
35
|
Oettle H, Neuhaus P, Hochhaus A, Hartmann
JT, Gellert K, Ridwelski K, Niedergethmann M, Zulke C, Fahlke J,
Arning MB, et al: Adjuvant chemotherapy with gemcitabine and
long-term outcomes among patients with resected pancreatic cancer:
The CONKO-001 randomized trial. JAMA. 310:1473–1481. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Uesaka K, Boku N, Fukutomi A, Okamura Y,
Konishi M, Matsumoto I, Kaneoka Y, Shimizu Y, Nakamori S, Sakamoto
H, et al: Adjuvant chemotherapy of S-1 versus gemcitabine for
resected pancreatic cancer: A phase 3, open-label, randomised,
non-inferiority trial (JASPAC 01). Lancet. 388:248–257. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Frese KK, Neesse A, Cook N, Bapiro TE,
Lolkema MP, Jodrell DI and Tuveson DA: nab-Paclitaxel potentiates
gemcitabine activity by reducing cytidine deaminase levels in a
mouse model of pancreatic cancer. Cancer Discov. 2:260–269. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Z, Li Y, Ahmad A, Banerjee S, Azmi
AS, Kong D and Sarkar FH: Pancreatic cancer: Understanding and
overcoming chemoresistance. Nat Rev Gastroenterol Hepatol. 8:27–33.
2011. View Article : Google Scholar
|
|
39
|
Rajabpour A, Rajaei F and Teimoori-Toolabi
L: Molecular alterations contributing to pancreatic cancer
chemoresistance. Pancreatology. 17:310–320. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mackey JR, Mani RS, Selner M, Mowles D,
Young JD, Belt JA, Crawford CR and Cass CE: Functional nucleoside
transporters are required for gemcitabine influx and manifestation
of toxicity in cancer cell lines. Cancer Res. 58:4349–4357.
1998.PubMed/NCBI
|
|
41
|
Ohhashi S, Ohuchida K, Mizumoto K, Fujita
H, Egami T, Yu J, Toma H, Sadatomi S, Nagai E and Tanaka M:
Down-regulation of deoxycytidine kinase enhances acquired
resistance to gemcitabine in pancreatic cancer. Anticancer Res.
28:2205–2212. 2008.PubMed/NCBI
|
|
42
|
Ono H, Murase Y, Yamashita H, Kato T,
Asano D, Ishikawa Y, Watanabe S, Ueda H, Akahoshi K, Ogawa K, et
al: RRM1 is mediated by histone acetylation through gemcitabine
resistance and contributes to invasiveness and ECM remodeling in
pancreatic cancer. Int J Oncol. 62:512023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Iwagami Y, Eguchi H, Nagano H, Akita H,
Hama N, Wada H, Kawamoto K, Kobayashi S, Tomokuni A, Tomimaru Y, et
al: miR-320c regulates gemcitabine-resistance in pancreatic cancer
via SMARCC1. Br J Cancer. 109:502–511. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hasegawa S, Eguchi H, Nagano H, Konno M,
Tomimaru Y, Wada H, Hama N, Kawamoto K, Kobayashi S, Nishida N, et
al: MicroRNA-1246 expression associated with CCNG2-mediated
chemoresistance and stemness in pancreatic cancer. Br J Cancer.
111:1572–1580. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
De Sousa Cavalcante L and Monteiro G:
Gemcitabine: Metabolism and molecular mechanisms of action,
sensitivity and chemoresistance in pancreatic cancer. Eur J
Pharmacol. 741:8–16. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang L, Li J, Zong L, Chen X, Chen K,
Jiang Z, Nan L, Li X, Li W, Shan T, et al: Reactive oxygen species
and targeted therapy for pancreatic cancer. Oxid Med Cell Longev.
2016:16167812016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Prudner BC, Rathore R, Robinson AM, Godec
A, Chang SF, Hawkins WG, Hirbe AC and Van Tine BA: Arginine
starvation and docetaxel induce c-Myc-Driven hENT1 surface
expression to overcome gemcitabine resistance in ASS1-Negative
tumors. Clin Cancer Res. 25:5122–5134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hioki M, Shimada T, Yuan T, Nakanishi T,
Tajima H, Yamazaki M, Yokono R, Takabayashi M, Sawamoto K, Akashita
G, et al: Contribution of equilibrative nucleoside transporters 1
and 2 to gemcitabine uptake in pancreatic cancer cells. Biopharm
Drug Dispos. 39:256–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Greenhalf W, Ghaneh P, Neoptolemos JP,
Palmer DH, Cox TF, Lamb RF, Garner E, Campbell F, Mackey JR,
Costello E, et al: Pancreatic cancer hENT1 expression and survival
from gemcitabine in patients from the ESPAC-3 trial. J Natl Cancer
Inst. 106:djt3472014. View Article : Google Scholar
|
|
50
|
Farrell JJ, Elsaleh H, Garcia M, Lai R,
Ammar A, Regine WF, Abrams R, Benson AB, Macdonald J, Cass CE, et
al: Human equilibrative nucleoside transporter 1 levels predict
response to gemcitabine in patients with pancreatic cancer.
Gastroenterology. 136:187–195. 2009. View Article : Google Scholar
|
|
51
|
Aughton K, Elander NO, Evans A, Jackson R,
Campbell F, Costello E, Halloran CM, Mackey JR, Scarfe AG, Valle
JW, et al: hENT1 predicts benefit from gemcitabine in pancreatic
cancer but only with low CDA mRNA. Cancers (Basel). 13:57582021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gupta R, Arora K, Mehrotra Arora N and
Kundu P: Significance of VLPs in Vlp-circRNA vaccines: A vaccine
candidate or delivery vehicle? RNA Biol. 21:17–28. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Patel M, Singh P, Gandupalli L and Gupta
R: Identification and evaluation of Survival-associated common
chemoresistant genes in cancer. Biomed Biotechnol Res J. 8:320–327.
2024. View Article : Google Scholar
|