Introduction
Esophageal squamous cell carcinoma (ESCC) is a
highly aggressive malignancy, with early lymphatic and hematogenous
metastasis. The treatment of ESCC primarily relies on classical
treatment modalities including surgery, radiotherapy and
chemotherapy, or a combination of these methods; however, the
outcome has not improved significantly (1). The poor prognosis of ESCC may improve
with a greater understanding of the molecular basis of this
disease, with a focus on molecular risk markers that may lead to
improved detection and treatment strategies.
ESCC develops through the accumulation of genetic
and epigenetic abnormalities, evolving from preinvasive lesions to
invasive esophageal cancer, with the malignant cells commonly
exhibiting abnormal histone expression. EHMT1 is an important
subunit of H3K9 methyltransferases. The highly similar euchromatic
H3K9 methyltransferases EHMT1 and EHMT2 (also referred to as GLP
and G9a, respectively) form a heteromeric complex and the loss of
either one significantly reduces mono- and dimethylation of H3K9, a
marker of silent euchromatin (2),
which is a procedure crucial for the transcription, signal
transduction, proliferation and differentiation of cells (3,4).
EHMT1 has also been recognized for its ability to methylate histone
H1.4, as well as other non-histone proteins, including itself
(5–7). EHMT1 binds to its H3K9me1 products
via ankyrin repeat domains, which contain a hydrophobic cage
present in methyllysine-binding modules of diverse folds.
Therefore, it is hypothesized that the disruption of EHMT1 may also
result in major alterations in gene expression patterns that may
contribute to cell canceration.
It was previously suggested that disordered histone
methylation may induce tumorigenesis (8). However, our knowledge of the
expression and clinical significance of EHMT1 in ESCC and
peripheral tissues is limited; therefore, thorough investigation of
the EHMT1 expression in ESCC and the normal peripheral esophageal
epithelium is required. In our study, we analyzed EHMT1 expression
in a cohort of 50 patients and demonstrated that EHMT1 expression
was significantly upregulated in squamous preinvasive lesions and
ESCC, with the expression of EHMT1 in ESCC being associated with
tumor grade, depth of invasion and lymph node metastasis. This
clinical case study verified that the expression of EHMT1 predicted
poor cancer-specific survival in human ESCC. This study also
demonstrated that the expression of EHMT1 may be a novel prognostic
biomarker for ESCC and a potential target for clinical
treatment.
Materials and methods
Clinical samples
A total of 50 patients who underwent resection for
ESCC between 2003 and 2007 at the Department of Thoracic Surgery,
the First Affiliated Hospital of China Medical University, were
included in this study. None of these patients had received
chemotherapy or radiotherapy prior to surgery. The tumor specimens
were either cut immediately after removal from the resected
esophagus, frozen in liquid nitrogen and stored at −80°C, or fixed
in 10% formalin and embedded in paraffin for histopathological
analysis. All the cases were independently classified by two
experienced pathologists as ESCC, according to the guidelines of
the World Health Organization. The criteria of the
tumor-node-metastasis (TNM) staging system were used to classify
the clinicopathological factors and clinical stages of esophageal
cancer (defined by the International Union Against Cancer TNM
Classification of Malignant Tumors, 7th edition, 2009). The
patients provided signed informed consent and were subjected to
close follow-up. The patient sample comprised 40 men and 10 women,
with a mean age of 63 years (range, 41–84 years) at the time of
surgery. A summary of the clinicopathological characteristics is
presented in Table I. The median
follow-up time after surgery was 30 months (range, 3–60
months).
 | Table I.Association between EHMT1 expression
and clinicopathological data of esophageal squamous cell
cancer. |
Table I.
Association between EHMT1 expression
and clinicopathological data of esophageal squamous cell
cancer.
| Clinicopathological
parameters | No. | EHMT1 expression
| χ2 | P-value |
|---|
| Positive | Negative | Positivity rate
(%) |
|---|
| Age (years) | | | | | 0.002 | 0.963 |
| ≤60 | 21 | 11 | 10 | 52.4 | | |
| >60 | 29 | 15 | 14 | 51.7 | | |
| Gender | | | | | 0.020 | 0.887 |
| Male | 40 | 21 | 19 | 52.5 | | |
| Female | 10 | 5 | 5 | 50.0 | | |
| Tumor grade | | | | | 12.426 | 0.002a |
| G1 | 14 | 6 | 8 | 42.9 | | |
| G2 | 20 | 6 | 14 | 30.0 | | |
| G3–4 | 16 | 14 | 2 | 87.5 | | |
| Depth of
invasion | | | | | 9.992 | 0.002a |
| T1–2 | 18 | 4 | 14 | 22.2 | | |
| T3–4 | 32 | 22 | 10 | 68.8 | | |
| Lymph node
metastasis | | | | | 14.753 | 0.001a |
| N0 | 28 | 8 | 20 | 28.6 | | |
| N1 | 16 | 14 | 2 | 87.5 | | |
| N2–3 | 6 | 4 | 2 | 66.7 | | |
| Tumor stage | | | | | 32.902 | 0.000a |
| I | 10 | 2 | 8 | 20.0 | | |
| II | 18 | 6 | 12 | 33.3 | | |
| III–IV | 22 | 18 | 4 | 81.8 | | |
This study was approved by the Human Research Ethics
Committee of China Medical University, which is accredited by the
National Council on Ethics in Human Research.
Immunohistochemistry
Immunohistochemical studies on EHMT1 were performed
on formalin-fixed, paraffin-embedded tissue sections obtained from
the 50 ESCC patients. The tissue sections were deparaffinized and
boiled in 0.01 mol/l sodium citrate buffer (pH=6.0) in a 1,000-Watt
microwave oven for 10 min to retrieve cell antigens. Rabbit
monoclonal anti-EHMT1 was used as primary antibody (1:100 dilution;
Abcam, Cambridge, UK). The tissue sections were
immunohistochemically stained using the avidin-biotin-peroxidase
method and counterstained with hematoxylin.
Evaluation of immunostaining for
EHMT1
The staining was scored by two independent
investigators who were blinded to the patient outcomes. The
sections were evaluated at low magnification (×100) to identify
areas of even EHMT1 staining. The percentage of positively stained
cells was calculated among >400 tumor cells. The expression of
EHMT1 was scored by staining intensity and percentage of cells
exhibiting nuclear staining as follows: negative expression,
negative or weak staining in all the tumor cells or moderate
staining in <25% of the tumor cells; positive expression,
moderate or strong staining in >25% of the tumor cells. Any
discrepancies were jointly re-evaluated by the investigators and a
consensus was obtained.
Quantitative reverse-transcription
polymerase chain reaction (qRT-PCR)
Total RNA was extracted from ESCC cells using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions and reverse-transcribed using the
SuperScript III RT-PCR system (Invitrogen) according to the
manufacturer’s protocol. cDNA (1 μl) was used for PCR
amplification. The cDNA templates included the GLP fragment (179
bp) with primers 5′-TTC AAG CAA TTT TCC TGT CT-3′ (forward) and
3′-CAT TAA TCC CAG CAC TTT GG-5′ (reverse) and the internal control
β-actin gene (314 bp) with primers 5′-TCC TGT GGC ATC CAC GAA
ACT-3′ (forward) and 3′-GAA GCA GCA TTT GCG GTG GAC GAT-5′
(reverse). The thermal cycle reactions were performed at annealing
temperatures of 57.5°C for EHMT1 and 55.5°C for β-actin, for a
total of 26 cycles. The products were electrophoresed on 1.2%
agarose gel containing ethidium bromide and visualized and
photographed under UV light.
Statistical analysis
Data were analyzed with the SPSS software, version
17.0 (SPSS Inc., Chicago, IL, USA) and are representative of at
least three independent experiments with similar results. The
correlation between the EHMT1 expression and the
clinicopathological parameters was assessed by the χ2
test and bivariate analysis. Survival curves were calculated by the
Kaplan-Meier product-limit estimate method and examined with the
log-rank test. The significance of multiple predictors of survival
was assessed by the Cox regression analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
EHMT1 expression in esophageal squamous
preinvasive lesions and ESCC
The immunohistochemical staining for EHMT1 was most
prominent in the nuclei of the ESCC cells. EHMT1 was also strongly
expressed in the proliferative esophageal epithelium. However, the
expression of EHMT1 was low in the normal esophageal epithelial
cells (Fig. 1A). The expression of
EHMT1 differed significantly between ESCC and the normal esophageal
squamous epithelium (Table
II).
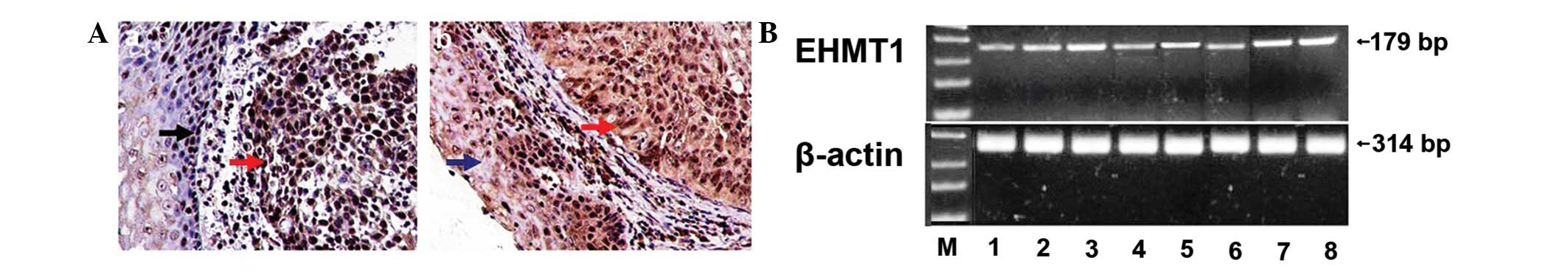 | Figure 1.EHMT1 expression in esophageal
squamous cell carcinoma (ESCC). (A) Immunohistochemical results:
(A-a) EHMT1 expression in normal esophageal epithelium (black
arrow) and ESCC (red arrow); (A-b) EHMT1 expression in atypical
hyperplastic esophageal epithelium (blue arrow) and ESCC (red
arrow); bar, 20 μm. (B) RT-PCR analysis of EHMT1 in ESCC.
mRNA expression levels in: lane M, marker; lane 1, normal
esophageal tissue; lane 2, well- and moderately differentiated ESCC
(G1–2); lane 3, poorly differentiated ESCC (G3–4); lane 4, tumor
depth of invasion T1–2; lane 5, tumor depth of invasion T3–4; lane
6, lymph node metastasis-negative; lane 7, lymph node
metastasis-positive (N1); lane 8, lymph node metastasis-positive
(N2–3). β-actin was used as internal control. |
 | Table II.EHMT1 expression in esophageal
squamous cell cancer and normal esophageal squamous epithelium. |
Table II.
EHMT1 expression in esophageal
squamous cell cancer and normal esophageal squamous epithelium.
| Group | No. | EHMT1 expression
| χ2 | P-value |
|---|
| Positive | Negative | Positivity rate
(%) |
|---|
| Esophageal
cancer | 50 | 26 | 24 | 52.0 | 9.361 | 0.002a |
| Normal esophageal
epithelium | 46 | 10 | 36 | 21.7 | | |
The RT-PCR analysis indicated that the expression of
EHMT1 was significantly stronger in cancer tissues compared to the
corresponding normal tissues. At the RNA level, the overexpression
of EHMT1 in human ESCC tissues was correlated with tumor grade,
depth of invasion and lymph node metastasis (Fig. 1B). These findings were in
concordance with the immunohistochemical data.
Association between EHMT1 expression and
clinicopathological findings of ESCC
The correlation between the expression of EHMT1 and
the clinicopathological characteristics of ESCC patients is
summarized in Table I. The results
demonstrated that the expression of the EHMT1 protein was not
correlated with age and gender. However, EHMT1 expression was
significantly correlated with tumor grade, depth of invasion, lymph
node metastasis and tumor stage (P<0.05).
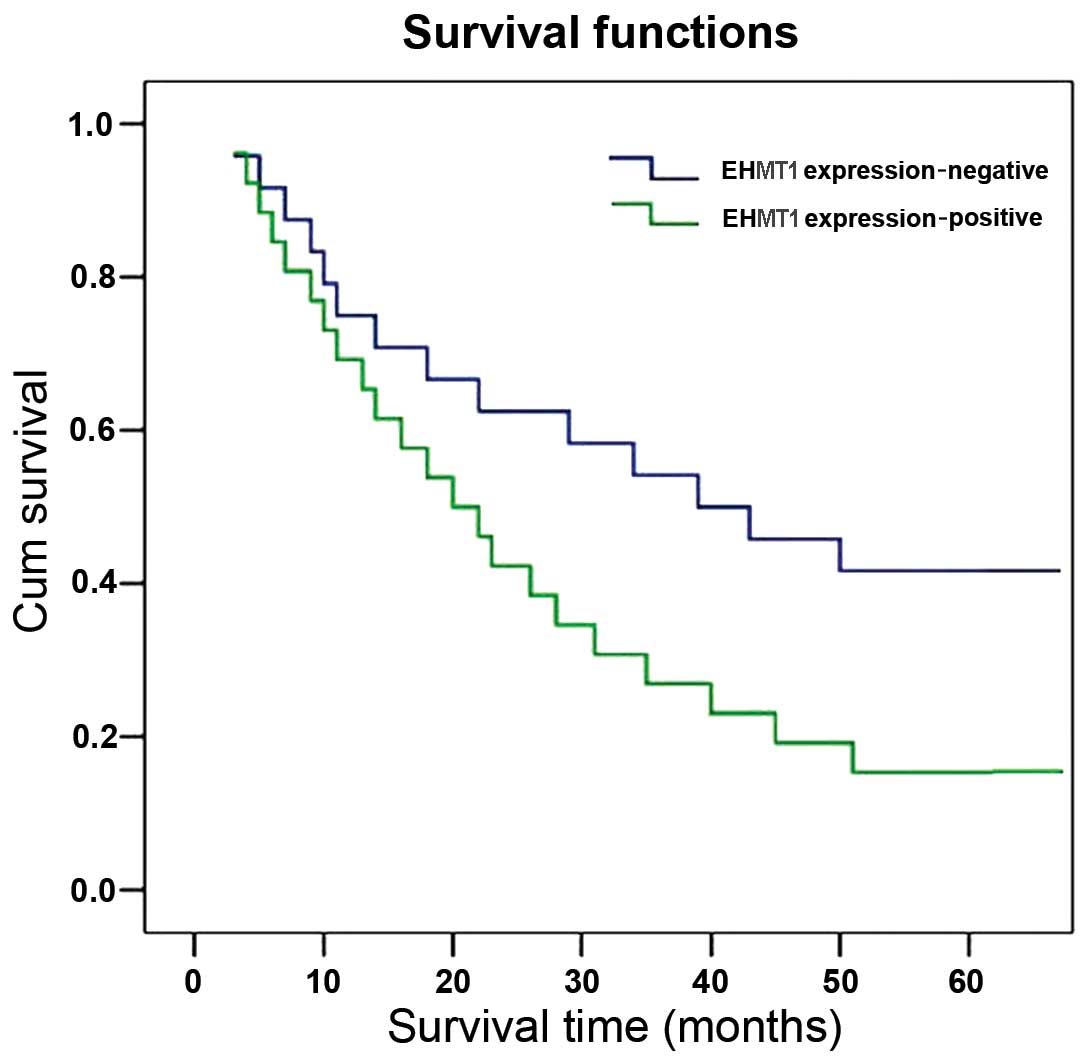
Survival analysis and prognostic
significance of EHMT1 expression
The correlation between survival and EHMT1
expression was evaluated in the 50 cases with operable ESCC. A
significant difference was observed when the patient cohort was
stratified by the level of EHMT1 expressions. Of note, patients
with ESCC who exhibited high expression levels of EHMT1 had a lower
survival rate compared to that of patients without high EHMT1
expression (χ2=3.922; P=0.048) (Fig. 2). The multiple predictors of
survival assessed by the Cox regression analysis identified EHMT1
expression as an independent prognostic factor for overall survival
in ESCC patients (hazard ratio=1.605; 95% confidence interval:
1.037–2.485; P=0.034) (unpublished data).
Discussion
EHMT1 is a subunit of histone methyltransferases.
Although previous studies identified EHMT1 as a regulator of tumor
progression (9), the
clinicopathological significance of EHMT1 expression in ESCC has
not been fully elucidated. In the present study, EHMT1 expression
was shown to be higher in ESCC and the proliferative esophageal
epithelium compared to that in normal esophageal tissues and its
expression was correlated with poor prognosis. To the best of our
knowledge, this is the first study on the association between EHMT1
and ESCC.
Histone H3K9 methyltransferases (EHMT1 and EHMT2),
originally identified by their ability to interact with histones in
cells through the methylation of heterochromatin, are considered to
be crucial in maintaining the balance state of cells. EHMT1/EHMT2
may catalyze the dimethylation of H3K9, a process that represses
gene transcription in euchromatic cells (10,11).
A previous study by Bird indicated that the methylation of H3K9 may
exert an effect on DNA methylation, directly or indirectly
(12). H3K9 methylation directs
CpNpG methylation and was shown to be necessary for DNA methylation
in fungi (13,14). DNA methylation is crucial in the
regulation of gene expression and chromatin organization within
normal eukaryotic cells. EHMT1/EHMT2 is also required for the
maintenance of DNA methylation at endogenous retrotransposons,
imprinted loci and other genes present in differentiated cells
(15). A recent study also
revealed that EHMT1 is required for the DNA methylation of cancer
germ-line and genomic DNA in mouse embryonic stem cells (16). The direct effect reverse to
methylation is demethylation; however, histone methylation is
considered to be irreversible and, once it occurs, such a change
may be present for the entire cell life span and may even be
transferred to the next generation. In cancer cells, alterations of
the global patterns of DNA methylation are considered to be normal
(17). DNA hypermethylation of
certain CIMP (colorectal tumors with a CpG island methylator
phenotype)-associated gene promoters was detected during the early
stages of colorectal tumorigenesis (18). Moreover, promoter DNA methylation
of the ERG gene is a common event in human prostate cancer
(19). It was also proven that
aberrant DNA methylation is associated with chronic myelogenous
leukemia progression and that DNA methylation may be a marker
associated with imatinib resistance (20). These results suggested an
association between histone methylation and cancer progression.
In our study, we selected 50 cases of patients with
operable ESCC. The results confirmed that the expression of EHMT1
was higher in ESCC and the atypical hyperplastic esophageal
epithelium compared to that in normal esophageal tissues, by using
immunohistochemistry and RT-PCR. EHMT1 may be a key molecular
marker for the progression of human ESCC. Moreover, EHMT1
expression was also associated with tumor grade, depth of invasion,
lymph node metastasis and tumor stage.
It was also verified that the EHMT2 expression
pattern in ESCC was similar to the EHMT1 expression pattern, using
immunohistochemistry with an anti-EHMT2 antibody (1:80 dilution; BD
Biosciences, Franklin Lakes, NJ, USA (unpublished data). This
confirmed that in ESCC or atypical hyperplastic esophageal squamous
epithelia, the expression level of each histone methyltransferase
subunit may rise aberrantly and the process of histone methylation
becomes disordered.
Our study also demonstrated that the expression of
EHMT1 was positively correlated with lower survival in ESCC.
Therefore, the expression of EHMT1 may serve as a prognostic factor
for predicting the outcome of ESCC patients. Thus, the detection of
EHMT1 aberrations may be a useful biomarker to identify ESCC
patients with poor prognosis.
In conclusion, our data indicated that EHMT1
expression is elevated in human ESCC and may play a significant
role in human ESCC progression. Our analysis of clinical studies
also demonstrated that the expression of EHMT1 may be a useful
biomarker predicting ESCC progression and prognosis.
Acknowledgements
This study was supported by the
National Nature Science Foundation of China (no. 81272605).
References
|
1.
|
Thallinger CM, Kiesewetter B, Raderer M
and Hejna M: Pre- and postoperative treatment modalities for
esophageal squamous cell carcinoma. Anticancer Res. 11:4609–4627.
2012.PubMed/NCBI
|
|
2.
|
Tachibana M, Ueda J, Fukuda M, Takeda N,
Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T and Shinkai Y:
Histone methyltransferases G9a and GLP form heteromeric complexes
and are both crucial for methylation of euchromatin at H3-K9. Genes
Dev. 19:815–826. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Collins R and Cheng X: A case study in
cross-talk: the histone lysine methyltransferases G9a and GLP.
Nucleic Acids Res. 38:3503–3511. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Chin HG, Esteve PO, Pradhan M, Benner J,
Patnaik D, Carey MF and Pradhan S: Automethylation of G9a and its
implication in wider substrate specificity and HP1 binding. Nucleic
Acids Res. 35:7313–7323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Rathert P, Dhayalan A, Murakami M, Zhang
X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X and Jeltsch
A: Protein lysine methyltransferase G9a acts on non-histone
targets. Nat Chem Biol. 4:344–346. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Pless O, Kowenz-Leutz E, Knoblich M,
Lausen J, Beyermann M, Walsh MJ and Leutz A: G9a-mediated lysine
methylation alters the function of CCAAT/enhancer-binding
protein-beta. J Biol Chem. 283:26357–26363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Fritsch L, Robin P, Mathieu JR, et al: A
subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a,
GLP, and SETDB1 participate in a multimeric complex. Mol Cell.
37:46–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Sampath SC, Marazzi I, Yap KL, Sampath SC,
Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM,
Chait BT and Tarakhovsky A: Methylation of a histone mimic within
the histone methyltransferase G9a regulates protein complex
assembly. Mol Cell. 27:596–608. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Lee SH, Kim J, Kim WH and Lee YM: Hypoxic
silencing of tumor suppressor RUNX3 by histone modification in
gastric cancer cells. Oncogene. 28:184–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Purcell DJ, Khalid O, Ou CY, et al:
Recruitment of coregulator G9a by Runx2 for selective enhancement
or suppression of transcription. J Cell Biochem. 113:2406–2414.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Purcell DJ, Jeong KW, Bittencourt D, et
al: A distinct mechanism for co-activator versus corepressor
function by histone methytransferase G9a in transcriptional
regulation. J Biol Chem. 286:41963–41971. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Bird A: Methylation talk between histones
and DNA. Science. 294:2113–2115. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Tamaru H, Zhang X, McMillen D, et al:
Trimethylated lysine 9 of histone H3 is a mark for DNA methylation
in Neurospora crassa. Nat Genet. 34:75–79. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Jackson JP, Lindroth AM, Cao X and
Jacobsen SE: Control of CpNpG DNA methylation by the KRYPTONITE
histone H3 methyltransferase. Nature. 416:556–560. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Ikegami K, Iwatani M, Suzuki M, et al:
Genome-wide and locus-specific DNA hypomethylation in G9a deficient
mouse embryonic stem cells. Gene Cells. 12:1–11. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Chen L, Li Z, Zwolinska AK, et al: MDM2
recruitment of lysine methyltransferases regulates p53
transcriptional output. EMBO J. 29:2538–2552. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Candelaria M, de la Cruz-Hernandez E,
Taja-Chayeb L, et al: DNA methylation-independent reversion of
gemcitabine resistance by hydralazine in cervical cancer cells.
PloS One. 7:e291812010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Ibrahim AE, Arends MJ, Silva AL, Wyllie
AH, Greger L, Ito Y, Vowler SL, Huang TH, Tavare S and Murrell A:
Sequential DNA methylation changes are associated with DNMT2B
over-expression in colorectal neoplastic progression. Gut.
60:499–508. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Schwartzman J, Mongoue-Tchokote S, Gibbs
A, et al: A DNA methylation microarray-based study identifies ERG
as a gene commonly methylated in prostate cancer. Epigenetics.
6:1248–1256. 2011. View Article : Google Scholar
|
|
20.
|
Jelinek J, Gharibyan V, Estecio MR, et al:
Aberrant DNA methylation is associated with disease progression,
resistance to imatinib and shortened survival in chronic
myelogenous leukemia. PloS One. 6:e221102011. View Article : Google Scholar : PubMed/NCBI
|