Contents
Introduction
Aptamers and SELEX
Clinical applications of aptamers in cancer
Conclusion and future perspectives
Introduction
Cancer is one of the leading causes of mortality
worldwide. A statistical survey conducted in 2008 reported ∼12.7
million cancer cases and 7.6 million cancer-related deaths
worldwide (1). An estimated total
of 1,529,560 new cancer cases and 569,490 cancer-related deaths
occurred in 2010 (2). Over the
last several decades, significant investments and efforts have
focused on battling cancer, making it a top priority in the
pharmaceutical industry and the global Institutes of Health. With
the development of novel anticancer agents with greater efficacy
and fewer side effects, accurate and efficient delivery of these
agents to the tumor sites is of utmost importance (3). In order to design an ideal and
effective anticancer drug, several aspects have to be optimized
simultaneously, including toxicity, side effects, targeting,
delivery and controlled release. Although a large number of
anticancer drugs have been approved by the Food and Drug
Administration (FDA) (4), the
majority are not molecularly targeted, which means that developing
novel drugs with higher efficacy but less toxicity and fewer side
effects is a major aim for cancer therapy.
Aptamers are small single-stranded (ss) DNA or RNA
oligonucleotides (with a molecular weight of 5-40 kDa), which fold
into well-defined three-dimensional (3D) structures due to various
intramolecular interactions (5).
Aptamers bind onto their targets with high affinity and specificity
and are traditionally generated through systematic evolution of
ligands by exponential enrichment (SELEX) (6,7).
Aptamers have been selected against a wide range of targets,
similar to antibodies, which may be crafted to bind onto multiple
and different targets (5,8), such as proteins, phospholipids,
sugars, nucleic acids and whole cells. However, in comparison to
antibodies, aptamers possess the following advantages: i) they are
of small size and lower complexity, with low immunogenicity; ii)
they are easier to synthesize and modify in vitro; iii) they
exhibit higher affinity and specificity for their targets; iv)
their structural flexibility enables aptamers to bind onto hidden
epitopes, which cannot be targeted by antibodies (9); and v) they exhibit higher stability,
with the potential to be stored easily until use (Table I). Over the last two decades, since
aptamers were first selected through SELEX, they have quickly
emerged as a novel and powerful class of ligands with an excellent
potential for diagnostic and therapeutic applications (10). As the use of aptamers has been
extended from the basic biology of cell processes and gene
regulation to therapeutic and diagnostic applications, numerous
patented aptamers are currently being tested in clinical trials and
were recently reviewed (9).
Pegaptanib (Pfizer, New York, NY, USA / EyeTech, Melville, NY,
USA), an aptamer that binds to human vascular endothelial growth
factor (VEGF), has been approved by the FDA for clinical use in
treating age-related macular degeneration (AMD) and has already
been proven to be a milestone for the applications of aptamer
technology. In this review, we focused on recent developments in
aptamer technology, as well as their applications in cancer
diagnosis and therapy.
 | Table I.Advantages of nucleic acid
aptamers. |
Table I.
Advantages of nucleic acid
aptamers.
| No. | Advantages |
|---|
| 1 | High binding
affinity and specificity |
| 2 | Ease of synthesis
and modification |
| 3 | Low immunogenicity
and no toxicity |
| 4 | Small size and
stable structure |
| 5 | Structural
flexibility and ease of storage |
Aptamers and SELEX
Overview of aptamers
By definition, aptamers are synthetic, highly
structured, small, typically <100 mer, ssDNA or RNA ligands. The
term ‘aptamer’ means ‘to fit’ (aptus) in Latin (6), which indicates two important
properties of aptamers: i) their ability to fold into complex
tertiary structures and recognize their targets with high affinity
(low nM to high pM equilibrium dissociation constants); and ii)
their specificity, somewhat analogous to antigen-antibody
interactions. Using this technique, a number of aptamers that
specifically recognize targets, such as metal ions (11–13),
organic dyes and amino acids (14–17),
antibiotics (18,19) and peptides (20,21),
as well as proteins of various sizes and functions (6,22,23),
whole cells (24–27), whole organisms (28), viruses (29) and bacteria (30), have been obtained (31–33).
The SELEX process is based on the ability of these small
oligonucleotides to fold into unique 3D structures that interact
with a target with high specificity and affinity through such
interactions as van der Waals surface contacts, hydrogen bonding
and base stacking. Starting with a library containing random RNA
sequences with ∼10 (34–36) varieties of RNA molecules, in
vitro binding, elution and reverse polymerase chain reaction
(PCR) amplification techniques allow for the selection of RNA
molecules that efficiently bind to a specific receptor or ligand
with high affinity (37,38). Conceptually, the marked specificity
and high affinity of aptamers to a wide variety of targets, coupled
with the ease of design and molecular engineering, as mentioned
above, have made them broadly popular and highly suitable for
development as clinical diagnostic and therapeutic agents in cancer
research (39–41). Pegaptanib (Pfizer/Eyetech), an
aptamer targeting VEGF, has already been approved by the FDA for
clinical use in treating AMD (42). A variety of aptamers against other
molecular targets are currently under clinical investigation
(10), with several more in the
pipeline.
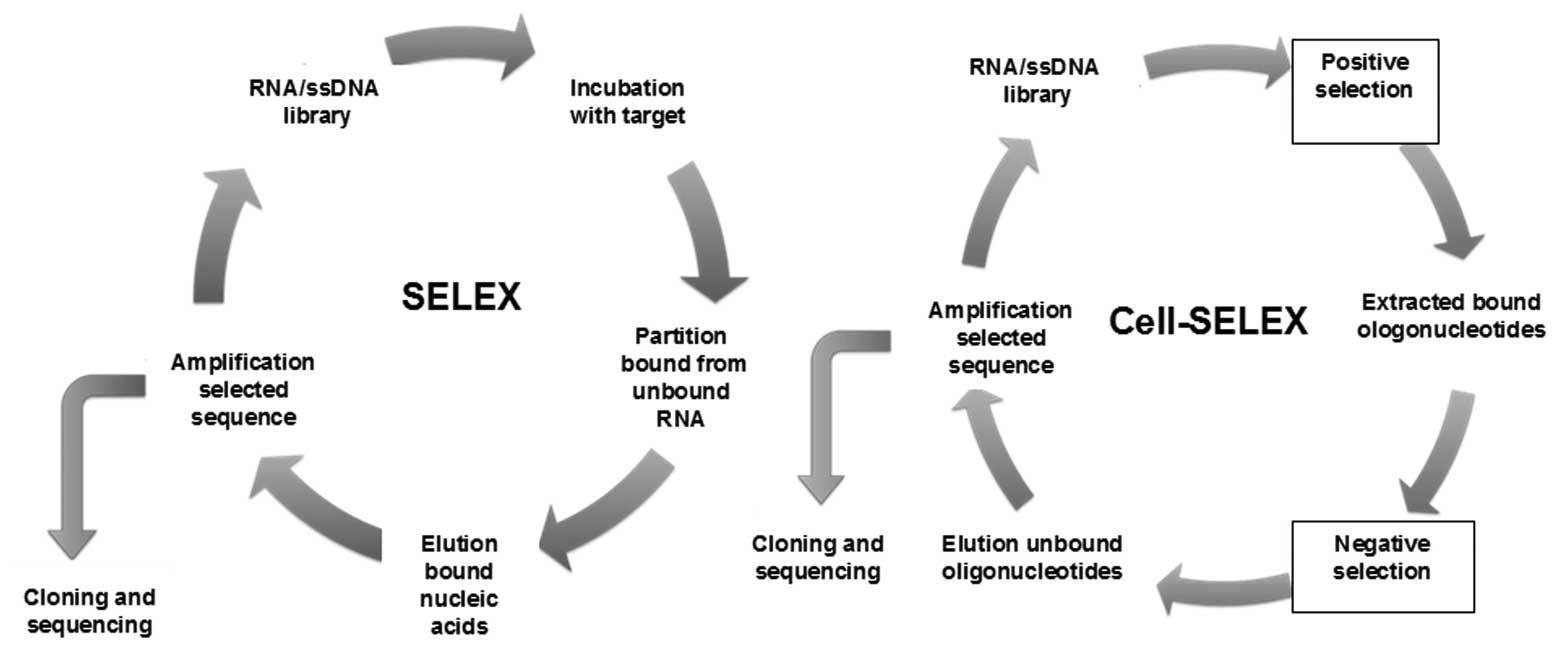
In vitro SELEX
Aptamers are identified through an in vitro
process, which specifically isolates aptamers for a target of
interest involving iterative rounds, termed as SELEX. Briefly, for
the SELEX process, as shown in Fig.
1, a random aptamer or oligonucleotide library pool (DNA or
RNA) is incubated with the target of interest, with heating and
cooling to promote formation of stable structures. After washing,
the bound sequences are eluted and incubated with a control target
(if negative selection is required) to remove sequences that
exhibit recognition with the control as well. The protein-bound
aptamers are then recovered. These sequences are amplified with PCR
or reverse transcription-PCR. ssRNA or DNA sequences representing
the recovered sequences are then generated from these PCR products
and used in the subsequent selection round. The process is repeated
until the pool is enriched for sequences that specifically
recognize the target. The enriched pool is cloned and then
sequenced to obtain the individual sequences known as aptamers
(7). As this is an in vitro
process, the selection conditions may be manipulated to obtain
aptamers with properties desirable for a particular purpose, i.e.,
aptamers that bind to a target at different temperatures and buffer
compositions may be used for different purposes, or modified
oligonucleotide bases which may be introduced to enhance
stability.
Cell-SELEX
The cancer cell surface represents a complex
environment with a vast array of potential targets of diagnostic
and therapeutic interest; therefore, the aptamers selected by SELEX
may exhibit low or no affinity towards these targets, due to
shielding of the aptamer binding domain. To overcome this
hindrance, a process called Cell-SELEX (cell-based selection of
aptamers specific to cancer cells) has been reported to isolate
aptamers from living cells as targets, such as cancer cells
(24). In Cell-SELEX, instead of
using purified protein targets, whole living cells are used as the
targets. To generate aptamers that specifically target cancer
cells, a library of ssDNA is used (24). This library, which has a random
sequence of 30–40 bases and a region flanked by primer sequences of
18–20 bases, is incubated with the target cells. The library and
forward primers are labeled with a fluorophore, so that the sense
strand of the PCR product for each round, which serves as the the
library for the next round, is also fluorescently labeled, enabling
the entire process to be monitored by flow cytometry. After
washing, the DNA sequences bound to the target cell surface are
collected and then incubated with the negative control cells. All
the DNA sequences that bind to the negative control cells are
removed. To avoid recognition of normal cells, the aptamers bound
to these non-specific proteins are removed. The remaining sequences
are amplified for the next round of selection. Generally, ∼20
rounds of Cell-SELEX are required to isolate aptamers with the
highest selective affinity to the target cells.
A panel of aptamer probes has been successfully
selected by Cell-SELEX (Fig. 1)
for several types of cancer cells, including lymphocytic leukemia,
myeloid leukemia, liver cancer, small- and non-small-cell lung
cancer cells (25–27). All these aptamers were found to
exhibit high affinity and specific selectivity for their targets.
Those results illustrated how aptamers may be selected without
specific knowledge of the molecular signature of the cell or the
number or type of proteins on the cell surface. Moreover, it is
possible to profile the molecular characteristics of the target
cancer type, which has been a main objective in the development of
cell-based aptamers for use in cancer diagnosis and treatment. The
applications of aptamers for cancer diagnosis and therapy through
cancer cell detection, cancer cell imaging and targeted drug
delivery are discussed below.
Clinical applications of aptamers in
cancer
Cancer is a class of diseases that originate from
mutations and alterations at the genetic and, subsequently, the
molecular level. However, at the precancerous and early stages of
cancer, detection is limited due to the low number of tumor cells
and molecular markers. Furthermore, to a certain degree, detection
may exert a negative effect on cancer therapy. Therefore,
determining the molecular characteristics of cancer, particularly
the characteristic proteins associated with a specific type of
cancer, may prove beneficial for clinical diagnosis and therapy
(43), particularly during the
early stage of tumorigenesis and cancer progression. By contrast,
the traditional diagnostic methods, such as computed tomography
(CT), magnetic resonance imaging (MRI) and positron emission
tomography with radiolabeled 2-fluoro-deoxyglucose, only assess
anatomical changes or non-specific glucose metabolism (44).
Aptamers selected by SELEX have shown the high
sensitivity and specificity required of bioprobes used for the
accurate and early diagnosis of tumors. With their high-recognition
specificity, aptamers can accurately distinguish between different
types and even subtypes of cancer cells (45). Moreover, aptamers targeting
wide-range receptors have been extensively adapted for disease
diagnosis and the delivery of therapeutic moieties into cells. In
2000, Hicke et al (46)
coined the term ‘escort aptamers’ and demonstrated that aptamers
may be used as tools to deliver therapeutic and diagnostic
reagents. A wide range of molecular species have been transported
intracellularly using these nanoscale delivery vehicles that
include small interfering RNAs (siRNAs), drugs, toxins, enzymes,
photodynamic molecules and radionuclides. Intracellular delivery of
these compounds may be achieved by employing aptamers for
internalized cell surface receptors. Furthermore, to achieve
delivery selectivity to the diseased cells, the cell surface
receptor selected is either a tumor biomarker or an aberrantly
expressed surface protein unique to the target cell.
Aptamer-mediated targeted detection and delivery increases the
positive prognosis and sensibility of early diagnosis,
correspondingly decreasing treatment costs due to the reduced
incidence of late-stage cancers and increased number of therapeutic
options, while reducing the side effects. The examples of
aptamer-mediated clinical applications in cancer, which are
discussed below, are summarized in Table II.
 | Table II.Clinical application of aptamers in
cancer. |
Table II.
Clinical application of aptamers in
cancer.
| Aptamers | Target | Nucleic acid | Clinical
applications | Refs. |
|---|
| A9, A10 | PSMA | RNA | Prostate cancer
imaging and therapy | (67–74) |
| TTA1 | Tenascin C | RNA | Cancer imaging | (44) |
| AS1411 | Nucleolin | DNA | Cancer imaging and
therapy | (79,80) |
| AptA, AptB | Mucin 1 | DNA | Cancer imaging and
therapy | (84–86) |
| NOX-A12 | CXCL12 | RNA | Hematological
cancer therapy | (88,89) |
Aptamer-based cancer cell detection and
imaging
The binding affinity of aptamers to their targets
enables the detection of cancer cells at low levels, resulting in
an early and sensitive diagnosis. In addition, DNA molecules have
the advantage of predictable structure and easy site-specific
chemical modification. Therefore, aptamers may be quickly and
reproducibly synthesized with fluorescent molecules and
nanoparticles (NPs) conjugated for targeted cells and tissue
detection and imaging.
It is well known that gold nanomaterials possess
unusual optical and electronic properties, high stability and
biological compatibility, controllable morphology and size
dispersion, and easy surface functionalization (47–51)
from the standpoints of engineering and application.
Aptamer-conjugated gold NPs (Apt-AuNPs) provide a powerful platform
to facilitate targeted recognition and detection. By using
Apt-AuNPs, a colorimetric assay for the direct detection of cancer
cells has been developed (52).
Aptamers specific to their targets on the cell membrane surface
were conjugated to gold NPs. These Apt-AuNPs were then targeted to
assemble on the cell surface membrane through the recognition of
the aptamers to their targets. The assembly of the Apt-AuNPs around
the cell surface causes a shift in the extinction spectra of the
particles when the AuNPs are in sufficient proximity for their
surface plasmon resonances to overlap. This assay exhibits a
significantly high sensitivity, with a detection limit for simple
absorbance of just 90 cells. When the number of target cells
increases to 1,000, the color change reaction may be observed by
the naked eye. This method demonstrates the potential of
aptamer-based analysis for rapid, simple, direct and sensitive
detection of cancer cells in clinical diagnosis. In addition, using
the two-photon scattering (TPS) technique, Lu et al
(53) captured the signal from
Apt-AuNP-based cell detection, resulting in the highly selective
and sensitive detection of the SK-BR-3 breast cancer cell line at a
level of 100 cells/ml, using multifunctional (monoclonal
anti-HER2/c-erb-2 antibody and S6 RNA aptamer) AuNP conjugates.
Oval-shaped AuNPs were introduced instead of spherical AuNPs. As
the particle aspect ratio increased, the TPS intensity change
became higher, resulting in improved sensitivity (53).
The research on the applications of
aptamer-conjugated fluorophores has advanced rapidly over the last
few years (54), including the use
of quantum-dot (QD)-conjugated aptamers to perform multiplex
detection of cancer cells (44).
In that study, three aptamers (TTA1, AS1411 and Muc1) were
conjugated to QDs with emission wavelengths of 605, 655 and 705 nm,
respectively; for the confocal microscopic analysis of multiplex
imaging of cancer, healthy and disease cell lines were incubated
with each QD-conjugate. As expected, fluorophore-labeled aptamers
were able to detect and differentiate between different types of
cancer cells and produced a visible fluorescence signal in the
presence of target cells, which indicated that aptamers have high
potential as a clinical diagnostic tool.
Gold nanorods (NRs) have been utilized as a platform
for multiple aptamer immobilization to detect cancer cells
(55). Through covalent linkages
of fluorophore-labeled aptamers on the NR surface, ≤80
fluorophore-labeled aptamers may be attached on a single NR,
causing a 26-fold higher affinity and >300-fold higher
fluorescence signal. In this case, the use of NRs as a nanoplatform
for multivalent binding of aptamers increases the signal as well as
and binding strength of these aptamers in cancer cell recognition.
As a consequence of the selective recognition properties of
aptamers, this technique may be used in clinical detection to
enhance binding affinity and signaling strength when the
concentration of target cells is relatively low.
With increasing detection sensitivity, a dye-doped
silica NPs-based method for sensitive and rapid detection of cancer
cells was constructed (56). To
achieve specific recognition, the biotinylated aptamers are
immobilized onto the surfaces of neutravidin-coated fluorescent NPs
(FNPs) to form aptamer-conjugated FNPs (Apt-FNPs), which have
demonstrated low background signal, high signal enhancement and
efficient functionalization. The FNPs are then functionalized with
polyethylene glycol (PEG) to prevent non-specific interactions with
neutravidin and allow universal binding with biotinylated
molecules. Multiplexed cancer cells may also be detected by
employing a combination of fluorescence resonance energy transfer
of FNPs and the conjugation of different aptamers (57). Furthermore, single-, dual- and
triple-dye-doped silica NPs were prepared according to this
technology (58). This method
provided an additional potential candidate for cancer cell
imaging.
In addition to cancer cell detection, improved
molecular imaging techniques together with novel near-infrared
fluorescence probes, which may also be attached to aptamers, allow
in vivo multiparameter 3D exploration of tumors, even deep
inside the tissue (59).
Aptamer-mediated technology is likely to gain applications in
clinical cancer diagnosis, with cancer cell detection as well as
cancer cell and tissue visualization. Fluorescein isothiocyanate
(FITC)-labeled aptamers were used for tissue staining and imaging
under confocal microscopy (60).
Compared to the negative control (FITC-labeled unselected library),
targeted tissue was recognized with fluorescence signal following
incubation with FITC-labeled aptamers, which indicated aptamers as
potential molecular probes for cancer visualized diagnosis. Other
methods were also developed for labeling aptamers for flow
cytometry applications (23,61).
Flow cytometric analysis was employed to compare binding of a
FITC-labeled DNA aptamer with a complex of mouse anti-human
neutrophil elastase (HNE) antibody and FITC-labeled rat antimouse
antibody to HNE-labeled beads. The results revealed that aptamers
and antibodies were equally efficient in HNE detection (61). Further studies demonstrated that,
based on aptamers' high specificity of target recognition,
fluorescein-labeled aptamers were selective in flow cytometric
experiments for binding to human recombinant CD4-stained mouse T
cells expressing human CD4, but did not detect control mouse T
cells lacking human CD4 (23).
For cancer cell imaging, the antiprostate-specific
membrane antigen (PSMA) aptamer was used. Due to its high affinity
and specificity, this aptamer is able to mediate cell-specific
endocytosis of the NPs upon molecular recognition between the
aptamer and its receptors. The synthesized aptamer-functionalized
QDs and super-paramagnetic iron oxide NPs were used for the
recognition of cell receptors as well as optical imaging or MRI
(62,63). The same concept was also applied to
functionalize gold NPs with anti-PSMA aptamer for CT. The results
demonstrated that the functionalized gold NPs may enhance the CT
image by 4-fold (64). For further
study in vivo, molecular imaging of tumors inside mice was
performed. TD05, an aptamer that specifically binds Ramos cells,
was modified with fluorophores, successfully exhibiting aptamer
fluorescence overlapping the tumor site in a mouse cancer model
under a whole-animal imaging system.
Overall, the studies discussed in this section
helped expedite the process of using aptamers in the clinical
setting for cancer diagnosis.
Aptamer-mediated therapy for cancer
Several aptamers, including pegaptanib and REG1,
have already been used for clinical treatment (42).
From a therapeutic point of view, a useful
therapeutic agent may stop being used when a more cost-effective,
safer or simply generic product is released. Aptamers, targeting a
broad range of receptors, with numerous ideal properties, such as
high specificity and affinity, easy modification, easy synthesis,
stability, low immunogenicity and toxicity, have emerged as
promising therapeutic reagents.
PSMA, a transmembrane protein that is highly
expressed in human prostate cancer cells and the vascular
endothelium, is an attractive target, as it is abundantly expressed
on the surface of prostate cancer cells, is known to internalize
certain ligands, possesses enzymatic properties as a glutamate
carboxypeptidase and is a verified target of a clinically applied
imaging agent for prostate cancer (65). In 2002, a 2′-fluorpyrimidine RNA
aptamer screen was performed and two stabilized RNA aptamers, A9
and A10, which were capable of binding PSMA with low nanomolar
affinity, were first identified (66). These aptamers have been applied to
several anticancer preclinical targeting studies of NPs, toxins,
therapeutics and siRNAs. siRNAs are double-stranded
oligonucleotides (21–23 bp) that bind specifically to cognate mRNA
and promote its degradation, effectively decreasing the expression
of the target gene product. To develop its ability, a siRNA has to
be efficiently delivered to the intracellular space in vivo.
For this purpose, an aptamer-siRNA chimera construct was generated
to combine the RNA aptamer A10, specific for PSMA, with a siRNA
against the survival factors polo-like kinase 1 (Plk-1) and B-cell
lymphoma 2 (Bcl-2), which are two antiapoptotic genes involved in
cell survival and are commonly overexpressed in human tumors. After
being injected at the site of a tumor, aptamer-Plk-1 or Bcl-2 siRNA
chimeras were shown to deliver the siRNAs effectively to
PSMA-expressing cells, inducing apoptosis in targeted cancer cells
and tumor regression in animal models of prostate cancer (67).
An improved treatment of prostate cancer was
aptamer-short hairpin RNA (shRNA) chimera, a PSMA-targeted
aptamer-conjugated shRNA, which was used to attenuate DNA-repair
pathways, resulting in tissue-selective sensitization to ionizing
radiation (IR) therapy (68).
A10-3-shRNAs targeting the catalytic subunit of the DNA protein
kinase (DNAPK) caused a selective reduction of DNAPK RNA and
protein in PSMA-positive cells, xenografts and human prostatic
tissues. Intratumorally, the administration of aptamer-targeted
DNAPK shRNAs combined with IR significantly and specifically
enhanced PSMA-positive tumor response to IR, leading to decreased
DNA-repair and cancer cell survival following chemotherapy or
radiation treatment.
The application of antitumor agents, such as
docetaxel, dextran, cisplatin, daunorubicin and doxorubicin, is
limited due to their side-effects and toxicity. The first report of
targeted drug delivery with NP-aptamer bioconjugates was described
by Farokhzad et al (69).
The anti-PSMA A10 aptamer, or its truncated version A10-3, was used
to target NPs, demonstrating that the A10-3 aptamer may be used to
target poly(lactic acid)-block PEG copolymer NPs to PSMA-positive
prostate cancer cells (69).
Furthermore, the A10-3 aptamer was modified by
poly(D,L-lactic-co-glycolic acid; PLGA) NPs to deliver docetaxel
(Dtxl); an antimitotic chemotherapy drug (70) to prostate tumors in vivo.
When compared with non-targeted NPs that lacked the anti-PSMA
aptamer, the Dtxl-NP-Apt conjugates mediated targeted uptake and
controlled release of drugs, resulting in potent efficacy and
reduced toxicity after intratumoral injection (71). In an extensive study, following
systemic administration, A10-3 aptamer induced tumor accumulation
of PLGA NPs by 3.7-fold when compared to the non-targeted particle
(72). Further investigations
demonstrated that the NP-aptamer bioconjugates may be precisely
engineered and formulated by controlling size, composition,
polydispersity, aptamer density and drug loading, thereby resulting
in the desired functions and biodistribution required for further
clinical development (72–74). Another polymeric formulation has
been used to deliver cisplatin to PSMA expressing tumors, whereas a
dosage of 0.3 mg/kg of aptamer-targeted cisplatin NPs was more
efficacious compared to a 1 mg/kg of free cisplatin (75). These preclinical studies
demonstrated the potential of aptamers as therapeutic reagents to
efficiently deliver drugs to targeted cancers. In addition, these
results also indicated aptamers as targeting ligands to alter the
biodistribution, tumor uptake and tumor retention of therapeutic NP
reagents.
The AS1411 aptamer against nucleolin, a Bcl-2
mRNA-binding protein expressed on the surface of several cancer
cell types, including those associated with acute myeloid leukemia
(AML), has demonstrated promising antineoplastic effects, which may
be attributed to interference with the multitude of
nucleolin-mediated cell processes. AS1411, a DNA aptamer, was shown
to interfere with DNA replication, causing S-phase arrest (76), as well as with the stabilization of
Bcl-2 mRNA, a known inhibitor of apoptosis, suggesting another
possible mechanism of action (77). The use of AS1411-conjugated NPs for
drug delivery was performed on MCF-7 breast cancer cells which
overexpress nucleolin, exhibiting increased cell death compared to
that of cancer cells that do not overexpress nucleolin. Moreover,
the extent of delivery and dosage may be controlled using
complementary DNA of the aptamer as an antidote (78). Furthermore, in vitro and
in vivo, AS1411 has demonstrated growth inhibitory effects
and antitumor activity against multiple cancer cell lines and tumor
tissues, respectively. The subsequent toxicity studies in rats and
dogs exhibited a favorable safety profile, prompting the initiation
of human clinical trials. Notably, internalization of the aptamer
results in a decrease in the expression of several
cancer-associated mRNAs and, therefore, reduced cell
proliferation.
Completed phase I trials demonstrated that AS1411
exhibited minimal toxicity in 17 patients with advanced solid
tumors. Moreover, there was evidence of promising clinical
activity, with 7 patients achieving disease stabilization and 1
patient with renal cell cancer achieving a complete response by 11
months (79). Phase II studies on
metastatic renal cell cancer and AML are currently underway. The
development and clinical trial data were reviewed by Mongelard and
Bouvet (80).
Mucin 1 (Muc1) is a glycoprotein found on the
surface of normal epithelial cells and functions as a barrier
against microorganisms and degradative enzymes (81). Muc1 is released into the
bloodstream and has been successfully used as a tumor marker for
bladder cancer. In addition, Muc1 is an attractive target, as its
overexpression has been associated with various types of cancer and
its distribution pattern changes on the cell surface (82,83).
DNA aptamers directed against Muc1 (AptA and AptB) were identified
in 2006 (84) and have since been
implemented in ex vivo imaging and radiotherapy (85). In subsequent studies, when such an
DNA aptamer was coupled to the light-activated photodynamic therapy
agent chlorine e6, it specifically delivered chlorine e6 into
epithelial cancer cells, with a significant enhancement in
efficiency (>500-fold increase) compared to that of the drug
alone, and achieved cancer-specific cytotoxicity (86). These results suggested the
anti-Muc1 aptamers may function as drug carriers to specific
epithelial cancer cells. As target QD scaffolds allow for labelling
of a single QD with multiple aptamers, the efficiency of binding
anti-Muc1 aptamers with QDs may be enhanced by avidity (87). In a mouse model bearing A2780/AD
human ovarian cancer xenografts, an increased amount of QD-Muc1
accumulated in the tumors compared to non-modified QD. This data
support that anti-Muc1 aptamer targeting may be a therapeutic
candidate for drug delivery when improved by the avidity associated
with NP decoration.
NOX-A12 is a 45-nt L-RNA aptamer, based on L-ribose,
which is clinically developed by NOXXON Pharma AG (Berlin, Germany)
(88). NOX-A12 was selected
against stromal cell-derived factor-1 (SDF-1), which binds to
chemokine (C-X-C motif) ligand 12, a chemokine that is involved in
tumor metastasis, angiogenesis, cell homing and tissue regeneration
(89). Inhibition of the SDF-1 by
NOX-A12 may prove useful in the treatment of several types of
cancer. NOX-A12 is currently in phase I clinical trials for the
treatment of lymphoma, multiple myeloma and hematopoietic stem cell
transplantation.
The ultimate aim in cancer therapy remains the
development of treatment modalities that effectively eliminate
cancer cells without affecting normal cells. Due to their
uniqueness and high specificity, aptamers and aptamer-based drug
delivery systems are promising tools for targeted cancer therapy.
Several more aptamers may soon be joining pegaptanib in the field
of cancer diagnosis and treatment.
Conclusion and future perspective
Aptamers have become increasingly important
molecular tools for diagnosis and therapeutics and aptamer-based
technology holds promise for the advancement of targeted cancer
diagnosis and therapy. Notably, due to their unique pharmacokinetic
properties, aptamers may be exploited for various purposes. One
highly attractive characteristic of aptamers is that they are
amenable to chemical modification and bioconjugation to various
moieties, such as NPs, imaging agents, siRNAs and therapeutic
drugs. As discussed in this review, aptamers were clinically proven
as inhibitors with great promise as targeting ligands for treating
or diagnosing cancer. Further maturation of the field of
nanotechnology and RNA interference may produce more sophisticated,
multifunctional constructs, capable of improved selective
cytotoxicity. Moreover, recent studies demonstrated improved
binding characteristics for multivalent aptamer constructs over
their monovalent counterparts; therefore, it is likely that this
will become a more popular approach for delivery applications. The
continued development of animal models for the evaluation of safety
and efficacy of these constructs is expected to lay the foundation
for their increased use in humans. Encouraging results from aptamer
clinical trials and successful outcomes, including the development
of pegaptanib sodium (Macugen), have further strengthened the
optimism surrounding their development and use as therapeutic and
diagnostic reagents. Although aptamers are relatively new to the
clinical setting, their versatility and numerous aptamer-based
therapeutic approaches discussed above suggest that this technology
may have a substantial impact on patient care in the following
years. Overall, aptamers offer a promising and unique technology
that may be applied in clinical diagnostics and therapeutics. Their
rapid development over the last two decades is notable, indicating
that aptamers may become a valuable tool for clinicians in the near
future.
Acknowledgements
This study was supported by a grant
from the Ministry of Science and Technology of China (no.
2010CB732405).
References
|
1.
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
3.
|
Rajendran L, Knolker HJ and Simons K:
Subcellular targeting strategies for drug design and delivery. Nat
Rev Drug Discov. 9:29–42. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Blagosklonny MV: Analysis of FDA approved
anticancer drugs reveals the future of cancer therapy. Cell Cycle.
3:1035–1042. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kanwar JR, Mohan RR, Kanwar RK, et al:
Applications of aptamers in nanodelivery systems in cancer, eye and
inflammatory diseases. Nanomedicine (Lond). 5:1435–1445. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Ellington AD and Szostak JW: In vitro
selection of RNA molecules that bind specific ligands. Nature.
346:818–822. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Tuerk C and Gold L: Systematic evolution
of ligands by exponential enrichment: RNA ligands to bacteriophage
T4 DNA polymerase. Science. 249:505–510. 1990. View Article : Google Scholar
|
|
8.
|
Keefe AD and Cload ST: SELEX with modified
nucleotides. Curr Opin Chem Biol. 12:448–456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Majumder P, Gomes KN and Ulrich H:
Aptamers: from bench side research towards patented molecules with
therapeutic applications. Expert Opin Ther Pat. 19:1603–1613. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Keefe AD, Pai S and Ellington A: Aptamers
as therapeutics. Nat Rev Drug Discov. 9:537–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Ciesiolka J and Yarus M: Small
RNA-divalent domains. RNA. 2:785–793. 1996.PubMed/NCBI
|
|
12.
|
Hofmann HP, Limmer S, Hornung V and
Sprinzl M: Ni2+-binding RNA motifs with an asymmetric
purine-rich internal loop and a G-A base pair. RNA. 3:1289–1300.
1997.
|
|
13.
|
Rajendran M and Ellington AD: Selection of
fluorescent aptamer beacons that light up in the presence of zinc.
Anal Bioanal Chem. 390:1067–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Geiger A, Burgstaller P, von der Eltz H,
et al: RNA aptamers that bind L-arginine with sub-micromolar
dissociation constants and high enantioselectivity. Nucleic Acids
Res. 24:1029–1036. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Connell GJ, Illangesekare M and Yarus M:
Three small ribooligonucleotides with specific arginine sites.
Biochemistry. 32:5497–5502. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Mannironi C, Scerch C, Fruscoloni P and
Tocchini-Valentini GP: Molecular recognition of amino acids by RNA
aptamers: the evolution into an L-tyrosine binder of a
dopamine-binding RNA motif. RNA. 6:520–527. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Harada K and Frankel AD: Identification of
two novel arginine binding DNAs. EMBO J. 14:5798–5811.
1995.PubMed/NCBI
|
|
18.
|
Wallis MG, Streicher B, Wank H, et al: In
vitro selection of a viomycin-binding RNA pseudoknot. Chem Biol.
4:357–366. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Wallace ST and Schroeder R: In vitro
selection and characterization of streptomycin-binding RNAs:
recognition discrimination between antibiotics. RNA. 4:112–123.
1998.PubMed/NCBI
|
|
20.
|
Nieuwlandt D, Wecker M and Gold L: In
vitro selection of RNA ligands to substance P. Biochemistry.
34:5651–5659. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Williams KP, Liu XH, Schumacher TN, et al:
Bioactive and nuclease-resistant L-DNA ligand of vasopressin. Proc
Natl Acad Sci USA. 94:11285–11290. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Mallikaratchy P, Stahelin RV, Cao Z, et
al: Selection of DNA ligands for protein kinase C-delta. Chem
Commun (Camb). 30:3229–3231. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Davis KA, Lin Y, Abrams B and Jayasena SD:
Staining of cell surface human CD4 with 2′-F-pyrimidine-containing
RNA aptamers for flow cytometry. Nucleic Acids Res. 26:3915–3924.
1998.
|
|
24.
|
Shangguan D, Li Y, Tang Z, et al: Aptamers
evolved from live cells as effective molecular probes for cancer
study. Proc Natl Acad Sci USA. 103:11838–11843. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Tang Z, Shangguan D, Wang K, et al:
Selection of aptamers for molecular recognition and
characterization of cancer cells. Anal Chem. 79:4900–4907. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Chen HW, Medley CD, Sefah K, et al:
Molecular recognition of small-cell lung cancer cells using
aptamers. ChemMedChem. 3:991–1001. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Sefah K, Tang ZW, Shangguan DH, et al:
Molecular recognition of acute myeloid leukemia using aptamers.
Leukemia. 23:235–244. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Lorger M, Engstler M, Homann M and
Goringer HU: Targeting the variable surface of African trypanosomes
with variant surface glycoprotein-specific, serum-stable RNA
aptamers. Eukaryot Cell. 2:84–94. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Tang Z, Parekh P, Turner P, et al:
Generating aptamers for recognition of virus-infected cells. Clin
Chem. 55:813–822. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Bruno JG and Kiel JL: In vitro selection
of DNA aptamers to anthrax spores with electrochemiluminescence
detection. Biosens Bioelectron. 14:457–464. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Shu D and Guo P: A viral RNA that binds
ATP and contains a motif similar to an ATP-binding aptamer from
SELEX. J Biol Chem. 278:7119–7125. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Clark SL and Remcho VT: Aptamers as
analytical reagents. Electrophoresis. 23:1335–1340. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Bouvet P: Determination of nucleic acid
recognition sequences by SELEX. Methods Mol Biol. 148:603–610.
2001.PubMed/NCBI
|
|
34.
|
Sablin EP and Fletterick RJ: Nucleotide
switches in molecular motors: structural analysis of kinesins and
myosins. Curr Opin Struct Biol. 11:716–724. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Shenton W, Pum D, Sleytr UB and Mann S:
Synthesis of cadmium sulphide superlattices using self-assembled
bacterial S-layers. Nature. 389:585–587. 1997. View Article : Google Scholar
|
|
36.
|
Yeakley JM, Fan JB, Doucet D, et al:
Profiling alternative splicing on fiber-optic arrays. Nat
Biotechnol. 20:353–358. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Klug SJ and Famulok M: All you wanted to
know about SELEX. Mol Biol Rep. 20:97–107. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Ciesiolka J, Gorski J and Yarus M:
Selection of an RNA domain that binds Zn2+. RNA.
1:538–550. 1995.PubMed/NCBI
|
|
39.
|
Nimjee SM, Rusconi CP, Harrington RA and
Sullenger BA: The potential of aptamers as anticoagulants. Trends
Cardiovasc Med. 15:41–45. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Levy-Nissenbaum E, Radovic-Moreno AF, Wang
AZ, et al: Nanotechnology and aptamers: applications in drug
delivery. Trends Biotechnol. 26:442–449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Thiel KW and Giangrande PH: Therapeutic
applications of DNA and RNA aptamers. Oligonucleotides. 19:209–222.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Dyke CK, Steinhubl SR, Kleiman NS, et al:
First-in-human experience of an antidote-controlled anticoagulant
using RNA aptamer technology: a phase 1a pharmacodynamic evaluation
of a drug-antidote pair for the controlled regulation of factor IXa
activity. Circulation. 114:2490–2497. 2006. View Article : Google Scholar
|
|
43.
|
Smith JE, Medley CD, Tang Z, et al:
Aptamer-conjugated nanoparticles for the collection and detection
of multiple cancer cells. Anal Chem. 79:3075–3082. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Kang WJ, Chae JR, Cho YL, et al: Multiplex
imaging of single tumor cells using quantum-dot-conjugated
aptamers. Small. 5:2519–2522. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Zhao Z, Xu L, Shi X, et al: Recognition of
subtype non-small cell lung cancer by DNA aptamers selected from
living cells. Analyst. 134:1808–1814. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Hicke BJ and Stephens AW: Escort aptamers:
a delivery service for diagnosis and therapy. J Clin Invest.
106:923–928. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Daniel MC and Astruc D: Gold
nanoparticles: assembly, supramolecular chemistry,
quantum-size-related properties, and applications toward biology,
catalysis, and nanotechnology. Chem Rev. 104:293–346. 2004.
View Article : Google Scholar
|
|
48.
|
Rosi NL, Giljohann DA, Thaxton CS,
Lytton-Jean AK, et al: Oligonucleotide-modified gold nanoparticles
for intracellular gene regulation. Science. 312:1027–1030. 2006.
View Article : Google Scholar
|
|
49.
|
Sperling RA, Rivera Gil P, Zhang F, et al:
Biological applications of gold nanoparticles. Chem Soc Rev.
37:1896–1908. 2008. View Article : Google Scholar
|
|
50.
|
Storhoff JJ, Lucas AD, Garimella V, et al:
Homogeneous detection of unamplified genomic DNA sequences based on
colorimetric scatter of gold nanoparticle probes. Nat Biotechnol.
22:883–887. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Grzelczak M, Perez-Juste J, Mulvaney P and
Liz-Marzan LM: Shape control in gold nanoparticle synthesis. Chem
Soc Rev. 37:1783–1791. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Medley CD, Smith JE, Tang Z, et al: Gold
nanoparticle-based colorimetric assay for the direct detection of
cancerous cells. Anal Chem. 80:1067–1072. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Lu W, Arumugam SR, Senapati D, et al:
Multifunctional oval-shaped gold-nanoparticle-based selective
detection of breast cancer cells using simple colorimetric and
highly sensitive two-photon scattering assay. ACS Nano.
4:1739–1749. 2010. View Article : Google Scholar
|
|
54.
|
Zhang J, Jia X, Lv XJ, et al: Fluorescent
quantum dot-labeled aptamer bioprobes specifically targeting mouse
liver cancer cells. Talanta. 81:505–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Huang YF, Chang HT and Tan W: Cancer cell
targeting using multiple aptamers conjugated on nanorods. Anal
Chem. 80:567–572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Estevez MC, O'Donoghue MB, Chen X and Tan
W: Highly fluorescent dye-doped silica nanoparticles increase flow
cytometry sensitivity for cancer cell monitoring. Nano Res.
2:448–461. 2009. View Article : Google Scholar
|
|
57.
|
Chen X, Estevez MC, Zhu Z, et al: Using
aptamer-conjugated fluorescence resonance energy transfer
nanoparticles for multiplexed cancer cell monitoring. Anal Chem.
81:7009–7014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Wang L and Tan W: Multicolor FRET silica
nanoparticles by single wavelength excitation. Nano Lett. 6:84–88.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Wessels JT, Busse AC, Mahrt J, et al: In
vivo imaging in experimental preclinical tumor research - a review.
Cytometry A. 71:542–549. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Shangguan D, Meng L, Cao ZC, et al:
Identification of liver cancer-specific aptamers using whole live
cells. Anal Chem. 80:721–728. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
Davis KA, Abrams B, Lin Y and Jayasena SD:
Use of a high affinity DNA ligand in flow cytometry. Nucleic Acids
Res. 24:702–706. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
62.
|
Bagalkot V, Zhang L, Levy-Nissenbaum E, et
al: Quantum dot-aptamer conjugates for synchronous cancer imaging,
therapy, and sensing of drug delivery based on bi-fluorescence
resonance energy transfer. Nano Lett. 7:3065–3070. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Wang AZ, Bagalkot V, Vasilliou CC, et al:
Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for
combined prostate cancer imaging and therapy. ChemMedChem.
3:1311–1315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Kim D, Jeong YY and Jon S: A drug-loaded
aptamer-gold nanoparticle bioconjugate for combined CT imaging and
therapy of prostate cancer. ACS Nano. 4:3689–3696. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65.
|
Bander NH: Technology insight: monoclonal
antibody imaging of prostate cancer. Nat Clin Pract Urol.
3:216–225. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
66.
|
Lupold SE, Hicke BJ, Lin Y and Coffey DS:
Identification and characterization of nuclease-stabilized RNA
molecules that bind human prostate cancer cells via the
prostate-specific membrane antigen. Cancer Res. 62:4029–4033.
2002.PubMed/NCBI
|
|
67.
|
McNamara JO II, Andrechek ER, Wang Y, et
al: Cell type-specific delivery of siRNAs with aptamer-siRNA
chimeras. Nat Biotechnol. 24:1005–1015. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
68.
|
Ni X, Zhang Y, Ribas J, et al:
Prostate-targeted radiosensitization via aptamer-shRNA chimeras in
human tumor xenografts. J Clin Invest. 121:2383–2390. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Farokhzad OC, Jon S, Khademhosseini A, et
al: Nanoparticle-aptamer bioconjugates: a new approach for
targeting prostate cancer cells. Cancer Res. 64:7668–7672. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Bleickardt E, Argiris A, Rich R, et al:
Phase I dose escalation trial of weekly docetaxel plus irinotecan
in patients with advanced cancer. Cancer Biol Ther. 1:646–651.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Farokhzad OC, Cheng J, Teply BA, et al:
Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy
in vivo. Proc Natl Acad Sci USA. 103:6315–6320. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Cheng J, Teply BA, Sherifi I, et al:
Formulation of functionalized PLGA-PEG nanoparticles for in vivo
targeted drug delivery. Biomaterials. 28:869–876. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
Farokhzad OC, Khademhosseini A, Jon S, et
al: Microfluidic system for studying the interaction of
nanoparticles and microparticles with cells. Anal Chem.
77:5453–5459. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Gu F, Zhang L, Teply BA, et al: Precise
engineering of targeted nanoparticles by using self-assembled
biointegrated block copolymers. Proc Natl Acad Sci USA.
105:2586–2591. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75.
|
Dhar S, Kolishetti N, Lippard SJ and
Farokhzad OC: Targeted delivery of a cisplatin prodrug for safer
and more effective prostate cancer therapy in vivo. Proc Natl Acad
Sci USA. 108:1850–1855. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76.
|
Ireson CR and Kelland LR: Discovery and
development of anti-cancer aptamers. Mol Cancer Ther. 5:2957–2962.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
77.
|
Soundararajan S, Chen W, Spicer EK, et al:
The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger
RNA in human breast cancer cells. Cancer Res. 68:2358–2365. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
78.
|
Cao Z, Tong R, Mishra A, et al: Reversible
cell-specific drug delivery with aptamer-functionalized liposomes.
Angew Chem Int Ed Engl. 48:6494–6498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
79.
|
Bates PJ, Laber DA, Miller DM, et al:
Discovery and development of the G-rich oligonucleotide AS1411 as a
novel treatment for cancer. Exp Mol Pathol. 86:151–164. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
80.
|
Mongelard F and Bouvet P: AS-1411, a
guanosine-rich oligo-nucleotide aptamer targeting nucleolin for the
potential treatment of cancer, including acute myeloid leukemia.
Curr Opin Mol Ther. 12:107–114. 2010.
|
|
81.
|
Gendler SJ: MUC1, the renaissance
molecule. J Mammary Gland Biol Neoplasia. 6:339–353. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
82.
|
Brockhausen I, Yang JM, Burchell J, et al:
Mechanisms underlying aberrant glycosylation of MUC1 mucin in
breast cancer cells. Eur J Biochem. 233:607–617. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
83.
|
Reis CA, David L, Seixas M, et al:
Expression of fully and under-glycosylated forms of MUC1 mucin in
gastric carcinoma. Int J Cancer. 79:402–410. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
84.
|
Ferreira CS, Matthews CS and Missailidis
S: DNA aptamers that bind to MUC1 tumour marker: design and
characterization of MUC1-binding single-stranded DNA aptamers.
Tumour Biol. 27:289–301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
85.
|
Borbas KE, Ferreira CS, Perkins A, Bruce
JI and Missailidis S: Design and synthesis of mono- and multimeric
targeted radio-pharmaceuticals based on novel cyclen ligands
coupled to anti-MUC1 aptamers for the diagnostic imaging and
targeted radiotherapy of cancer. Bioconjug Chem. 18:1205–1212.
2007. View Article : Google Scholar
|
|
86.
|
Ferreira CS, Cheung MC, Missailidis S, et
al: Phototoxic aptamers selectively enter and kill epithelial
cancer cells. Nucleic Acids Res. 37:866–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
87.
|
Savla R, Taratula O, Garbuzenko O and
Minko T: Tumor targeted quantum dot-mucin 1 aptamer-doxorubicin
conjugate for imaging and treatment of cancer. J Control Release.
153:16–22. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
88.
|
Nolte A, Klussmann S, Bald R, et al:
Mirror-design of L-oligonucleotide ligands binding to L-arginine.
Nat Biotechnol. 14:1116–1119. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
89.
|
Sayyed SG, Hagele H, Kulkarni OP, et al:
Podocytes produce homeostatic chemokine stromal cell-derived
factor-1/CXCL12, which contributes to glomerulosclerosis, podocyte
loss and albuminuria in a mouse model of type 2 diabetes.
Diabetologia. 52:2445–2454. 2009. View Article : Google Scholar
|