Introduction
Nasopharyngeal carcinoma (NPC) is a common type of
cancer in Southern China, whereas its incidence in Western
countries is relatively low (1).
Due to its deep-seated anatomic location and relative
radiosensitivity, radiotherapy (RT) is the treatment of choice for
primary NPC. Advanced RT techniques, such as intensity-modulated RT
(IMRT) have achieved improvements the locoregional control of the
tumour and reduced the incidence of complications (2). However, radiation-induced fibrosis
remains one of the major late complications, regardless of the
treatment method (2,3). Since different patients may present
with different degrees of skin fibrosis despite receiving identical
treatment, it was hypothesized that the severity of such
complications may be genetically determined. Previous studies
investigated the association of single-nucleotide polymorphisms
(SNPs) in genes involved in DNA repair, such as X-ray repair
cross-complementing protein 1 (XRCC1) rs25487 (c.1196A>G,
p.Gln399Arg) and X-ray repair cross-complementing protein 3
(XRCC3) rs861539 (c.722C>T, p.Thr241Met) with late
complications in various types of cancer (4–18).
The identification of particular functional variants
in cancer patients prior to the initiation of any treatment is
likely to improve the prediction of the severity of
radiation-induced complications in individual patients, leading to
improved patient care and customization of treatment protocols.
Since the majority of studies were conducted in Caucasian
populations and allelic frequencies vary between ethnic groups,
little is known regarding the association between genetic variants
and post-RT complications in Chinese patients. The previously
published studies mainly focused on particular functional variants
of candidate genes; however, common genetic variants were
overlooked. Therefore, this preliminary study aimed to evaluate the
association of radiation-induced fibrosis with XRCC1 and
XRCC3 functional variants and possible haplotypes in Chinese
NPC patients.
Materials and methods
Subject recruitment
A total of 120 Chinese NPC patients, including 70.8%
men and 29.2% women, aged ≥18 years, without distant metastasis,
who were treated with conventional RT (CRT) or IMRT, were recruited
during their follow-up between 2010 and 2011 at the Department of
Clinical Oncology, Queen Mary Hospital, Hong Kong (Table I). RT was administered at 2 Gy per
fraction, 5 fractions per week for 6–7 weeks. The patients who
received CRT and IMRT generally received a total dose of 66–68 Gy
and 70–76 Gy to the tumour, respectively. The patients were
classified as ‘cases’ if they presented with persistent ≥grade 1
fibrosis at the time of the follow-up, according to the radiation
morbidity scoring criteria published by the Radiation Therapy
Oncology Group. Patients without significant fibrotic changes
(grade 0) for ≥2-years post-RT were classified as ‘controls’. All
the patients signed a written informed consent form prior to
enrolment.
 | Table ISummary clinical characteristics of
recruited patients. |
Table I
Summary clinical characteristics of
recruited patients.
| Clinical
variables | Controls (n=91) | Cases (n=29) | P-value |
|---|
| Age, years [mean
(SD)] | 52.60 (10.46) | 55.10 (9.05) | NS |
| Follow-up, years
[mean (SD)] | 8.13 (5.57) | 12.38 (5.15) | <0.01 |
| Gender, n | | | <0.05 |
| Male | 69 | 16 | |
| Female | 22 | 13 | |
| TNM stage, n | | | NS |
| I | 16 | 0 | |
| II | 26 | 6 | |
| III | 29 | 15 | |
| IV | 16 | 4 | |
| Unavailable | 4 | 4 | |
| Radiotherapy, n | | | <0.01 |
| CRT | 26 | 19 | |
| IMRT | 65 | 10 | |
| Chemotherapy, n | 46 | 15 | NS |
Tag SNP selection and genotyping
The selection of the tag SNPs of XRCC1 and
XRCC3 was performed with Tagger software (Cambridge, MA,
USA) using the International HapMap Project data for Han Chinese
subjects (release 27, phase II+III, Feb 9) (19,20).
Tag SNPs were selected using a pairwise tagging algorithm,
r2=0.8 and a minor allele frequency (MAF) of ≥0.2.
Previously reported SNPs were also included in the analysis. In
order to capture any upstream and downstream regulatory elements of
the candidate gene, tag SNPs were selected from 3 kb at the start
and the end of the candidate region. A total of 12 tag SNPs were
selected and were genotyped using restriction fragment length
polymorphism analysis (RFLP) or unlabeled probe melting
analysis.
Genotyping method
For the DNA analysis of each patient, 6 ml venous
blood was collected in ethylenediaminetetraacetic acid (EDTA)
tubes. DNA was extracted using a FlexiGene DNA kit (Qiagen, Hilden,
Germany). The concentration of DNA was measured using NanoDrop™
1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, MA,
USA). The DNA concentration was adjusted to 10 ng/μl using
Tris-EDTA buffer for polymerase chain reaction (PCR). A total of 10
μl reaction mixture was used for both RFLP and unlabeled probe
melting analysis. Each reaction mixture contained 10 ng of genomic
DNA, 0.2 mM of each deoxynucleotide triphosphate, 0.3 U of
HotStarTaq Plus DNA polymerase with 1X PCR buffer [containing
Tris-Cl, KCl, (NH4)2SO4 and 15 mM
MgCl2, pH 8.7] (Qiagen) and specific optimized primers
and MgCl2 concentrations.
Statistical analysis
Genotyped data were analyzed using the PLINK
statistical package (http://pngu.mgh.harvard.edu/~purcell/plink), version
1.07 (21). The genotypes in the
control and case groups were tested for deviation from the
Hardy-Weinberg equilibrium (HWE) using the Fisher’s exact test and
the odds ratio (OR) for each genotype was calculated. Analysis of
single markers was performed using the PLINK toolset. Gender, age,
treatment regimen and other clinical factors were assessed by
multivariate logistic regression analysis. Analyses of all the
possible haplotypes using the sliding window approach were also
performed using PLINK. Empirical P-values (Pemp) for
single markers, as well as haplotype analysis, were generated by
multiple comparisons based on 10,000 permutations. Genotype data
from published studies were extracted to perform combined genotype
analysis. Combined genotype analysis was performed with the Meta
package, version 2.5.1 in R version 2.15.1 for Windows (http://cran.r-project.org/web/packages/meta/index.html)
(22).
Results
Group comparison
Of the 120 patients, 45 received CRT and 75 received
IMRT, whereas 61 patients were treated with chemotherapy. A total
of 29 patients suffered from ≥grade 1 neck fibrosis that persisted
for ≥2 years. There were significantly more patients who received
CRT rather than IMRT in the case group compared to those in the
control group (P<0.01). There were also significant differences
in the mean follow-up duration (P<0.01) and in the number of men
and women (P<0.05) between the control and case groups. There
were no significant differences in the number of patients who
received chemotherapy, tumor stage and mean age between the two
groups (P>0.05). All the abovementioned clinical factors that
may affect the severity of radiation-induced fibrosis were used as
covariates in the logistic regression analysis.
Statistical analysis
All the tag SNPs were successfully genotyped and
were in HWE (P>0.05), except the control group of tag SNPs
rs861544 (P=0.003). This tag SNP was included in the analysis,
since ~1 significant result could be obtained due to random chance
with 12 comparisons when α=0.05.
Single-marker multivariate logistic regression
analysis revealed that only the T allele of rs861539 was associated
with increased risk of fibrosis [asymptotic P-value
(Pasym)=0.0116; OR=3.88]. However, a significance level
was lost after multiple comparisons (Pemp=0.0632). There
was no significant association between fibrosis and the remaining
tag SNPs in the single-marker multivariate logistic regression
analysis (Table II).
 | Table IISummary statistics of selected tag
SNPs in the XRCC1 and XRCC3 genes and results of
single-marker analysis. |
Table II
Summary statistics of selected tag
SNPs in the XRCC1 and XRCC3 genes and results of
single-marker analysis.
| | | | Genotype counts
11/12/22 | MAF | | | | | |
|---|
| | | |
|
| | | | | |
|---|
| Gene | SNPa | A1 | A2 | Control | Case | Control | Case | ORb | L95 | U95 |
Pasym |
Pemp |
|---|
| XRCC1 | rs3213282
(XRCC1.S1) | C | G | 4/35/52 | 2/11/16 | 0.2363 | 0.2586 | 1.19 | 0.53 | 2.65 | 0.6975 | 0.9899 |
| rs12611088
(XRCC1.S2) | T | C | 7/33/51 | 1/11/17 | 0.2582 | 0.2241 | 0.59 | 0.25 | 1.36 | 0.2345 | 0.6137 |
| rs1001581
(XRCC1.S3) | A | G | 11/46/34 | 4/13/12 | 0.3736 | 0.3621 | 0.67 | 0.32 | 1.39 | 0.2900 | 0.7165 |
| rs3213344
(XRCC1.S4) | G | C | 8/32/51 | 2/11/16 | 0.2637 | 0.2586 | 1.34 | 0.64 | 2.82 | 0.4430 | 0.8889 |
| rs1799782
(XRCC1.S5) | T | C | 8/31/52 | 2/11/16 | 0.2582 | 0.2586 | 1.40 | 0.67 | 2.94 | 0.3833 | 0.8289 |
| rs25487
(XRCC1.S6) | A | G | 6/35/50 | 1/13/15 | 0.2582 | 0.2586 | 0.80 | 0.36 | 1.79 | 0.6099 | 0.9705 |
| XRCC3 | rs1799794
(XRCC3.S1) | G | A | 17/50/24 | 6/10/13 | 0.4615 | 0.3793 | 0.65 | 0.32 | 1.31 | 0.2408 | 0.5839 |
| rs861530
(XRCC3.S2) | G | A | 19/52/20 | 8/13/8 | 0.4945 | 0.5000 | 1.08 | 0.55 | 2.13 | 0.8371 | 0.9992 |
| rs3212090
(XRCC3.S3) | A | G | 15/44/32 | 2/16/11 | 0.4066 | 0.3448 | 0.68 | 0.33 | 1.38 | 0.2959 | 0.6733 |
| rs12432907
(XRCC3.S4) | A | G | 18/47/26 | 7/11/11 | 0.4560 | 0.4310 | 0.82 | 0.42 | 1.62 | 0.5821 | 0.9502 |
| rs861539
(XRCC3.S5) | T | C | 1/8/82 | 0/8/21 | 0.0550 | 0.1379 | 3.88 | 1.25 | 12.07 |
0.0116c | 0.0632 |
| rs861544
(XRCC3.S6) | T | C | 10/59/22 | 3/18/8 | 0.4341 | 0.4138 | 0.94 | 0.43 | 2.05 | 0.8795 | 0.9999 |
A total of 42 sliding windows were generated for
XRCC1 and XRCC3, with 21 sliding windows created for
each gene. No association was found in any of the sliding windows
following multiple comparisons (omnibus test
Pemp>0.05) (Table
III).
 | Table IIISummary of sliding window haplotype
analysis of all the possible sizes based on omnibus test of the
XRCC1 and XRCC3 genes. |
Table III
Summary of sliding window haplotype
analysis of all the possible sizes based on omnibus test of the
XRCC1 and XRCC3 genes.
| | | | Most significant
omnibus test |
|---|
| | | |
|
|---|
| Gene | NSNP | First SW | Last SW | SW |
Pasym |
Pemp |
|---|
| XRCC1 | 2 |
XRCC1.S1..XRCC1.S2 |
XRCC1.S5..XRCC1.S6 |
XRCC1.S4..XRCC1.S5 | 0.4500 | 0.9159 |
| 3 |
XRCC1.S1..XRCC1.S3 |
XRCC1.S4..XRCC1.S6 |
XRCC1.S2..XRCC1.S4 | 0.2924 | 0.8450 |
| 4 |
XRCC1.S1..XRCC1.S4 |
XRCC1.S3..XRCC1.S6 |
XRCC1.S2..XRCC1.S5 | 0.2925 | 0.8450 |
| 5 |
XRCC1.S1..XRCC1.S5 |
XRCC1.S2..XRCC1.S6 |
XRCC1.S2..XRCC1.S6 | 0.0877 | 0.5960 |
| 6 |
XRCC1.S1..XRCC1.S6 |
XRCC1.S1..XRCC1.S6 |
XRCC1.S1..XRCC1.S6 | 0.2633 | 0.8891 |
| XRCC3 | 2 |
XRCC3.S1..XRCC3.S2 |
XRCC3.S5..XRCC3.S6 |
XRCC3.S5..XRCC3.S6 | 0.0147 | 0.1944 |
| 3 |
XRCC3.S1..XRCC3.S3 |
XRCC3.S4..XRCC3.S6 |
XRCC3.S1..XRCC3.S3 | 0.0561 | 0.3531 |
| 4 |
XRCC3.S1..XRCC3.S4 |
XRCC3.S3..XRCC3.S6 |
XRCC3.S1..XRCC3.S4 | 0.0301 | 0.3231 |
| 5 |
XRCC3.S1..XRCC3.S5 |
XRCC3.S2..XRCC3.S6 |
XRCC3.S2..XRCC3.S6 | 0.1132 | 0.6833 |
| 6 |
XRCC3.S1..XRCC3.S6 |
XRCC3.S1..XRCC3.S6 |
XRCC3.S1..XRCC3.S6 | 0.0339 | 0.4571 |
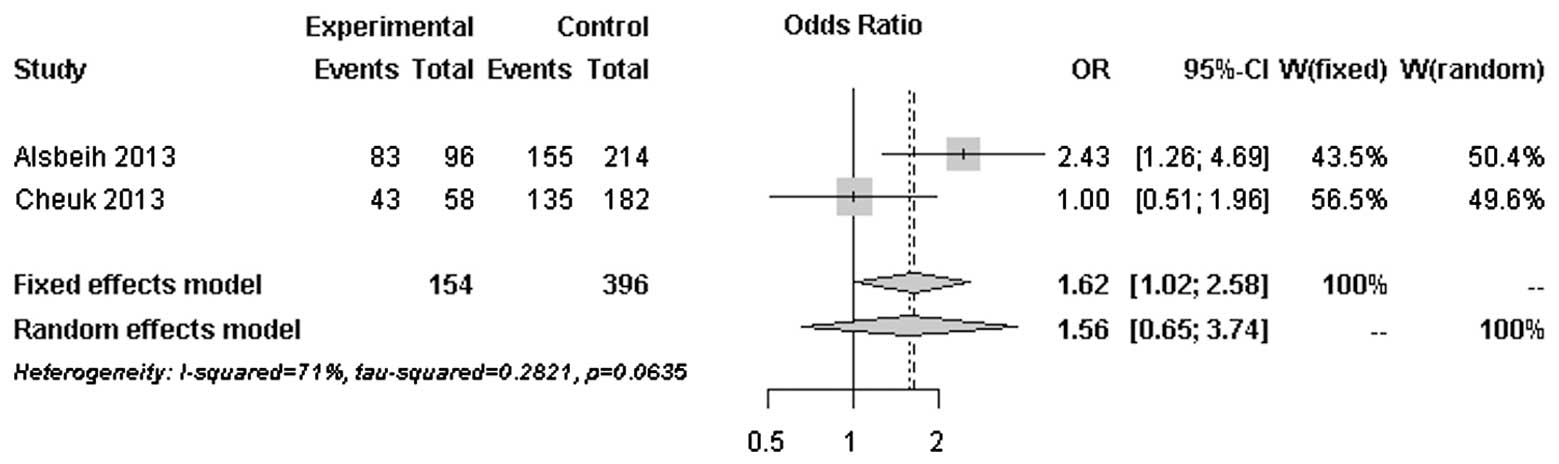
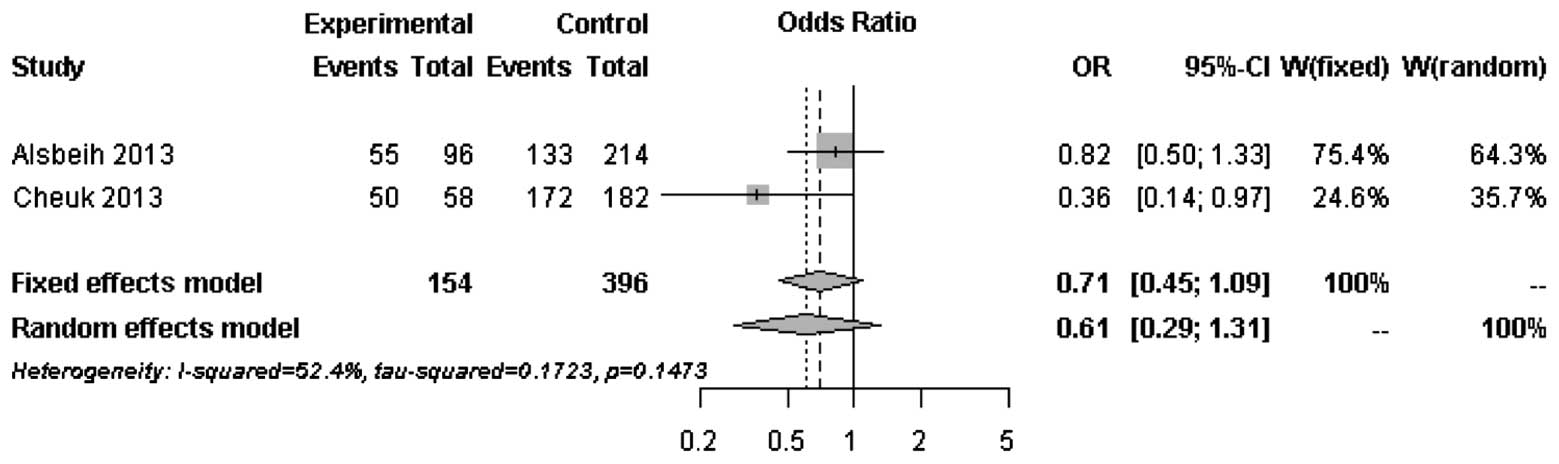
The forest plots of rs25487 from XRCC1 and
rs861539 from XRCC3 in NPC patients are shown in Figs. 1 and 2. The percentage of between-study
variation due to heterogeneity is presented as I2
(23). There was no significant
difference for the two SNPs analyzed by the random effects model
(P>0.05). However, a significant difference was observed when
the fixed effects model was used for rs25487 (P=0.0415).
Discussion
In this retrospective study, the association of DNA
repair genes with the development of post-RT fibrosis in Chinese
NPC patients was investigated. Selected tag SNPs, as well as
previously reported functional SNPs that were associated with late
complications, were determined to obtain comprehensive information
on common genetic variants of the XRCC1 and XRCC3
genes and their association with radiation-induced fibrosis.
Additional haplotype analysis was performed in order to investigate
the effects of all the possible haplotypes on radiation-induced
fibrosis. No significant association was found in the SNPs in
either single-marker or haplotype analysis following multiple
comparisons.
Radiation-induced fibrosis, unlike normal wound
healing, is a complex process that its endpoint is the result of
accumulated chronic inflammatory reactions (24). Since DNA is the critical target of
ionizing radiation damage, it is hypothesized that functional
variants in DNA repair genes may affect the DNA repair capacity and
lead to variations in radiation sensitivity. XRCC1 and
XRCC3 are two of the genes that are involved in DNA repair.
The association of functional variants of these genes with
radiation-induced late complications was previously investigated
with inconsistent results (25).
The majority of the published studies included breast and prostate
cancer patients in Caucasian populations. No association with
rs25487 of XRCC1 and rs861539 of XRCC3 was
identified. Our results were in line with findings reported by
other studies with relatively large sample sizes (16,17).
However, our results were in contrast to those reported by 3
studies performed in Saudi Arabian NPC patients by Alsbeih et
al (4,5,18).
Our results indicated a trend of possible association between
rs861539 of XRCC3 and radiation-induced fibrosis in Chinese
NPC patients, but not rs25487 of XRCC1. In order to
investigate the heterogeneity between studies with a similar study
design, forest plots of rs25487 and rs861539 were constructed based
on genotyped data from our study and the most recent study by
Alsbeih et al (18).
Moderate to significant heterogeneity was observed
for rs25487 and rs861539. The discrepancies between studies may be
due to variations in the study design, length of follow-up period
and allele frequencies in different ethnic groups. The MAFs of
rs25487 obtained by Alsbeih et al (18) and the present study were 0.232 and
0.258, respectively. The MAFs of rs861539 obtained by Alsbeih et
al (18) and the present study
were 0.394 and 0.075, respectively. While the deviation of allele
frequencies between the two studies was small for rs25487, an
increased risk of developing fibrosis was not identified by the
present study. The median follow-up time was 8 years in the present
study and 40 months in the study by Alsbeih et al (18). Since late toxicities may progress
over time several years after post-RT, the length of the follow-up
is significantly associated with the development of fibrosis.
Normal tissue radiosensitivity is a complex
phenomenon. Unlike truncated proteins, such as ataxia
telangiectasia mutated, particular functional variants in selected
genes may not be sufficient to cause severe radiosensitivity. Apart
from specific functional variants, the role of other common
variants of candidate genes and their effect have not been clearly
determined. This study aimed to investigate the association of
common and functional variants of DNA repair genes with fibrosis in
NPC patients. We captured certain common variants and constructed
haplotype structures of all possible sizes using tag SNPs, in order
to obtain more information on the effects of DNA repair genes. One
of the major limitations of our study was the relatively small
sample size. A small sample size may not have enough power to
detect SNPs with small and modest effects. Genotyped SNPs with
small and modest effects were possibly not the causal SNPs of this
phenomenon, but served as surrogates of other SNPs in high linkage
disequilibrium located in distant regions. In addition, the effect
sizes of common variants are likely to be small, in contrast to the
large effect caused by rare variants (26,27).
Therefore, individual common variants may not be ideal universal
biomarkers for identification of radiosensitivity.
Two large cohort studies focusing on
radiation-induced toxicities developed in breast and prostate
cancers were recently published (26,28).
One study by Barnett et al (26), involving 1,613 breast and prostate
cancer patients, captured common variants using tag SNPs with
MAF<0.05 and functional SNPs in 92 genes and found no
association in any of the SNPs. Furthermore, a replication study by
Talbot et al (28),
involving 2,036 patients from three cohorts, captured 43 candidate
SNPs in 35 genes and found that TNF-α may be associated with
increased risk of radiation toxicities in breast cancer patients.
No previously reported associations were found to be significant in
those studies. The negative findings of the studies indicated that
previously published SNPs may not exert clinical effects
individually and significant SNPs may have small effect sizes.
The first genome-wide association analysis focused
on the association of African-American prostate cancer patients
with the risk of radiation-induced erectile dysfunction suggested
that cancer type and ethnicity may be genetic predictors (29). Based on our findings, the
low-frequency variant rs861539 may be an ethnic group-specific
marker in predicting radiation-induced toxicities in Chinese NPC
patients. The amount of radiation dose received by NPC patients is
generally different from that received by breast and prostate
cancers patients, regardless of the different RT methods.
Non-irradiated cells were shown to exhibit similar inflammatory
responses as irradiated cells (30). In addition to genetic factors,
epigenetic factors may also affect individual radiosensitivity.
Large-scale studies should also be conducted in combined ethnic
groups with all the types of cancer that require RT as standard
treatment, in order to investigate the effects of common and
functional variants. With the advancements in technology, the
genetic profiles of individual patients may be obtained to assess
the risk effect or perform a combined analysis. In addition,
meta-analyses may be performed to assess the risk of
radiation-induced toxicities in NPC patients and obtain more cancer
type-specific information. The Standardized Total Average Toxicity
score can be used in future studies to enhance data pooling
(31). Since normal tissue
radiosensitivity may be the result of the combined effects of genes
involved in different cellular pathways, functional study focus on
the performance of DNA repair proteins following irradiation with
respect to genotypes and epigenetic changes may be conducted.
Casual common or functional variants may be used as pre-treatment
biomarkers for identifying highly radiosensitive cancer patients,
leading to the customization of treatment protocols and improved
treatment outcomes in the future.
References
|
1
|
Wei WI and Sham JS: Nasopharyngeal
carcinoma. Lancet. 365:2041–2054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kam MK, Teo PM, Chau RM, et al: Treatment
of nasopharyngeal carcinoma with intensity-modulated radiotherapy:
the Hong Kong experience. Int J Radiat Oncol Biol Phys.
60:1440–1450. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee AW, Law SC, Ng SH, et al:
Retrospective analysis of nasopharyngeal carcinoma treated during
1976–1985 late complications following megavoltage irradiation. Br
J Radiol. 65:918–928. 1992.
|
|
4
|
Alsbeih G, Al-Harbi N, Al-Hadyan K,
El-Sebaie M and Al-Rajhi N: Association between normal tissue
complications after radiotherapy and polymorphic variations in
TGFB1 and XRCC1 genes. Radiat Res. 173:505–511. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alsbeih GA, El-Sebaie MM, Al-Rajhi NM, et
al: Association between XRCC1 G399A polymorphism and late
complications to radiotherapy in Saudi head and neck cancer
patients. J Egypt Natl Canc Inst. 20:302–308. 2008.PubMed/NCBI
|
|
6
|
Andreassen CN, Alsner J, Overgaard J, et
al: TGFB1 polymorphisms are associated with risk of late normal
tissue complications in the breast after radiotherapy for early
breast cancer. Radiother Oncol. 75:18–21. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Andreassen CN, Alsner J, Overgaard M and
Overgaard J: Prediction of normal tissue radiosensitivity from
polymorphisms in candidate genes. Radiother Oncol. 69:127–135.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Azria D, Ozsahin M, Kramar A, et al:
Single nucleotide polymorphisms, apoptosis, and the development of
severe late adverse effects after radiotherapy. Clin Cancer Res.
14:6284–6288. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brem R, Cox DG, Chapot B, et al: The XRCC1
-77T->C variant: haplotypes, breast cancer risk, response to
radiotherapy and the cellular response to DNA damage.
Carcinogenesis. 27:2469–2474. 2006.
|
|
10
|
Burri RJ, Stock RG, Cesaretti JA, et al:
Association of single nucleotide polymorphisms in SOD2, XRCC1 and
XRCC3 with susceptibility for the development of adverse effects
resulting from radiotherapy for prostate cancer. Radiat Res.
170:49–59. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Damaraju S, Murray D, Dufour J, et al:
Association of DNA repair and steroid metabolism gene polymorphisms
with clinical late toxicity in patients treated with conformal
radiotherapy for prostate cancer. Clin Cancer Res. 12:2545–2554.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giotopoulos G, Symonds RP, Foweraker K, et
al: The late radiotherapy normal tissue injury phenotypes of
telangiectasia, fibrosis and atrophy in breast cancer patients have
distinct genotype-dependent causes. Br J Cancer. 96:1001–1007.
2007. View Article : Google Scholar
|
|
13
|
Moullan N, Cox DG, Angèle S, Romestaing P,
Gerard JP and Hall J: Polymorphisms in the DNA repair gene XRCC1,
breast cancer risk, and response to radiotherapy. Cancer Epidemiol
Biomarkers Prev. 12:1168–1174. 2003.PubMed/NCBI
|
|
14
|
Suga T, Iwakawa M, Tsuji H, et al:
Influence of multiple genetic polymorphisms on genitourinary
morbidity after carbon ion radiotherapy for prostate cancer. Int J
Radiat Oncol Biol Phys. 72:808–813. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zschenker O, Raabe A, Boeckelmann IK, et
al: Association of single nucleotide polymorphisms in ATM, GSTP1,
SOD2, TGFB1, XPD and XRCC1 with clinical and cellular
radiosensitivity. Radiother Oncol. 97:26–32. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang-Claude J, Ambrosone CB, Lilla C, et
al: Genetic polymorphisms in DNA repair and damage response genes
and late normal tissue complications of radiotherapy for breast
cancer. Br J Cancer. 100:1680–1686. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Popanda O, Marquardt JU, Chang-Claude J
and Schmezer P: Genetic variation in normal tissue toxicity induced
by ionizing radiation. Mutat Res. 667:58–69. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alsbeih G, El-Sebaie M, Al-Harbi N,
Al-Hadyan K, Shoukri M and Al-Rajhi N: SNPs in genes implicated in
radiation response are associated with radiotoxicity and evoke
roles as predictive and prognostic biomarkers. Radiat Oncol.
8:1252013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Bakker PI, Yelensky R, Pe’er I, Gabriel
SB, Daly MJ and Altshuler D: Efficiency and power in genetic
association studies. Nat Genet. 37:1217–1223. 2005.PubMed/NCBI
|
|
20
|
International HapMap Consortium. The
International HapMap Project. Nature. 426:789–796. 2003. View Article : Google Scholar
|
|
21
|
Purcell S, Neale B, Todd-Brown K, et al:
PLINK: a tool set for whole-genome association and population-based
linkage analyses. Am J Hum Genet. 81:pp. 559–575. 2007, http://pngu.mgh.harvard.edu/~purcell/plink.
Accessed, July 19, 2013
|
|
22
|
Schwarzer G: Meta: An R package for
meta-analysis. R News. 7:pp. 40–45. 2007, http://cran.r-project.org/web/packages/meta/index.html.
Accessed, September 8, 2013
|
|
23
|
Higgins JPT and Green S: Cochrane Handbook
for Systematic Reviews of Interventions, version 5.10. The Cochrane
Collaboration; 2011, http://handbook.cochrane.org.
Accessed July 27, 2013
|
|
24
|
Bentzen SM: Preventing or reducing late
side effects of radiation therapy: radiobiology meets molecular
pathology. Nat Rev Cancer. 6:702–713. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Andreassen CN and Alsner J: Genetic
variants and normal tissue toxicity after radiotherapy: a
systematic review. Radiother Oncol. 92:299–309. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barnett GC, Coles CE, Elliott RM, et al:
Independent validation of genes and polymorphisms reported to be
associated with radiation toxicity: a prospective analysis study.
Lancet Oncol. 13:65–77. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
West CM, Dunning AM and Rosenstein BS:
Genome-wide association studies and prediction of normal tissue
toxicity. Semin Radiat Oncol. 22:91–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Talbot CJ, Tanteles GA, Barnett GC, et al:
A replicated association between polymorphisms near TNFα and risk
for adverse reactions to radiotherapy. Br J Cancer. 107:748–753.
2012.PubMed/NCBI
|
|
29
|
Kerns SL, Ostrer H, Stock R, et al:
Genome-wide association study to identify single nucleotide
polymorphisms (SNPs) associated with the development of erectile
dysfunction in African-American men after radiotherapy for prostate
cancer. Int J Radiat Oncol Biol Phys. 78:1292–1300. 2010.
View Article : Google Scholar
|
|
30
|
Lorimore SA, Coates PJ and Wright EG:
Radiation-induced genomic instability and bystander effects:
inter-related nontargeted effects of exposure to ionizing
radiation. Oncogene. 22:7058–7069. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Barnett GC, West CM, Coles CE, et al:
Standardized total average toxicity score: a scale- and
grade-independent measure of late radiotherapy toxicity to
facilitate pooling of data from different studies. Int J Radiat
Oncol Biol Phys. 82:1065–1074. 2012. View Article : Google Scholar
|