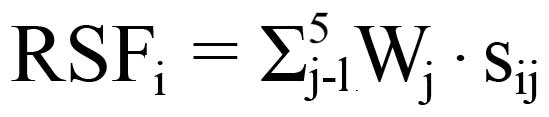Introduction
Cervical cancer is the third most commonly diagnosed
cancer and the fourth leading cause of cancer-related mortality in
women worldwide, accounting for 9% (529,800) of total new cancer
cases and 8% (275,100) of total cancer deaths among women in 2008
(1). Cervical cancer is also the
major cause of death in women of reproductive age (2–4). Cervical
cancer is characterized by a long period of preclinical disease
progressing through a number of well-defined premalignant stages.
The transition of normal epithelium to preneoplastic cervical
intraepithelial neoplasia (CIN) and invasive cervical cancer may
require >10 years (5). The overall
survival was reported to be ~80–90% for stage Ib, 70–80% for stage
II, 60% for stage III and 15% for stage IVa disease (6). From a clinical perspective, if detected
prior to the point of progression to invasive disease, a variety of
treatment options are available and the disease is almost certainly
curable. Unfortunately, the early stages of cervical cancer are
usually asymptomatic. When common signs and symptoms (such as
vaginal bleeding and/or discharge and pelvic or back pain) appear,
the disease is usually at an advanced stage. Furthermore, a number
of these symptoms are non-specific and may represent a variety of
different conditions (7).
Cervical carcinoma in situ is confined to the
epithelial layer; therefore, the Papanicolaou test (Pap smear)
cannot effectively detect the lesion. A technology assessment of
cervical cytology concluded that conventional Pap smear screening
had a specificity of 98%, but sensitivity of only 51% (8). Pap smear screening is not highly
effective at detecting adenocarcinoma, or its precursors.
Colposcopy and random biopsies may diagnose some early-stage
cervical cancer patients, although the invasiveness of these
diagnostic procedures limits their efficacy. CIN is asymptomatic
and essentially unrecognizable on gross inspection or palpation
(9). Squamous cell carcinoma (SCC)
antigen and carbohydrate antigen 125 (CA125) are two serum tumor
biomarkers commonly used in clinical practice to detect and monitor
cervical cancer; however, neither of these two makers is specific
to this malignancy. In addition, both markers have a poor
sensitivity for early-stage cervical cancer (26 and 23﹪,
respectively) (10,11).
Specific and sensitive non-invasive biomarkers for
the detection of human epithelial malignancy are urgently required
to reduce the worldwide morbidity and mortality of cervical cancer.
MicroRNAs (miRNAs) are 19–24-nt non-coding RNAs that are frequently
deregulated in cancer and have shown great promise as tissue-based
markers for cancer classification (12–14).
Recent studies have also demonstrated that the differential
expression of tissue miRNAs in cervical cancer may be of
substantial diagnostic and prognostic value (15,16).
The surgical collection of tissue samples is an
invasive procedure. By contrast, serum sample collection is an easy
and cost-effective method. We systematically demonstrated that the
unique expression patterns of these circulating miRNAs are
correlated with certain human diseases (17–23).
However, the global miRNA pattern in the sera of cervical cancer
patients has not yet been determined.
By analyzing the genome-wide miRNA expression
profile using Solexa sequencing and the stem-loop miRNA
quantitative polymerase chain reaction (qPCR) assay, we aimed to
determine a unique panel of miRNAs that are differentially
expressed in the serum of cervical cancer patients.
Materials and methods
Study design, patients and
controls
All the samples were collected from consenting
individuals according to the protocols approved by the Ethics
Committee of the First Affiliated Hospital of Nanjing Medical
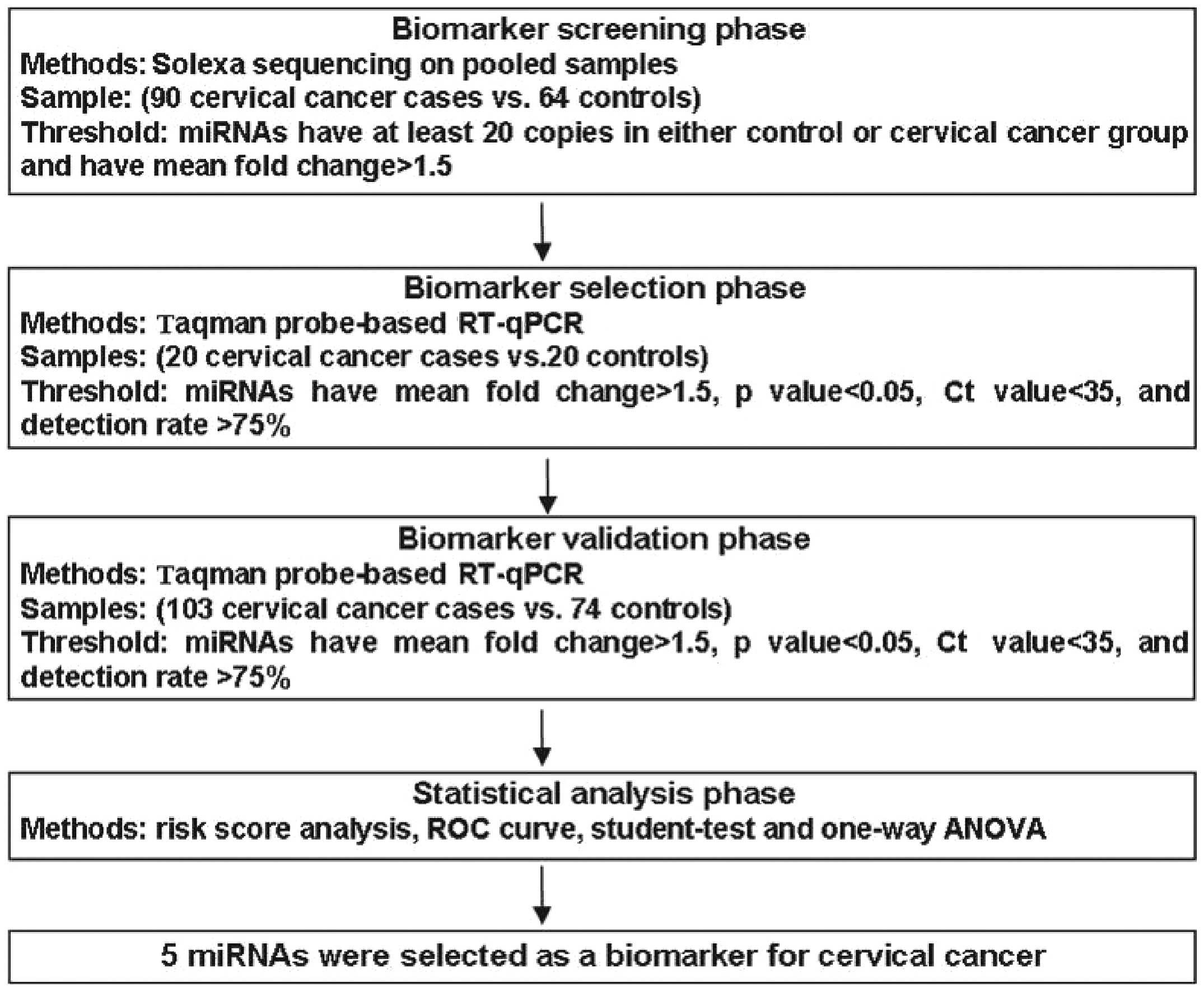
University (Nanjing, China). A multistage case-control study was
designed to identify a serum miRNA profile for cervical cancer
(Fig. 1). In total, 213 patients with
primary cervical cancer and 158 control subjects were enrolled in
our study. In the initial screening stage, cervical cancer serum
samples pooled from 90 cervical cancer patients and control donors
pooled from 64 normal samples were subjected to Solexa sequencing
to identify the miRNAs. We subsequently performed a confirmation
analysis with a hydrolysis probe-based reverse transcription-qPCR
(RT-qPCR) assay to refine the number of serum miRNAs in the
cervical cancer signature (123 cervical cancer and 94 control
samples). This analysis was performed in two phases: i) Training
set, serum samples from 20 cervical cancer patients and 20
controls; and ii) validation set, an additional 103 cervical cancer
serum samples and 74 normal subjects. All the patients were
diagnosed with cervical carcinoma and treated at the Jinling
Hospital between March, 2010 and December, 2011. Blood samples were
collected prior to any therapeutic procedures, such as surgery,
chemotherapy or radiotherapy. The results were histopathologically
confirmed following surgical resection of the tumors and tumor
staging was performed according to criteria of the International
Federation of Gynecology and Obstetrics. For patients who were
unsuitable for surgical management, the histopathological
characteristics and tumor stage were confirmed by histobiopsy and
imaging technology. Control participants were recruited from a
large pool of individuals undergoing a routine health checkup at
the Jinling Hospital (Nanjing, China). The demographics and
clinical characteristics of the patients in the training and
validation sets are listed in Table
I. The controls were matched to the patients by age and
ethnicity. None of the healthy controls had previously been
diagnosed with malignancy.
 | Table I.Demographic and clinical
characteristics of the patients and control individuals in the
training and validation sets. |
Table I.
Demographic and clinical
characteristics of the patients and control individuals in the
training and validation sets.
| Variables | Cases, no. (%)
(n=123) | Controls, no. (%)
(n=94) | P-value |
|---|
| Age, years |
|
|
|
| (mean ± SD) | 46.0±8.6 | 47.8±7.5 | 0.350a |
|
≥55 | 20 (16.2) | 18 (19.1) | 0.303b |
|
45–54 | 61 (50.0) | 53 (56.4) |
|
|
<45 | 42 (33.8) | 23 (24.5) |
|
| Marital status |
|
| 0.892b |
|
Married | 123 (100) | 93 (98.9) |
|
|
Unmarried | 0 (0.0) | 1 (1.1) |
|
| Menopausal
status |
|
| 0.248b |
|
Postmenopausal | 27 (21.9) | 18 (19.1) |
|
|
Premenopausal | 77 (62.6) | 53 (56.4) |
|
|
Unknown | 19 (15.5) | 23 (24.5) |
|
| FIGO stage |
|
| – |
| 0
(CIN) | 38 (31.4) |
|
|
| I | 59 (47.0) |
|
|
| II | 24 (19.6) |
|
|
|
III | 2 (2) |
|
|
| IV | 0 (0) |
|
|
| Histological
differentiation |
|
| – |
|
Moderate or high | 39 (31.7) |
|
|
|
Poor | 25 (20.6) |
|
|
|
CIN | 38 (30.9) |
|
|
|
Undetermined | 21 (16.8) |
|
|
| Tumor
histology |
|
| – |
|
Adenocarcinoma | 8 (6.5) |
|
|
|
SCC | 114 (92.7) |
|
|
| Clear
cell carcinoma | 1 (0.8) |
|
|
| Significant cardiac
dysfunction |
|
| 0.813b |
|
Yes | 3 (2.4) | 1 (1.1) |
|
| No | 120 (97.6) | 93 (98.9) |
|
| Hypertension |
|
| 0.106b |
|
Yes | 10 (8.1) | 1 (1.1) |
|
| No | 113 (91.9) | 93 (98.9) |
|
| Neurological
disease or diabetes |
|
| 0.599b |
|
Yes | 1 (0.8) | 0 (0.0) |
|
| No | 122 (99.2) | 94 (100) |
|
Sample processing and RNA
extraction
Serum separation was performed by centrifugation of
the blood samples at 3,000 × g for 10 min, followed by a 15-min
high-speed centrifugation at 12,000 × g. Subsequently, the
supernatant sera were stored at −80°C until further analysis.
For the Solexa sequencing assay, equal volumes of
sera from 90 patients with cervical cancer (0.67 ml each) and 64
controls with similar age and gender distributions (0.94 ml each)
were pooled separately to form the case and control sample pools
(Table II). TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) was used
according to the manufacturer's instructions with minor
modifications to extract total RNA from each pool of serum samples
(~60 ml). The aqueous phase was subjected to three steps of acid
phenol/chloroform purification to eliminate protein residues prior
to isopropyl alcohol precipitation. The resulting RNA pellet was
dissolved in 20 µl diethylpyrocarbonate-treated water and stored at
−80°C until further analysis.
 | Table II.Information of cervical cancer
patients and healthy controls in Solexa sequencing. |
Table II.
Information of cervical cancer
patients and healthy controls in Solexa sequencing.
| Variables | Cervical cancer
cases, no. (%) (n=90) | Controls, no. (%)
(n=64) |
|---|
| Age, years |
|
|
| (mean ± SD) | 46.0±9.3 | 46.0±7.1 |
|
≥55 | 24 (26.5) | 13 (20.3) |
|
45–54 | 42 (47.0) | 27 (42.2) |
|
<45 | 24 (26.5) | 24 (37.5) |
| Histological
differentiation |
|
|
|
Poor | 9 (10.5) |
|
|
Moderate | 50 (55.3) |
|
|
High | 31 (34.2) |
|
| FIGO stage |
|
|
| 0
(CIN) | 33 (36.4) |
|
| I | 45 (50.0) |
|
| II | 11 (12.1) |
|
|
III | 1 (1.5) |
|
For the RT-qPCR assay, total RNA was extracted from
100 µl serum with a one-step phenol/chloroform purification
protocol. In brief, 100 µl serum was mixed with 200 µl acid phenol,
200 µl chloroform and 300 µl diethylpyrocarbonate-treated water.
The mixture was vortex-mixed vigorously and incubated at room
temperature for 15 min. Following phase separation, the aqueous
layer was mixed with 40 µl sodium acetate (3 mol/l, pH 5.3) and 800
µl isopropyl alcohol. This solution was stored at −20°C for 1 h.
The RNA pellet was collected by centrifugation at 16,000 × g for 20
min at 4°C. The resulting RNA pellet was washed once with 750 ml/l
ethanol and dried for 10 min at room temperature. Finally, the
pellet was dissolved in 20 µl of ribonuclease-free water and stored
at −80°C until further analysis.
Solexa sequencing and in silico
analysis
First, total RNA was extracted as mentioned above.
Using PAGE purification, all the small RNA molecules (<30 bp)
were isolated. After ligating a pair of adaptors to their 5′ and 3′
ends, the small RNA molecules were amplified for 17 cycles and then
~90-bp fragments were isolated from agarose gels. The Illumina
genome analyzer (Illumina, San Diego, CA, USA) was used for cluster
generation and sequencing analysis according to the manufacturer's
instructions. Finally, the reads were processed for in
silico analysis as previously described (23).
RT-qPCR
Briefly, total RNA (2 µl) was reverse-transcribed to
cDNA using AMV reverse transcriptase (Takara Biomedical Technology,
Dalian, China) and the stem-loop RT primer (Applied Biosystems,
Foster City, CA, USA). qPCR was performed using TaqMan miRNA probes
(Applied Biosystems) on the Applied Biosystems 7300 Sequence
Detection system. All the reactions were run in triplicate and the
Ct values were determined using the fixed threshold settings. U6
and 5S rRNA are degraded in serum samples and there is no current
consensus on housekeeping miRNAs for RT-qPCR analysis of serum
miRNAs; furthermore, our group observed that the expression level
of the mixture of let-7i, −7g and −7d is rather stable in human
serum (24). Therefore, the miRNA
expression level was normalized to the mixture of let-7i, −7j and
−7d in our study. The relative expression levels of target miRNAs
were determined by the 2−ΔΔCt equation, in which ΔCt was
calculated as follows: ΔCt = CtmiR-of-interest -
Ctlet-7. The miRNA expression levels were also
normalized to the serum volume in this study.
SCC antigen and CA125
determination
The serum levels of SCC antigen and CA125 were
measured by chemiluminescence immunoassay using an ARCHITECT™
i2000SR Access Immunoassay system (Abott, Lake Forest, IL,
USA).
Statistical analysis
Quantitative data are presented as means ± standard
deviation. Statistical significance was determined using the
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference. For each miRNA, we
constructed a receiver operating characteristic (ROC) curve and
calculated the area under the ROC curve (AUC) to evaluate the
specificity and sensitivity of cervical cancer prediction. We
performed a risk score analysis to evaluate the associations
between cervical cancer and serum miRNA expression level. The risk
score of each miRNA, denoted as s, was set to 1 if the expression
level was higher than the upper 95% confidence interval (95% CI)
for the corresponding miRNA level in controls and to 0 otherwise. A
risk score function (RSF) to predict cervical cancer risk was
defined according to a linear combination of the expression level
for each miRNA. For example, the RSF for sample i using information
from 5 miRNAs was calculated as follows:
In this equation, sij is the risk score
for miRNA j on sample i and Wj is the weight of the risk
score of miRNA j. To determine Ws, five univariate
logistic regression models were fitted using the disease status
with each of the risk scores. The regression coefficient of each
risk score was used as the weight to indicate the contribution of
each miRNA to the RSF. Frequency tables and ROC curves were then
used to evaluate the diagnostic effects of the profiling and to
determine the appropriate cut-off point. All the statistical
analyses were performed with Statistical Analysis system software,
version 9.1.3 (SAS Institute, Cary, NC, USA).
Results
Clinical characteristics of the
patients
All the patients enrolled in the present study had
been clinically and pathologically diagnosed with cervical cancer.
There was no significant difference in the distribution of age,
marital and menopausal status between the cancer patients and the
controls. In general, cervical cancer patients and controls had no
other diseases, including significant cardiac dysfunction,
hypertension, neurological disorders or diabetes at the time of
blood sample collection (Table
I).
Solexa sequencing of serum miRNAs in
cervical cancer
The Solexa data revealed that miRNAs were the major
components of small RNAs (<30 bp) in the serum. The expression
of a miRNA was considered ‘significantly altered’ only if ≥20
copies were detected by Solexa sequencing, together with a
>1.5-fold change in its expression level between the patient and
control groups. Based on these criteria, the 12 miRNAs found to be
differentially expressed in cervical cancer were further analyzed
by RT-qPCR (Table III).
 | Table III.Differentially expressed microRNAs
(miRNAs) in cervical cancer patient serum samples compared to
controls, as determined by Solexa sequencing. |
Table III.
Differentially expressed microRNAs
(miRNAs) in cervical cancer patient serum samples compared to
controls, as determined by Solexa sequencing.
| miRNAs | Copy no. in
cervical cancer | Copy no. in
controls | Cervical
cancer/healthy |
|---|
| hsa-miR-21 | 677 | 101 | 6.70 |
| hsa-miR-29a | 1,159 | 253 | 4.58 |
| hsa-miR-486-5p | 23,699 | 9,509 | 2.49 |
| hsa-miR-25 | 114 | 46 | 2.49 |
|
hsa-miR-146b-5p | 346 | 159 | 2.18 |
| hsa-miR-423-3p | 52 | 26 | 2.00 |
| hsa-miR-140-3p | 455 | 235 | 1.94 |
| hsa-miR-101 | 61 | 32 | 1.90 |
| hsa-miR-26b | 36 | 21 | 1.72 |
| hsa-miR-191 | 179 | 109 | 1.64 |
| hsa-miR-29c | 61 | 37 | 1.64 |
| hsa-miR-200a | 32 | 21 | 1.50 |
Confirmation of miRNA production by
RT-qPCR analysis
We used the RT-qPCR assay to confirm the expression
of candidate miRNAs. In the training set, miRNAs were measured in a
separate set of individual serum samples from 20 cervical cancer
patients and 20 healthy controls; only miRNAs with a mean change
>1.5-fold and a P<0.05 were selected for further analysis
(Fig. 1). This phase generated a list
of 5 miRNAs that had a significant differential expression pattern,
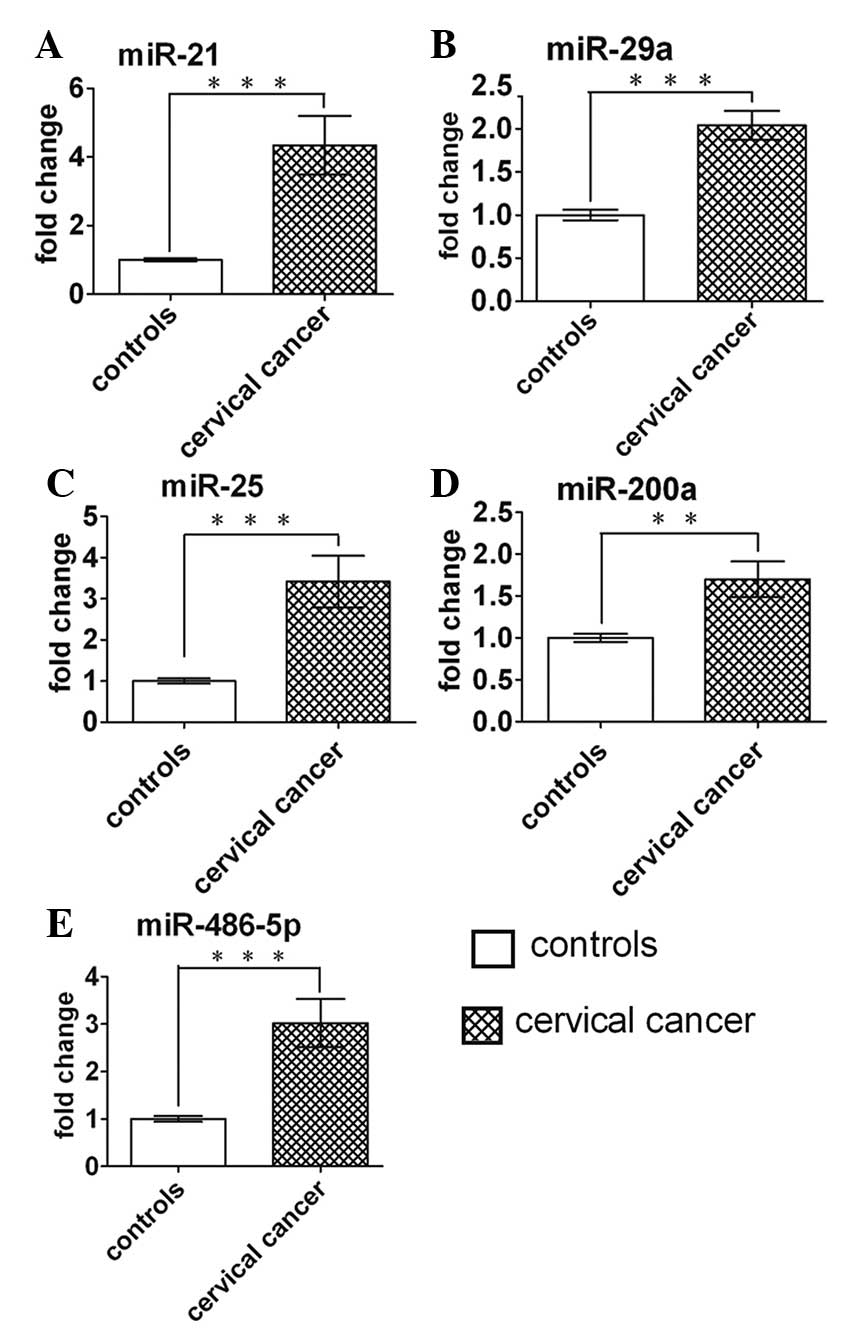
namely miR-21, −29a, −200a, −25 and −486-5p. Compared to their
levels in control samples, these 5 miRNAs were increased by 3.99-,
1.70-, 1.88-, 2.12- and 2.62-fold, respectively, in cervical cancer
samples (Table IV).
 | Table IV.Differentially expressed serum
microRNAs (miRNAs) in cervical cancer cases compared to controls in
the training and validation sets. |
Table IV.
Differentially expressed serum
microRNAs (miRNAs) in cervical cancer cases compared to controls in
the training and validation sets.
|
| Training set
(fold-change ± SD) | Validation set
(fold-change ± SD) |
|---|
|
|
|---|
| miRNAs | Controls
(n=20) | Cervical cancer
(n=20) | P-value | Controls
(n=74) | Cervical cancer
(n=103) | P-value |
|---|
| miR-21 |
1±0.06 |
3.99±0.82 |
4.8×10−3 |
1±0.06 |
4.42±1.00 |
3.8×10−3 |
| miR-29a |
1±0.07 |
1.70±0.35 |
3.5×10−2 |
1±0.05 |
2.04±0.17 |
3.9×10−6 |
| miR-200a |
1±0.06 |
1.88±0.44 |
3.3×10−2 |
1±0.05 |
1.65±0.24 |
9.3×10−3 |
| miR-25 |
1±0.16 |
2.12±0.44 |
1.1×10−2 |
1±0.07 |
3.63±0.72 |
3.6×10−3 |
| miR-486-5p |
1±0.07 |
2.62±0.72 |
1.8×10−2 |
1±0.09 |
3.18±0.72 |
3.0×10−3 |
These 5 miRNAs were further examined by RT-qPCR in a
larger cohort including 103 cervical cancer patients and 74 matched
controls. The miRNA expression pattern alterations in the
validation set were consistent with those in the training set
(Table IV). The differences in
concentration for the 5 miRNAs in 123 cervical cancer patients and
94 control subjects enrolled in the training and validation sets
are shown in Fig. 2.
Risk score and ROC curve analysis
To further evaluate the diagnostic value of the
5-miRNA profiling system, we used a risk score formula to calculate
RSF for cervical cancer and control samples. The samples were
ranked according to their RSF and then divided into a high-risk
group (predicted cervical cancer cases) and a low-risk group
(control individuals). The frequency table and the ROC curves were
then used to evaluate the diagnostic effect of the 5-miRNA
profiling system and determine the appropriate cut-off point.
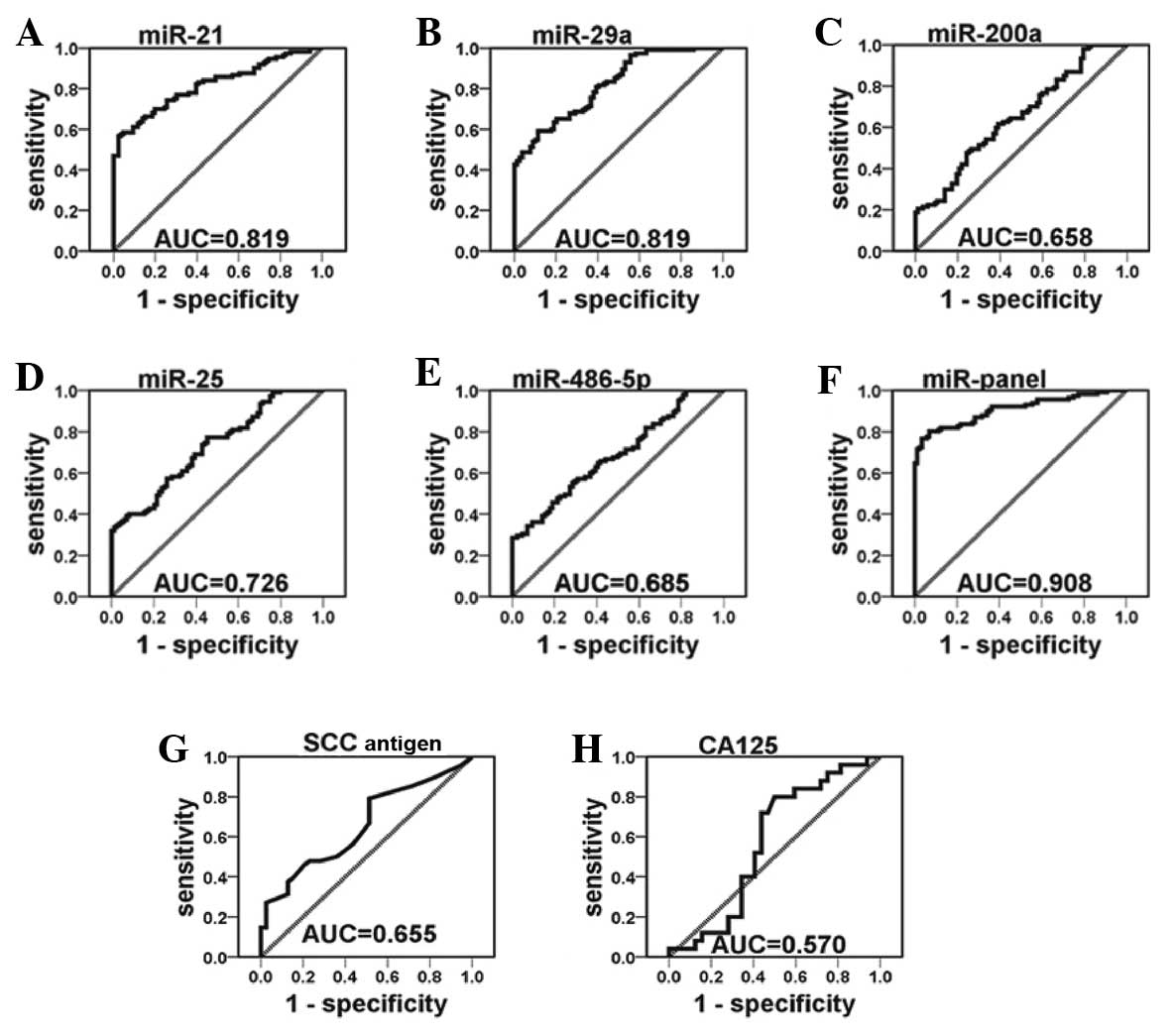
The ROC curves constructed to compare the relative
concentrations of the 5 miRNAs in the cervical cancer patients and
healthy controls yielded the following AUCs: miR-21, 0.819 (95% CI:
0.762–0.876); miR-29a, 0.819 (95% CI: 0.762–0.876); miR-25, 0.726
(95% CI: 0.656–0.795); miR-200a, 0.658 (95% CI: 0.575–0.728) and
miR-486-5p, 0.685 (95% CI: 0.610–0.759) (Fig. 3A–E). Among the 5 miRNAs investigated,
miR-21 displayed the highest sensitivity and specificity for
cervical cancer diagnosis. To illustrate the contribution of
individual serum miRNAs to the AUC of the ROC curve, we established
ROC curves to evaluate the diagnostic value of each miRNA for
differentiating between cervical cancer cases and controls. We
found that the subsequent addition of each of the 5 miRNAs
incrementally improved the sensitivity and specificity of the
miRNA-based biomarkers in discriminating cervical cancer cases from
controls (Fig. 3F). The AUC value of
the combination of 5 miRNAs [0.908 (95% CI: 0.868–0.948)], was
markedly higher compared with that of the SCC antigen [0.655 (95%
CI: 0.541–0.770)] and CA125 [0.570 (95% CI: 0.418–0.722)] (Fig.3G and H). With an optimal cut-off value,
in which the sum of the sensitivity and specificity was maximal,
the specificity was 88.6 and the sensitivity was 81.0%. The
positive and negative predictive values were 0.90 and 0.78,
respectively (Table V). The results
indicated that the 5-serum miRNA signature is more reliable
compared with any single miRNA-based assay, the SCC antigen or
CA125 in the diagnosis of cervical cancer.
 | Table V.Risk score analysis of cervical
cancer and control subjects on the 5-microRNA profile. |
Table V.
Risk score analysis of cervical
cancer and control subjects on the 5-microRNA profile.
| Group | 0–0.50 | 0.50–2.00 | PPV | NPV |
|---|
| Control | 83 | 11 | – | 0.78 |
| Cervical
cancer | 23 | 100 | 0.90 | – |
Altered serum miRNAs by different
stage and differentiation type
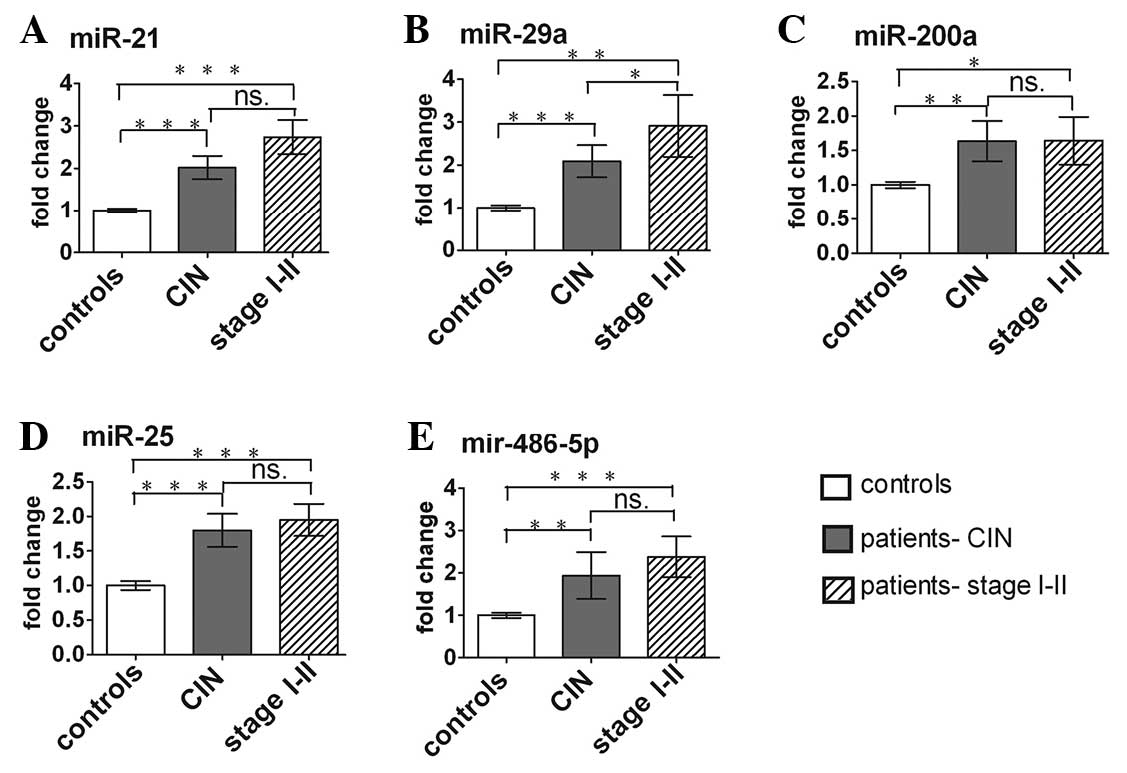
The expression levels of the 5 serum miRNAs in
cervical cancer patients at different stages and different
differentiation types were analyzed. The serum miRNAs in the
cervical cancer patients increased significantly in CIN cases
compared with normal controls. The fold-change of miR-21, −29a,
−200a, −25 and −486-5p was 2.02, 2.10, 1.64, 1.84 and 1.94,
respectively (Fig. 4). As shown in
Fig. 4B, the serum miR-29a level was
progressively higher from earlier-stage disease (CIN) to later
stages (I and II). In addition, miR-29a and miR-200a also exhibited
a distinct upregulation in poorly differentiated cases compared
with patients with well- or moderately differentiated tumors.
Discussion
miRNAs in human serum and other body fluids remain
stable after being subjected to harsh conditions, under which most
RNAs would be degraded (25–27). Possible explanations for the
remarkable stability of miRNAs in the serum are that they are
protected by binding proteins or microvesicles and that they may be
chemically modified (e.g., methylation) (26,27).
Several studies have suggested that active secretion by cells is a
major source of serum miRNAs. Furthermore, recent studies revealed
the novel genetic exchange between cells using miRNAs, either in
microvesicles (≤1 µm) or in small membrane vesicles of endocytic
origin, referred to as exosomes (50–100 nm) (28–30). The
secreted miRNAs contained in exosomes may also be transferred from
tumor cells to tumor cells, or from tumor cells to normal cells,
indicating that an oncogene may be propagated horizontally through
exosomes (31). In addition to this
high stability, the characteristics of miRNAs, such as
tissue-specific miRNA signatures and the availability of several
copies per cell, indicate potential advantages as biomarkers
compared with other nucleic acids, such as circulating DNA and mRNA
(32,33). Thus, previous findings support that
circulating miRNAs may be used as non-invasive diagnostic
markers.
Despite differences in pathogenesis, tumors share
common characteristics, such as unlimited proliferation and rapid
metastasis. The upregulation of certain miRNAs is likely to be
observed in the sera of patients with tumors. In this study, we
systematically demonstrated that serum miR-21, −29a, −25, −200a and
−486-5p may serve as a non-invasive, accurate biomarkers for
cervical cancer diagnosis. The ROC curves indicated that a panel of
5 miRNAs has great potential as a more sensitive and specific
diagnostic test compared with any single miRNA-based assay, the SCC
antigen and CA125 for cervical cancer.
The functional study of miRNAs in tumor tissue may
also be helpful for evaluating serum miRNAs as indicators of
various types of cancer. Hu et al (16) suggested that miR-200a is potentially
involved in tumor control by regulating cancer cell metastasis.
Zhang et al (34) demonstrated
that miR-25 directly regulates apoptosis by targeting Bim in
ovarian cancer. miR-486-5p may function as a novel tumor suppressor
miRNA in gastric cancer and its anti-oncogenic activity may involve
the direct targeting and inhibition of olfactomedin 4 (35). Such findings suggested that serum
miRNAs may play a pivotal and general role as signaling molecules
in physiological and pathological events.
Patients with CIN may undergo extensive
hysterectomy, which is likely curative in these patients. We
further focused on whether these miRNAs may be used as diagnostic
markers for early cervical cancer. These 5 miRNAs were clearly
upregulated in cervical cancer patient serum samples compared with
control samples and exhibited an average ~1.8-fold change in
patients with CIN, whereas the Pap smear is relatively inefficient.
More importantly, miR-29a and miR-200a may be of value for clinical
monitoring and prognosis. The results suggested a potential
application in diagnosing cervical cancer at an early stage, even
at the precancerous lesion stage.
In 1995 the World Health Organization declared human
papillomavirus (HPV) as a known carcinogen for cervical cancer, as
the DNA of mucosal high-risk HPV types was detected in almost all
cervical cancers (36). The
elucidation of unidentified genetic alterations due to HPV and
miRNA interactions may shed more light on the mechanistic
underpinnings of HPV-induced oncogenesis (37). HPVs exhibit oncogenic properties, at
least in part by reshaping the milieu of cellular miRNAs and
miR-29a is associated with HPV E6/E7 expression in vivo at
the pre-neoplastic stage (38).
Hence, our results may also be helpful in investigating the
mechanisms underlying the development of HPV-infected malignant
neoplasms in the future.
In summary, our study identified a panel of 5 serum
miRNAs as a signature for cervical cancer detection at its early
stages. miR-29a and miR-200a were also clearly upregulated in
poorly differentiated cases compared with patients with well- or
moderately differentiated tumors. These results may provide impetus
for the clinical value of serum miRNAs in predicting the prognosis
of cervical cancer.
Acknowledgements
This study was supported by National Natural Science
Foundation of China (grant nos. 30570731, 30871195, 81070653,
81270907, 81370926, J1103512 and J1210026).
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
Pap
|
Papanicolaou test
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
CIN
|
cervical intraepithelial neoplasia
|
|
SCC
|
squamous cell carcinoma
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the curve
|
|
RSF
|
risk score function
|
|
95% CI
|
95% confidence interval
|
|
HPV
|
human papillomavirus
|
References
|
1
|
Siegel R, Naishadham D, Jemal A, et al:
Cancer statistics, 2012. CA Cancer J Clin. 62:10–29. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Almonte M, Bruni L, et al:
Burden and trends of type-specific human papillomavirus infections
and related diseases in the Latin America and Caribbean region.
Vaccine. 26:(Suppl 11). L1–L15. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mathew A and George PS: Trends in
incidence and mortality rates of squamous cell carcinoma and
adenocarcinoma of cervix - worldwide. Asian Pac J Cancer Prev.
10:645–650. 2009.PubMed/NCBI
|
|
4
|
Vizcaino AP, Moreno V, Bosch FX, et al:
International trends in incidence of cervical cancer: II.
Squamous-cell carcinoma. Int J Cancer. 86:429–435. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Richart RM: A modified terminology for
cervical intraepithelial neoplasia. Obstet Gynecol. 75:131–133.
1990.PubMed/NCBI
|
|
6
|
Mayr NA, Small W Jr and Gaffney DK:
Cervical cancerDecision Making in Radiation Oncology. Lu JJ and
Brady LW: 2. Springer-Verlag; Berlin Heidelberg: pp. 661–701.
2011
|
|
7
|
Kyrgiou M and Mahmood SI: Invasive cancer
of the cervix. Obstet Gynecol Reprod Med. 20:147–154. 2010.
View Article : Google Scholar
|
|
8
|
Wright TC Jr, Schiffman M, Solomon D, et
al: Interim guidance for the use of human papillomavirus DNA
testing as an adjunct to cervical cytology for screening. Obstet
Gynecol. 103:304–309. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baker JJ: Conventional and liquid-based
cervicovaginal cytology: a comparison study with clinical and
histologic follow-up. Diagn Cytopathol. 27:185–188. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Avall-Lundqvist EH, Sjövall K, Nilsson BR,
et al: Prognostic significance of pretreatment serum levels of
squamous cell carcinoma antigen and CA 125 in cervical carcinoma.
Eur J Cancer. 28A:1695–1702. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bolli JA, Doering DL, Bosscher JR, et al:
Squamous cell carcinoma antigen: clinical utility in squamous cell
carcinoma of the uterine cervix. Gynecol Oncol. 55:169–173. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee JW, Choi CH, Choi JJ, et al: Altered
microRNA expression in cervical carcinomas. Clin Cancer Res.
14:2535–2542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pereira PM, Marques JP, Soares AR, et al:
MicroRNA expression variability in human cervical tissues. PLoS
One. 5:e117802010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu X, Schwarz JK, Lewis JS Jr, et al: A
microRNA expression signature for cervical cancer prognosis. Cancer
Res. 70:1441–1448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang C, Wang C, Chen X, et al:
Identification of seven serum microRNAs from a genome-wide serum
microRNA expression profile as potential noninvasive biomarkers for
malignant astrocytomas. Int J Cancer. 132:116–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li C, Fang Z, Jiang T, et al: Serum
microRNAs profile from genome-wide serves as a fingerprint for
diagnosis of acute myocardial infarction and angina pectoris. BMC
Med Genomics. 6:162013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li LM, Hu ZB, Zhou ZX, et al: Serum
microRNA profiles serve as novel biomarkers for HBV infection and
diagnosis of HBV-positive hepatocarcinoma. Cancer Res.
70:9798–9807. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Hu Z, Wang W, et al:
Identification of ten serum microRNAs from a genome-wide serum
microRNA expression profile as novel noninvasive biomarkers for
nonsmall cell lung cancer diagnosis. Int J Cancer. 130:1620–1628.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu R, Zhang C, Hu Z, et al: A
five-microRNA signature identified from genome-wide serum microRNA
expression profiling serves as a fingerprint for gastric cancer
diagnosis. Eur J Cancer. 47:784–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Wang C, Chen X, et al: Expression
profile of microRNAs in serum: a fingerprint for esophageal
squamous cell carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Li Q, Wang J, et al:
Identification and characterization of novel amphioxus microRNAs by
Solexa sequencing. Genome Biol. 10:R782009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Liang H, Guan D, et al: A
Combination of Let-7d, Let-7g and Let-7i serves as a stable
reference for normalization of serum microRNAs. PLoS ONE.
8:e79652(Epub ahead of print). doi. View Article : Google Scholar : 2013.PubMed/NCBI
|
|
25
|
Chim SS, Shing TK, Hung EC, et al:
Detection and characterization of placental microRNAs in maternal
plasma. Clin Chem. 54:482–490. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: a novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cocucci E, Racchetti G and Meldolesi J:
Shedding microvesicles: artefacts no more. Trends Cell Biol.
19:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ghosh AK, Secreto CR, Knox TR, et al:
Circulating microvesicles in B-cell chronic lymphocytic leukemia
can stimulate marrow stromal cells: implications for disease
progression. Blood. 115:1755–1764. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Valadi H, Ekström K, Bossios A, et al:
Exosome-mediated transfer of mRNAs and microRNAs is a novel
mechanism of genetic exchange between cells. Nat Cell Biol.
9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: a new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Calin GA and Croce CM: MicroRNA-cancer
connection: the beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang H, Zuo Z, Lu X, et al: MiR-25
regulates apoptosis by targeting Bim in human ovarian cancer. Oncol
Rep. 27:594–598. 2012.PubMed/NCBI
|
|
35
|
Oh HK, Tan AL, Das K, et al: Genetic loss
of miR-486 regulates tumor progression and the OLMF4 antiapoptotic
factor in gastric cancer. Clin Cancer Res. 17:2657–2667. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Walboomers JM, Jacobs MV, Manos MM, et al:
Human papillomavirus is a necessary cause of invasive cervical
cancer worldwide. J Pathol. 189:12–19. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Martinez I, Gardiner AS, Board KF, et al:
Human papillomavirus type 16 reduces the expression of microRNA-218
in cervical carcinoma cells. Oncogene. 27:2575–2582. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Wang F, Xu J, et al: Progressive
miRNA expression profiles in cervical carcinogenesis and
identification of HPV-related target genes for miR-29. J Pathol.
224:484–495. 2011. View Article : Google Scholar : PubMed/NCBI
|