Introduction
The role of the insulin-like growth factor 1
receptor (IGF1R) in the pathogenesis of malignant epithelial
tumours, including non-small cell lung cancer (NSCLC), has been
well-characterised (1–5). Activation of this receptor pathway
promotes tumour growth by inhibition of apoptosis, transformation,
metastasis and induction of angiogenesis through vascular
endothelial growth factor (6–10).
IGF1R is frequently overexpressed in NSCLC patients,
however, there is controversy over its significance as a prognostic
marker. Certain studies have shown no correlation between high
IGF1R expression and patient survival (3,4);
conversely, other studies have demonstrated that high IGF1R
expression was associated with nodal metastasis, recurrence and a
significantly poorer overall survival (OS) rate in NSCLC patients
(11,12). A recent meta-analysis suggests IGF1R
positive expression as an adverse factor for disease-free survival
(DFS) in NSCLC patients and reports the correlation to smoking
status and tumour size, but there was no significant association
between IGF1R expression and OS on univariate or multivariate
analysis (13). Ludovini et al
(14) reported that high
co-expression of IGF1R and epidermal growth factor receptor (EGFR)
is associated with a shorter DFS in resected NSCLC patients and a
trend towards a poorer OS. We have previously shown that high
co-expression of EGFR and IGF1R correlates with poor patient
prognosis in resected NSCLC (15).
Following the success of other targeted therapies,
such as the EGFR inhibitors, IGF1R also emerged as an attractive
therapeutic target. Although the results from early clinical trials
targeting IGF1R showed certain promise, larger randomized phase III
trials have not shown a clear clinical benefit of targeting this
pathway in combination with chemotherapy (16). The results of these trials and others
involving targeted agents have demonstrated the importance of
identifying predictive biomarkers to select the appropriate patient
population who will benefit from treatment.
Somatic mutations in the kinase domain of a receptor
can cause the cell to become highly dependent on the constitutively
active receptor signalling pathway. These aberrations can also
cause conformational changes that can impact on the binding
capabilities of a therapeutic agent targeting this region (17–22). As
many as 2,412 single-nucleotide polymorphisms (SNPs) have been
identified in IGF1R and several have been associated with a
cancer risk (22–26). A common polymorphism of the
IGFIR gene (G1013A) has been shown to modify the risk of
obesity for esophageal adenocarcinoma and, in combination with a
polymorphism in IGF2R (G1619A), is an independent prognostic
factor in advanced NSCLC (27–29). A
previous study has shown that the IGF1 pathway polymorphisms are
potential predictive/prognostic molecular markers for cetuximab
efficacy in wild-type KRAS colorectal cancer patients
(30). These polymorphisms may
activate crosstalk between the IGF1R and EGFR signalling pathways.
A study by Deming et al (31)
on genetic variation in patients with breast cancer found that SNP
rs951715 within the IGF1R gene was associated with breast
cancer survival in postmenopausal women. Another polymorphism, SNP
rs2229765, appears to be a silent mutation with no correlation to
survival rate in breast cancer patients and thus far has no
association with any epidemiological traits. In a retrospective
study of 304 NSCLC patients who underwent curative pulmonary
resection, 1 silent mutation in exon 16 and 3 intronic mutations
were detected within the IGF1R gene but did not correlate to
IGF1R protein expression (32).
Identifying functional polymorphisms in the IGF1R
pathway could be used to select patients who may benefit from
IGF1R-targeted agents. Thus far, there have been few reports of
SNPs or somatic mutations in the IGF1R tyrosine kinase (TK)
domain. Therefore, the aim of the present study was to screen NSCLC
patients, who had undergone lung tumour resection surgery, for gene
aberrations in the IGF1R TK domain.
Materials and methods
Subjects
This is a retrospective study in which a database of
all the patients who underwent curative-intent surgical resection
of a primary tumour at St. James's Hospital, Dublin (Republic of
Ireland) between February 2001 and February 2005, was analysed. A
cohort of 198 stage I–III NSCLC patients, staged according to the
International System of Staging for Lung Cancer (33), was randomly selected from the
database. Information on baseline demographics, clinicopathological
characteristics and surgical approach was collected following a
review of clinical notes and histopathology reports. Outcome data,
including peri-operative mortality and long-term survival, were
updated prospectively. Patient characteristics are detailed in
Table I. The study was approved by
the St. James's Hospital Ethics Committee. Controls (n=866) were
ascertained with written informed consent from the Trinity Biobank
and represented blood donors from the Irish Blood Transfusion
Service recruited in the Republic of Ireland. Individuals taking
regular prescribed medication are excluded from blood donation in
the Republic of Ireland and donors are not financially
remunerated.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
|
Characteristics | Cases, n | Controls, n |
|---|
| Total | 198 | 866 |
| Gender |
|
|
|
Male | 125 | 296 |
|
Female | 73 | 570 |
| Smoking
history |
|
|
|
Former | 130 | ND |
|
Current | 57 | ND |
|
Never | 11 | ND |
| Histology |
|
|
|
SCC | 94 | ND |
|
ADC | 84 | ND |
|
Other | 20 | ND |
| Age, years |
|
|
|
<50 | 28 | 698 |
|
>50 | 170 | 168 |
DNA extraction
DNA was extracted from 3×10-µm formalin-fixed
paraffin-embedded sections from NSCLC patients and prepared using
the QIAamp® DNA kit. DNA was extracted from control blood samples
using the Gentra Autopure system (Qiagen, Hilden, Germany).
PCR amplification of exons 16–20
The primer sequences used for the PCR reactions are
outlined in Table II. The following
PCR conditions were used: 1X GoTaq® Green Master mix (Promega
Corp., Madison, WI, USA), 5 µM of forward and reverse primers and
100 ng of DNA made up to a volume of 50 µl with dH20. A
pre-PCR heat step of 94°C for 5 min was carried out to activate the
enzyme and the DNA was amplified for 35 cycles at 94°C (1 min),
56°C (1 min) and 72°C (1 min), and at 72°C (10 min) after the last
cycle. A portion of the PCR product was electrophoresed on a 1.4%
agarose gel to verify product integrity. PCR products were purified
using the Qiagen QIAquick PCR purification kit. The DNA was
measured using the NanoDrop 1000 spectrophotometer.
 | Table II.Primer sequences used for
amplification of IGF1R RTK 1–6. |
Table II.
Primer sequences used for
amplification of IGF1R RTK 1–6.
| Exon | Primer sequence
(5′-3′) |
|---|
| RTK1 | F:
GGCTTGTTTCTGTACCTGCT |
|
| R:
AGCCAAGAACATACTGGGAG |
| RTK2 | F:
ACAACACAGGCATCAGCAAG |
|
| R:
GACACAGCATTTCCTTGCAG |
| RTK3 | F:
CTCGAAAGAAATTGGCATGG |
|
| R:
TCTCCAGGGGCAGACTAATG |
| RTK4 | F:
CTGCTCCAGCGTGTGACTCT |
|
| R:
GAGCTAAAGCTGGCAACGGG |
| RTK5 | F:
CTGCTCGGGATGTAAGAAGT |
|
| R:
CTCCTAATCTCCTGTGACCC |
| RTK6 | F:
CGTACGAGGTAAACAGGAG |
|
| R:
AGCTTGTTCTCCTCGCTGTA |
Sanger sequencing of PCR products
Sanger sequencing reactions were set up as follows:
2 µl BigDye® Terminator mix v3.1, 50 ng DNA, 5 µM forward or
reverse primers and 2 µl sequencing buffer, diluted to 20 µl with
water. A positive control was also set up to ensure the efficiency
of the sequencing reaction (1 µl pGem, 2 µl M13 primer, 2 µl
BigDye® Terminator mix v3.1 and 2 µl sequencing buffer). The pGem
and BigDye® Terminator mix v3.1 were sourced from Applied
Biosystems (Warrington, UK). Sequencing was performed on a 3130×l
genetic analyser (Applied Biosystems, Foster City, CA, USA) and
sequencing files were analysed using the BioEdit v 7.0.8 (Ibis
Biosciences, Carlsbad, CA, USA).
SNP genotyping assay
A TaqMan® SNP genotyping assay (Applied Biosystems)
that detected the SNP at codon 3179A>G (rs2229765) in exon 16 of
the TK domain by quantitive PCR was also used to screen patients.
TaqMan® SNP Genotyping Assay 5′ nuclease technology uses two
allele-specific TaqMan® MGB probes and a PCR primer pair to detect
the specific SNP target. The probes and primers uniquely align with
the genome, enabling TaqMan® genotyping technology to provide
unmatched specificity. Genotype data for rs2229765 was generated on
the control samples using the Genome-Wide Human SNP Array 6.0
(Affymetrix, Inc., Santa Clara, CA, USA) (34).
Immunohistochemistry
IGF1R immunohistochemical analysis was performed on
4-µm slides cut from 22 tissue microarrays and mounted on
Superfrost Plus glass slides (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The entire staining procedure was performed on
an automated immunohistochemistry device [BenchMark XT; Ventana
Medical Systems, Inc. (VMS), Tucson, AZ, USA] following the
manufacturer's instructions. In brief, slides were deparaffinized
on the thermopads using the Ez Prep reagent (950-102; VMS). For
epitope recovery, the standard CC1 buffer, a citric-acid-based
antigen retrieval solution, was used (950-102; VMS). All the
subsequent washing and blocking steps followed standard protocols.
Slides were incubated with the prediluted primary antibody
(monoclonal rabbit anti-IGF1R, clone G11; VMS) for 16 min at 37°C.
Negative controls included identically processed slides in which
the primary antibody was replaced by accordingly diluted non-immune
rabbit IgG (Abcam, Cambridge, UK; 27478). Positive controls
included identically treated paraffin slides of a H322M xenograft,
an IGF1R-overexpressing NSCLC cell line. Detection of primary
antibody binding was performed using the ultraView Universal DAB
Detection kit (760-091; VMS). After the diaminobenzidine (DAB)
reaction was developed, slides were counterstained with
haematoxylin II (760-500; VMS), dehydrated in a serial dilution of
ethanol, transferred in xylene and mounted with Eukitt (O. Kindler
GmbH, Freiburg im Breisgau, Germany).
Statistical analysis
The software package SPSS v16.0 (SPSS Inc., Chicago,
IL, USA) was used to perform the statistical analysis.
χ2 test, Cox regression analysis, Kaplan-Meier analysis
and the log-rank test were used to illustrate the significance of
various clinical characteristics. Assumption of the proportional
hazard was tested for all covariates. P<0.05 was considered to
indicate a statistically significant difference.
Results
Initially, 100 NSCLC patients were screened for the
presence of mutations, deletions or SNPs in exons 16–20 of the TK
domain of IGF1R. The polymorphism (rs2229765) located on
exon 16 of the IGF1R gene (GenBank accession NM_000875),
consisting of a G to A transition at nucleotide 3174 but not
leading to an amino acid change (Glu->Glu) at position 1043
(E1043E) (GenBank accession NP_000876), was identified (Table III). No other mutations, deletions
or SNPs were found in the TK domain. A further 98 NSCLC patients
were screened for the presence of the same SNP (rs2229765) and
compared to the control disease-free individuals (n=866). The
control patient DNA used came from the Trinity Biobank in the
Institute of Molecular Medicine, Trinity College Dublin. These
results where subsequently correlated to patient
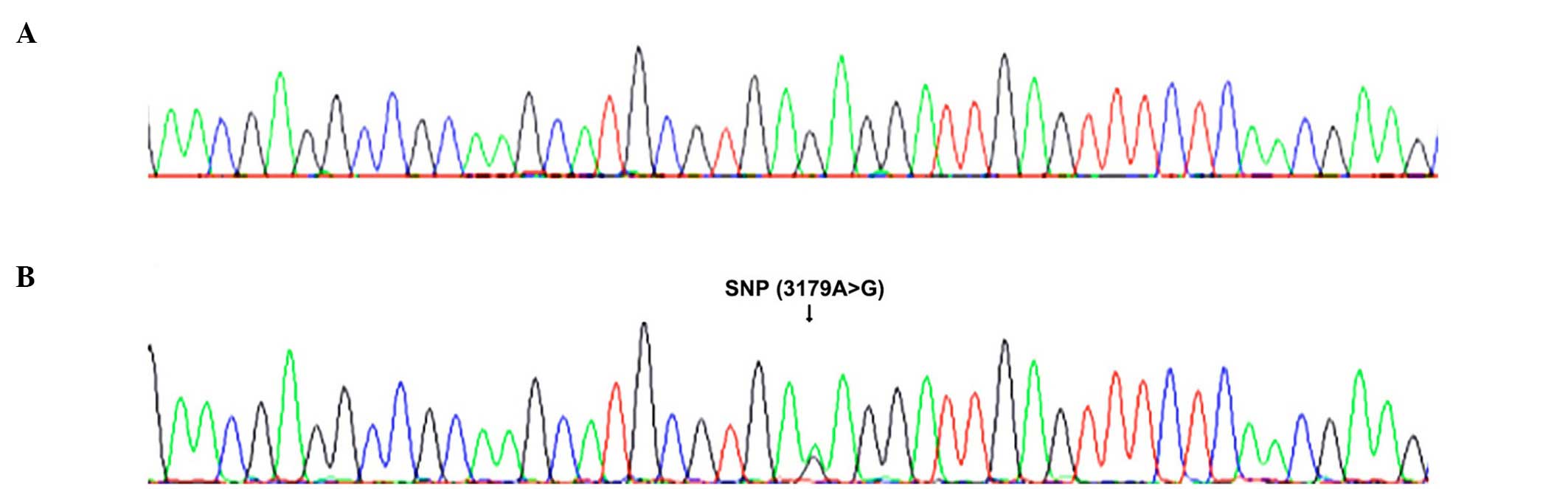
survival/pathological data. Example sequence traces from a
homozygous G/G patient and a heterozygous A/G patient are shown in
Fig. 1A and B, respectively.
 | Table III.Characteristics for the SNP
rs2229765, evaluated in the present study. |
Table III.
Characteristics for the SNP
rs2229765, evaluated in the present study.
|
Characteristics | SNP |
|---|
| Gene name | IGF1R |
| Alleles,
major>minor | G>A |
| SNP reference
ID | rs2229765 |
| Position in
gene | Exon 16 |
| Codon | Glu1013Glu |
The heterozygous GA genotype was found in 53.5 and
49.4% of NSCLC patients and controls, respectively (Table IV). The dominant GG genotype was
identified in 28.8 and 30.3% of NSCLC patients and control
individuals, respectively, while the recessive AA genotype was
found in 17.7 and 20.2% of NSCLC patients and control individuals,
respectively. No significant difference was found in the genotype
(P=0.5487) frequency between cases and controls. The A allele was
identified in 44.4 and 44.9% of NSCLC patients and control
individuals, respectively, while the G allele was identified in
55.5 and 55% of NSCLC patients and control individuals,
respectively. No significant difference was identified in the
allelic (P=0.9082) frequency.
 | Table IV.Genotypic and allelic frequency of
SNP rs2229765 in NSCLC and control patients. |
Table IV.
Genotypic and allelic frequency of
SNP rs2229765 in NSCLC and control patients.
| Frequency | NSCLC patients
(n=198), n (%) | Control patients
(n=866), n (%) | P-value |
|---|
| Genotype |
|
|
|
| AA | 35
(17.7) | 175 (20.2) | 0.5487 |
| GA | 106 (53.5) | 428 (49.4) |
|
| GG | 57
(28.8) | 263 (30.3) |
| Allele |
|
|
|
| A | 176 (44.4) | 778 (44.9) | 0.9082 |
| G | 220 (55.5) | 954 (55.0) |
From the overall patient and control cohorts,
patients >70 years were excluded as there were no matched
controls available for this age group. Age and gender matching
ensures that any difference between cases and controls is
disease-related. The genotypic and allelic frequencies were
subsequently examined in 95 NSCLC patients and 95 age- and
gender-matched control individuals. When patients were age- and
gender-matched the GA genotype was identified in 56.8 and 48.4% of
NSCLC and control patients, respectively (Table V). The dominant GG genotype was
identified in 22.1 and 29.4% of NSCLC patients and control
individuals, respectively, while the recessive AA genotype was
identified in 21.0 and 22.1% of NSCLC patients and control
individuals, respectively. No significant difference was identified
in the genotype (P=0.4351) frequency. The A allele was identified
in 49.4 and 46.3% of NSCLC patients and control individuals,
respectively, while the G allele was identified in 50.5 and 53.6%
of NSCLC patients and control individuals, respectively. No
significant difference was found in the allelic (P=0.2636)
frequency.
 | Table V.Genotypic and allelic frequency of
SNP rs2229765 in the IGF1R TK domain of NSCLC and control
patients (age- and gender-matched). |
Table V.
Genotypic and allelic frequency of
SNP rs2229765 in the IGF1R TK domain of NSCLC and control
patients (age- and gender-matched).
| Frequency | NSCLC patients
(n=95), n (%) | Control patients
(n=95), n (%) | P-value |
|---|
| Genotype |
|
|
|
| AA | 20 (21.0) | 21 (22.1) | 0.4351 |
| GA | 54 (56.8) | 46 (48.4) |
|
| GG | 21 (22.1) | 28 (29.4) |
|
| Allele |
|
|
|
| A | 94 (49.4) | 88
(46.3) | 0.2636 |
| G | 96 (50.5) | 102 (53.6) |
|
IGF1R expression and survival data were available
for 100 NSCLC patients. IGF1R expression was compared to the
results of the genotype distribution in NSCLC patients using
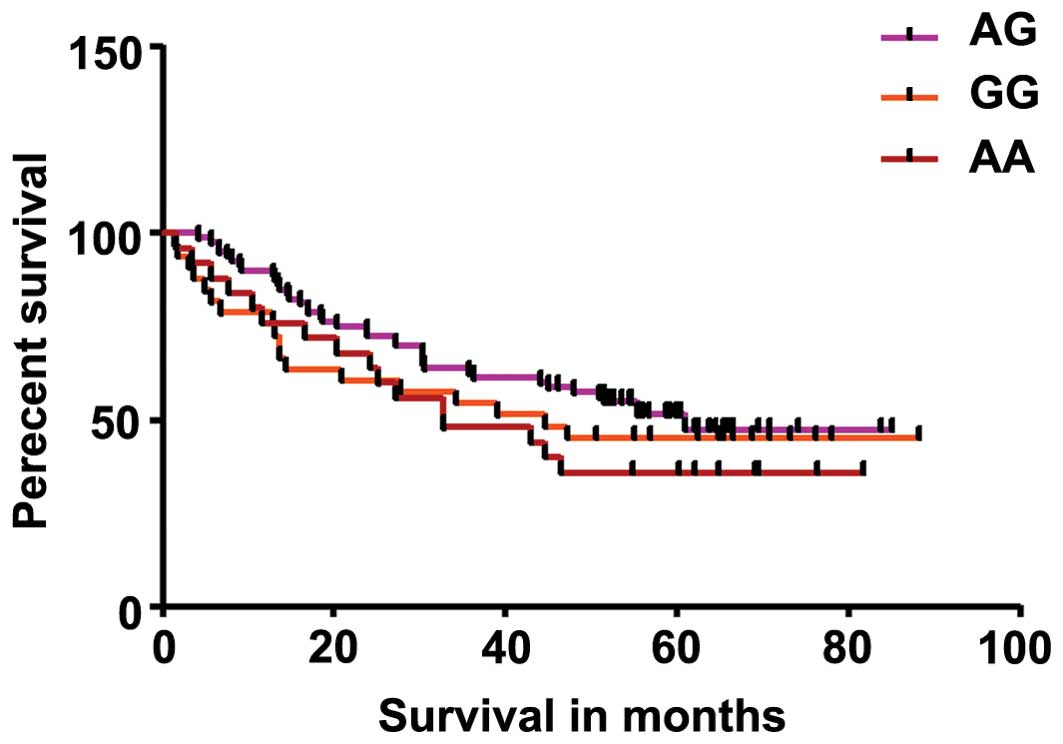
Kaplan-Meier analysis. Results showed no significant difference in
genotype distribution (Fig. 2) or
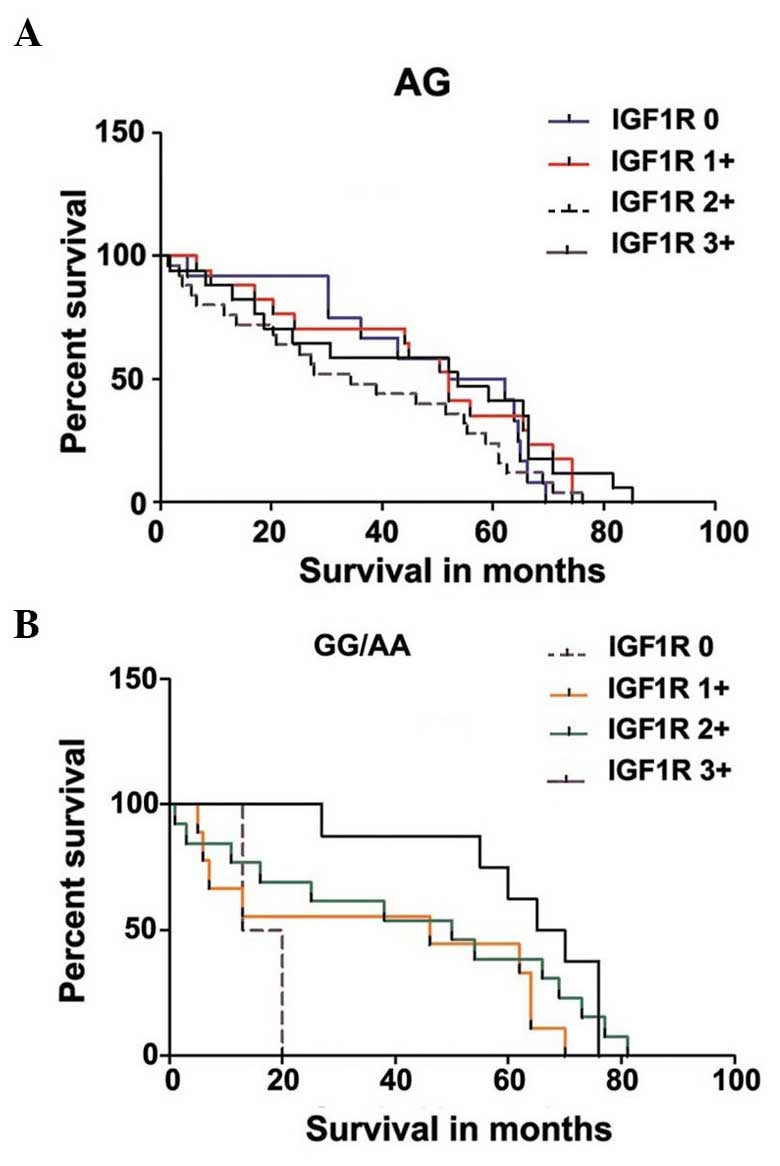
IGF1R expression (Fig. 3A and B) in
NSCLC patients. Analysis of genotype distribution in NSCLC patients
showed that the P-value was 0.3564 while the P-value for trend was
0.1514 (Fig. 2). Analysis correlating
IGF1R expression to genotype distribution in NSCLC patients showed
that the P-value was 0.3005 while the P-value for trend was 0.7099
in patients with heterozygous genotype (Fig. 3A), and that the P-value was 0.1164
while the P-value for trend was 0.0277 for patients with homozygous
dominant and homozygous recessive genotype (Fig. 3B).
Discussion
A number of previous studies have identified factors
that may influence sensitivity to IGF1R inhibitors. Kim et
al (35) evaluated the
anti-proliferation effect of figitumumab in gastric and
hepatocellular cancer cell lines and showed that the level of
N-linked glycosylated IGF1R/IR heterodimeric receptor is highly
associated with sensitivity to anti-IGF1R antibody in cancer cells.
Another study demonstrated that IGF1R TK inhibitors (TKIs)
exhibited significant antitumor activity in NSCLC cells with
wild-type EGFR and KRAS compared to those with mutations in these
genes (36).
Although SNPs have been reported in IGF1R,
there is limited knowledge regarding the frequency of gene
aberrations in the TK domain of this gene. Patients who harbour
point mutations and deletions within exons 18–21 of the EGFR
TK domain are known to have increased sensitivity to EGFR
TKIs. Therefore, the mutation status of the IGF1R TK domain
may also influence the binding of IGF1R TKIs to this
receptor and therefore influence patient response to targeted
therapy.
The present study investigated the frequency of gene
aberrations in the IGF1R TK domain in NSCLC patients and
whether such changes may influence IGF1R expression or survival
rate. Initially, 100 NSCLC patients were screened for the presence
of mutations and or deletions in exons 16–21 of the IGF1R TK
domain. No non-synonymous SNPs or deletions were detected in any of
the 100 patients screened. A synonymous SNP (rs2229765) was
identified in the coding region of exon 16. In order to strengthen
the power of the study, a further 98 NSCLC patients were screened
for the presence of the SNP (rs2229765) and the frequency was
compared to control disease-free individuals (n=866). No
significance was found in the genotype (P=0.5487) or the allelic
(P=0.9082) frequency (Table IV).
When patients were age- and gender-matched, no
significance was identified in the genotype (P=0.4351) or the
allelic (P=0.2636) frequency (Table
V). The Kaplan-Meier survival analysis showed no significant
survival advantage between the different genotypes in NSCLC
patients (Fig. 2). There was also no
significant difference when IGF1R expression was correlated
to heterozygous genotype distribution in NSCLC patients. A
significant difference for trend (P<0.05) was observed when
IGF1R expression was correlated to combine the homozygous dominant
and homozygous recessive genotype distribution in NSCLC patients
(Fig. 3A and B).
The role of the SNP, rs2229765, has been examined in
several diseases such as stroke breast cancer and type II diabetes.
It is a synonymous mutation that encodes a change in the DNA
sequence without altering the resultant protein sequence. These
silent SNPs are presumed to be not significant but they may
represent genetic markers for functional molecular alterations, as
recent studies have revealed that through various mechanisms these
synonymous SNPs may affect gene function and phenotype. Silent SNPs
have been linked to >40 diseases that are a result of a genetic
abnormality (37).
The possible role of this SNP has been investigated
in numerous studies. According to FASTSNP, it is predicted that SNP
rs2229765 may affect splicing regulation. It has been shown to
affect the susceptibility to ischemic stroke in the Chinese
population (38) and is associated
with higher plasma concentrations of circulating IGF1R. In a study
by Bonafè et al (39),
polymorphic variants of the IGF1 response pathway genes, including
IGF1R (G/A, codon 1013), phosphoinositol 3-kinase (T/C, 359 bp;
A/G, 303 bp), insulin receptor substrate-1 (G/A, codon 972) and
FOXO1A (T/C, 97,347 bp), were examined to observe whether
they are involved in systemic IGF1 regulation and human longevity.
It was found that subjects carrying at least an A allele at
IGF1R have low free plasma IGF1 levels and live longer. A
study performed in breast cancer identified that SNP rs2229765 had
no association with breast cancer survival and that it appears to
be a silent mutation (31).
Therefore, thus far it has not been associated with any
epidemiological traits. In another study, single SNP analysis
revealed a significant association of SNP rs2229765 with percent
and absolute mammographic density; increased numbers of the G
allele increased the least squares means of mammographic density
(40). The possible role of this
polymorphism has also been examined in type II diabetes, which
revealed no association with reduced birth weight, insulin
sensitivity index or type II diabetes in a Danish population
(41). These results also suggest
that the rs2229765 polymorphism leads to a silent mutation.
A previous study investigated whether germline
polymorphisms of the IGF1-pathway are associated with the response
to cetuximab in wild-type KRAS drug-refractory metastatic
colorectal cancer patients (mCRC) (28). Tissue samples from 130 drug-refractory
mCRC patients enrolled in a phase II clinical trial of cetuximab
monotherapy (IMC-0144) were used for the study. Analyses revealed
that 5 IGF-pathway SNPs were significantly associated with
progression-free survival and/or OS. Patients harbouring the
IGF1 rs2946834 A/A genotype had a 50% overall response rate,
while patients with the A/G genotype had 0%. This indicates that
IGF1-pathway polymorphisms may predict cetuximab efficacy in
wild-type KRAS mCRC patients.
Identifying functional polymorphisms in IGF1R
and its pathway could be used to select patients that would benefit
from IGF1R-targeted therapy resulting in more accurate treatment
for individuals with improved effectiveness and reduced toxicities.
In the present cohort of 100 NSCLC patients, no non-synonymous SNPs
were detected in the IGF1R TK domain. There was no significant
association between SNP rs2229765 and IGF1R expression or
patient survival. This data indicates that the IGF1R TK
domain does not appear to be as susceptible to mutations as
EGFR. Therefore, as opposed to EGFR, it will not be
necessary to screen for mutations in IGF1R to predict
response to targeted therapy.
Acknowledgements
Genetic data on the control samples were generated
by the International Schizophrenia Consortium.
References
|
1
|
Larsson O, Girnita A and Girnita L: Role
of insulin-like growth factor 1 receptor signalling in cancer. Br J
Cancer. 92:2097–2101. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Riedemann J and Macaulay VM: IGF1R
signalling and its inhibition. Endocr Relat Cancer. 13:(Suppl 1).
S33–S43. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cappuzzo F, Tallini G, Finocchiaro G,
Wilson RS, Ligorio C, Giordano L, Toschi L, Incarbone M, Cavina R,
Terracciano L, et al: Insulin-like growth factor receptor 1 (IGF1R)
expression and survival in surgically resected non-small-cell lung
cancer (NSCLC) patients. Ann Oncol. 21:562–567. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee CY, Jeon JH, Kim HJ, Shin DH, Roh TW,
Ahn CM and Chang YS: Clinical significance of insulin-like growth
factor-1 receptor expression in stage I non-small-cell lung cancer:
Immunohistochemical analysis. Korean J Intern Med. 23:116–120.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li R, Pourpak A and Morris SW: Inhibition
of the insulin-like growth factor-1 receptor (IGF1R) tyrosine
kinase as a novel cancer therapy approach. J Med Chem.
52:4981–5004. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rochester MA, Riedemann J, Hellawell GO,
Brewster SF and Macaulay VM: Silencing of the IGF1R gene enhances
sensitivity to DNA-damaging agents in both PTEN wild-type and
mutant human prostate cancer. Cancer Gene Ther. 12:90–100. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gong Y, Yao E, Shen R, Goel A, Arcila M,
Teruya-Feldstein J, Zakowski MF, Frankel S, Peifer M, Thomas RK, et
al: High expression levels of total IGF-1R and sensitivity of NSCLC
cells in vitro to an anti-IGF-1R antibody (R1507). PLoS One.
4:e72732009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vanamala J, Reddivari L, Radhakrishnan S
and Tarver C: Resveratrol suppresses IGF-1 induced human colon
cancer cell proliferation and elevates apoptosis via suppression of
IGF-1R/Wnt and activation of p53 signaling pathways. BMC Cancer.
10:2382010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hellawell GO, Turner GD, Davies DR,
Poulsom R, Brewster SF and Macaulay VM: Expression of the type 1
insulin-like growth factor receptor is up-regulated in primary
prostate cancer and commonly persists in metastatic disease. Cancer
Res. 62:2942–2950. 2002.PubMed/NCBI
|
|
10
|
Samani AA, Yakar S, LeRoith D and Brodt P:
The role of the IGF system in cancer growth and metastasis:
Overview and recent insights. Endocr Rev. 28:20–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamamoto T, Oshima T, Yoshihara K, Nishi
T, Arai H, Inui K, Kaneko T, Nozawa A, Adachi H, Rino Y, et al:
Clinical significance of immunohistochemical expression of
insulin-like growth factor-1 receptor and matrix
metalloproteinase-7 in resected non-small cell lung cancer. Exp
Ther Med. 3:797–802. 2012.PubMed/NCBI
|
|
12
|
Nakagawa M, Uramoto H, Oka S, Chikaishi Y,
Iwanami T, Shimokawa H, So T, Hanagiri T and Tanaka F: Clinical
significance of IGF1R expression in non-small-cell lung cancer.
Clin Lung Cancer. 13:136–142. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao S, Qiu Z, He J, Li L and Li W:
Insulin-like growth factor receptor 1 (IGF1R) expression and
survival in non-small cell lung cancer patients: A meta-analysis.
Int J Clin Exp Pathol. 7:6694–6704. 2014.PubMed/NCBI
|
|
14
|
Ludovini V, Bellezza G, Pistola L,
Bianconi F, Di Carlo L, Sidoni A, Semeraro A, Del Sordo R,
Tofanetti FR, Mameli MG, et al: High coexpression of both
insulin-like growth factor receptor-1 (IGFR-1) and epidermal growth
factor receptor (EGFR) is associated with shorter disease-free
survival in resected non-small-cell lung cancer patients. Ann
Oncol. 20:842–849. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gately K, Forde L, Cuffe S, Cummins R, Kay
EW, Feuerhake F and O'Byrne KJ: High coexpression of both EGFR and
IGF1R correlates with poor patient prognosis in resected
non-small-cell lung cancer. Clin Lung Cancer. 15:58–66. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Langer CJ, Novello S, Park K, Krzakowski
M, Karp DD, Mok T, Benner RJ, Scranton JR, Olszanski AJ and Jassem
J: Randomized, phase III trial of first-line figitumumab in
combination with paclitaxel and carboplatin versus paclitaxel and
carboplatin alone in patients with advanced non-small-cell lung
cancer. J Clin Oncol. 32:2059–2066. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Eberhard DA, Johnson BE, Amler LC, Goddard
AD, Heldens SL, Herbst RS, Ince WL, Jänne PA, Januario T, Johnson
DH, et al: Mutations in the epidermal growth factor receptor and in
KRAS are predictive and prognostic indicators in patients with
non-small-cell lung cancer treated with chemotherapy alone and in
combination with erlotinib. J Clin Oncol. 23:5900–5909. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gazdar AF, Shigematsu H, Herz J and Minna
JD: Mutations and addiction to EGFR: The Achilles ‘heal’ of lung
cancers? Trends Mol Med. 10:481–486. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weinstein IB, Joe A and Felsher D:
Oncogene addiction. Cancer Res. 68:3077–3080. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Al-Zahrani A, Sandhu MS, Luben RN,
Thompson D, Baynes C, Pooley KA, Luccarini C, Munday H, Perkins B,
Smith P, et al: IGF1 and IGFBP3 tagging polymorphisms are
associated with circulating levels of IGF1, IGFBP3 and risk of
breast cancer. Hum Mol Genet. 15:1–10. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feik E, Baierl A, Hieger B, Führlinger G,
Pentz A, Stättner S, Weiss W, Pulgram T, Leeb G, Mach K, et al:
Association of IGF1 and IGFBP3 polymorphisms with colorectal polyps
and colorectal cancer risk. Cancer Causes Control. 21:91–97. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Verheus M, McKay JD, Kaaks R, Canzian F,
Biessy C, Johansson M, Grobbee DE, Peeters PH and van Gils CH:
Common genetic variation in the IGF-1 gene, serum IGF-I levels and
breast density. Breast Cancer Res Treat. 112:109–122. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lönn S, Rothman N, Shapiro WR, Fine HA,
Selker RG, Black PM, Loeffler JS, Hutchinson AA and Inskip PD:
Genetic variation in insulin-like growth factors and brain tumor
risk. Neuro Oncol. 10:553–559. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Alencar SA and Lopes JCD: A
comprehensive in silico analysis of the functional and structural
impact of SNPs in the IGF1R gene. J Biomed Biotechnol.
2010:7151392010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
MacDonald K, Porter GA, Guernsey DL, Zhao
R and Casson AG: A polymorphic variant of the insulin-like growth
factor type I receptor gene modifies risk of obesity for esophageal
adenocarcinoma. Cancer Epidemiol. 33:37–40. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li HR, Chen YS, Shao H, Han LL and Zhang
XE: Association between polymorphisms of insulin-like growth factor
receptor gene and susceptibility to non-small-cell lung cancer in
Fujian Chinese. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 29:300–305.
2012.(In Chinese). PubMed/NCBI
|
|
29
|
Chen Y, Shao H, Li H, Han L and Zhang X:
Relationship of insulin-like growth factor receptor single
nucleotide polymorphism (SNP) with platinum-based chemotherapy
outcomes in advanced non-small cell lung cancer. Zhongguo Fei Ai Za
Zhi. 15:65–71. 2012.(In Chinese). PubMed/NCBI
|
|
30
|
Winder T, Zhang W, Yang D, Ning Y, Bohanes
P, Gerger A, Wilson PM, Pohl A, Mauro DJ, Langer C, et al: Germline
polymorphisms in genes involved in the IGF1 pathway predict
efficacy of cetuximab in wild-type KRAS mCRC patients. Clin Cancer
Res. 16:5591–5602. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Deming SL, Ren Z, Wen W, Shu XO, Cai Q,
Gao YT and Zheng W: Genetic variation in IGF1, IGF-1R, IGFALS, and
IGFBP3 in breast cancer survival among Chinese women: A report from
the Shanghai Breast Cancer Study. Breast Cancer Res Treat.
104:309–319. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Reinmuth N, Kloos S, Warth A, Risch A,
Muley T, Hoffmann H, Thomas M and Meister M: Insulin-like growth
factor 1 pathway mutations and protein expression in resected
non-small cell lung cancer. Hum Pathol. 45:1162–1168. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mountain CF: The international system for
staging lung cancer. Semin Surg Oncol. 18:106–115. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Purcell SM, Wray NR, Stone JL, Visscher
PM, O'Donovan MC, Sullivan PF and Sklar P: International
Schizophrenia Consortium: Common polygenic variation contributes to
risk of schizophrenia and bipolar disorder. Nature. 460:748–752.
2009.PubMed/NCBI
|
|
35
|
Kim JG, Kang MJ, Yoon YK, Kim HP, Park J,
Song SH, Han SW, Park JW, Kang GH, Kang KW, et al:
Heterodimerization of glycosylated insulin-like growth factor-1
receptors and insulin receptors in cancer cells sensitive to
anti-IGF1R antibody. PLoS One. 7:e333222012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim WY, Prudkin L, Feng L, Kim ES,
Hennessy B, Lee JS, Lee JJ, Glisson B, Lippman SM, Wistuba II, et
al: Epidermal growth factor receptor and K-Ras mutations and
resistance of lung cancer to insulin-like growth factor 1 receptor
tyrosine kinase inhibitors. Cancer. 118:3993–4003. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hunt R, Sauna ZE, Ambudkar SV, Gottesman
MM and Kimchi-Sarfaty C: Silent (synonymous) SNPs: Should we care
about them? Methods Mol Biol. 578:23–39. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheng J, Liu J, Li X, Peng J, Han S, Zhang
R, Xu Y and Nie S: Insulin-like growth factor-1 receptor
polymorphism and ischemic stroke: A case-control study in Chinese
population. Acta Neurol Scand. 118:333–338. 2008.PubMed/NCBI
|
|
39
|
Bonafè M, Barbieri M, Marchegiani F,
Olivieri F, Ragno E, Giampieri C, Mugianesi E, Centurelli M,
Franceschi C and Paolisso G: Polymorphic variants of insulin-like
growth factor I (IGF-I) receptor and phosphoinositide 3-kinase
genes affect IGF-I plasma levels and human longevity: Cues for an
evolutionarily conserved mechanism of life span control. J Clin
Endocrinol Metab. 88:3299–3304. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Diorio C, Brisson J, Bérubé S and Pollak
M: Genetic polymorphisms involved in insulin-like growth factor
(IGF) pathway in relation to mammographic breast density and IGF
levels. Cancer Epidemiol Biomarkers Prev. 17:880–888. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rasmussen SK, Lautier C, Hansen L, Echwald
SM, Hansen T, Ekstrøm CT, Urhammer SA, Borch-Johnsen K, Grigorescu
F, Smith RJ, et al: Studies of the variability of the genes
encoding the insulin-like growth factor I receptor and its ligand
in relation to type 2 diabetes mellitus. J Clin Endocrinol Metab.
85:1606–1610. 2000. View Article : Google Scholar : PubMed/NCBI
|