Introduction
Oral cancer is one of the most common cancers
worldwide, with an unclear pathogenesis; the oral squamous cell
carcinoma (OSCC) subtype accounts for >90% of all oral cancers
(1). While increasing evidence
suggests that environmental factors, as well as chemical
carcinogens (including tobacco and alcohol) are likely etiological
factors that contribute to the development of OSCC, only a small
minority of individuals exposed to these carcinogens will
subsequently develop head and neck SCC (2,3). Oral
carcinogenesis is widely recognized as a stepwise process, and the
involvement of genetic alterations and host polymorphisms may be
important to its development (4). The
identification of a predictive model of risk polymorphisms may
facilitate early diagnosis and the understanding of disease
progression in a subset of cancer patients (4).
The human mouse double minute 2 (MDM2) gene is an
important negative regulator of the p53 suppressor gene, promoting
the degradation of p53 through its E3 ubiquitin ligase activity
(5). A functional single-nucleotide
polymorphism (SNP), rs2279744, is located at nucleotide 309 in the
first intron of the MDM2 promoter region, consisting of a change
from T to G. This mutation has been named SNP309 (6). SNP 309T>G is known to enhance the
binding affinity of the transcriptional activator SP1, contributing
to increased expression of MDM2 and subsequent attenuation of the
p53 tumor suppressor pathway (6).
MDM2 SNP309 and its association with cancer susceptibility has been
identified and extensively investigated in a number of cancer types
(7,8).
To date, numerous previous studies have investigated
the association between MDM2 SNP309 and OSCC risk (9–15).
However, the results remain inconsistent and ambiguous, partly due
to the relatively small sample sizes of the independent studies and
sampling effects. Meta-analyses allow stronger conclusions for
identifying certain models of risk markers, which may help with
screening, early diagnosis and/or therapy in the clinical setting
(16–18). Therefore, a meta-analysis of all
eligible studies was performed, in order to achieve a more precise
estimation of this association, and to investigate the source of
heterogeneity and any potential bias in this method.
Materials and methods
Literature search strategy
The electronic databases PubMed, Web of Science,
Embase and China National Knowledge Infrastructure were searched
for relevant publications to be included in the present
meta-analysis, without restriction on language and publication year
(until January 10, 2015). The following search terms were used:
(‘murine double minute 2’ OR ‘MDM2’) AND (‘polymorphisms’ OR
‘variants’) AND (‘carcinoma’ OR ‘cancer’ OR ‘malignancy’ OR
‘neoplasm’ OR ‘tumour’ OR ‘tumor’) AND (‘oral’). Relevant articles
were reviewed to evaluate their appropriateness for inclusion in
the meta-analysis. Additional relevant publications were identified
through the references cited in the articles selected or review
articles on this topic.
Inclusion and exclusion criteria
The inclusion criteria for eligible articles were as
follows: i) Case-control studies on humans; ii) evaluation of MDM2
rs2279744 or 309T>G polymorphism and OSCC risk; iii) sufficient
genotype data to estimate an odds ratio (OR) and 95% confidence
interval (CI); and iv) histologically confirmed diagnosis of OSCC.
The exclusion criteria were as follows: i) Not a case-control
study; ii) no usable data reported; and iii) duplicate or
overlapping data.
Data extraction
According to the selection criteria, all relevant
crude data were extracted from each eligible article independently
by two researchers, and the inconsistencies were discussed until a
consensus was obtained. The following items were extracted from
each article: First author name, year of publication, country of
origin, ethnicity, genotyping method, source of control
(population- or hospital-based), number of cases and controls,
characteristics of cancer cases and controls, and genotype
frequencies for cases and controls.
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) of the control
group in each study was measured using either the χ2 or
Fisher's exact test. The degree of association between MDM2 SNP309
and OSCC risk was determined using the OR with 95% CI. In the
overall and subgroup meta-analysis, pooled ORs and 95% CIs for the
heterozygote (TG vs. TT), homozygote (GG vs. TT), dominant (TG+GG
vs. TT) and recessive models (GG vs. TG+TT) were calculated. The
allele comparison (G vs. T) was conducted as an additive mode. The
statistical significance of the pooled OR was evaluated using the
Z-test, and the heterogeneity of the ORs was tested by
χ2-based Q-test and I2 statistics. If the
result of heterogeneity test was P>0.1, ORs were pooled
according to the fixed-effects model (Mantel-Haenszel). Otherwise,
the random-effects model (DerSimonian and Laird) was used.
Furthermore, the Egger's test and Begg's funnel plot were used to
estimate the potential publication bias. All statistical analyses
were performed with the software Stata 10.0 (StataCorp LP, College
Station, TX, USA) and Review Manager 5.0 (Cochrane Informatics and
Knowledge Management, London, UK), using two-sided P-values.
Results
Study characteristics
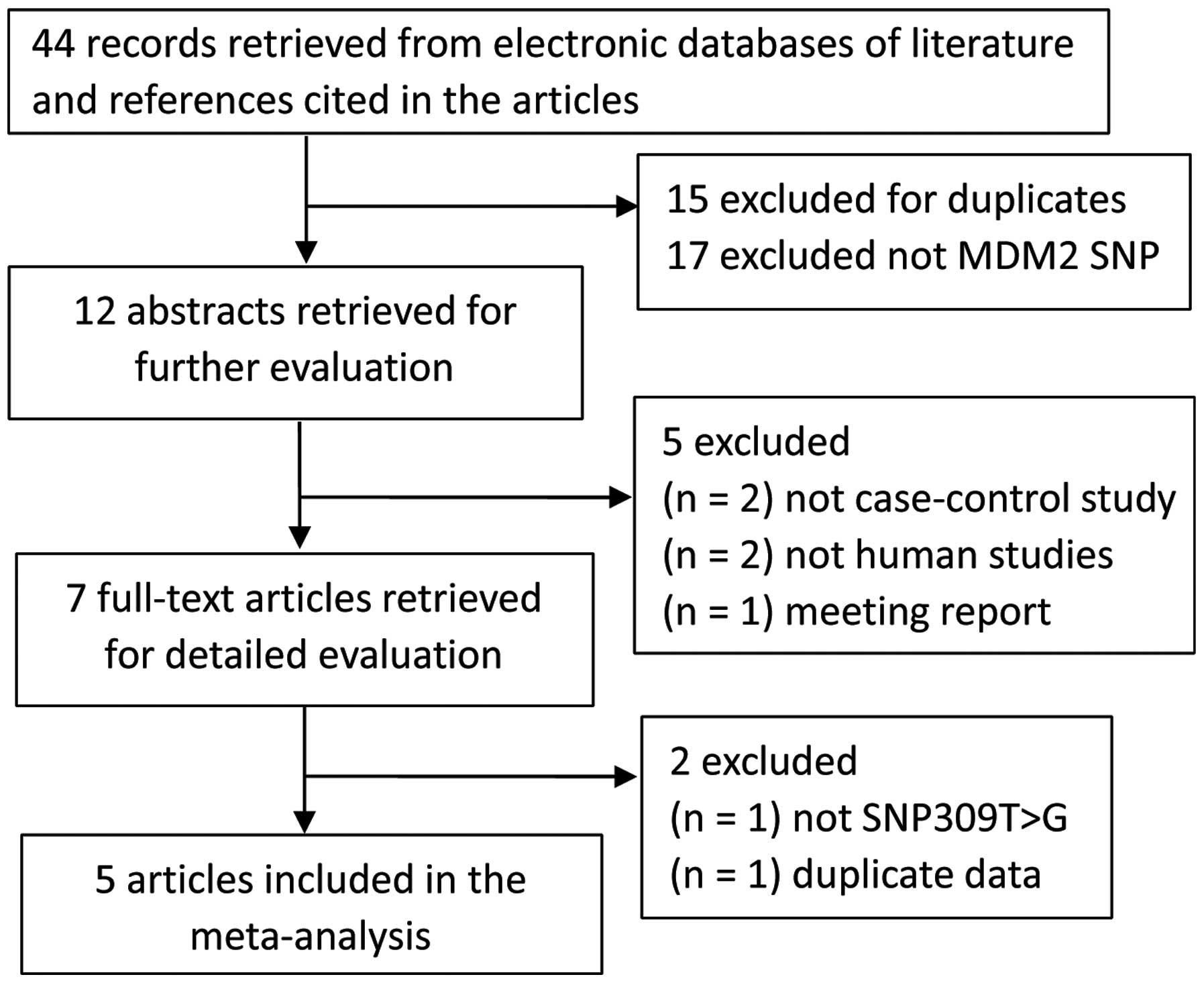
Of 44 potentially relevant articles, 37 were
gradually excluded (Fig. 1), and 7
full-text articles were selected for detailed evaluation from our
search of the published literature. During the extraction of data,
one study by Canova et al (14) that was not relevant to MDM2 309T>G
(rs2279744) polymorphism was excluded; in addition, one study by
Wang et al (15) that included
overlapping data from the authors' colleague was excluded according
to the inclusion and exclusion criteria. Therefore, 5 eligible
articles, including a total of 1,369 OSCC cases and 2,167 controls,
were ultimately included in the final meta-analysis (Fig. 1). The characteristics of the eligible
studies are summarized in Table I.
All were case-control studies, with Asian and Caucasian subjects.
There were 2 studies with hospital-based and 3 studies with
population-based controls. A classic polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP) assay
was performed in 4 of the 5 studies. The detailed variant genotype
distributions of MDM2 SNP309 for OSCC cases and controls in the
individual studies are listed in Table
I.
 | Table I.Study characteristics of the
meta-analysis. |
Table I.
Study characteristics of the
meta-analysis.
|
| Sample (n) | Cases | Controls |
|---|
|
|
|
|
|
|---|
| Author, year
(Refs.) | Country | Ethnicity | Source of
controls | Genotyping | HWE of controls | Cases | Controls | TT | TG | GG | TT | TG | GG |
|---|
| Tu et al, 2008
(9) | Taiwan | Asian | Hospital | PCR-RFLP | 0.582 | 189 | 116 | 44 | 93 | 52 | 29 | 55 | 32 |
| Huang et al,
2009 (10) | Taiwan | Asian | Population | MALDI-TOF | 0.286 | 351 | 1272 | 80 | 176 | 95 | 274 | 653 | 345 |
| Misra et al,
2009 (11) | India | Asian | Population | PCR-RFLP | 0.042 | 297 | 328 | 70 | 147 | 80 | 59 | 181 | 88 |
| Hamid et al,
2009 (12) | Malaysia | Asian | Hospital | PCR-RFLP | 0.997 | 207 | 116 | 48 | 104 | 55 | 30 | 58 | 28 |
| Chen et al,
2009 (13) | USA | Caucasian | Population | PCR-RFLP | 0.835 | 325 | 335 | 146 | 132 | 47 | 112 | 165 | 58 |
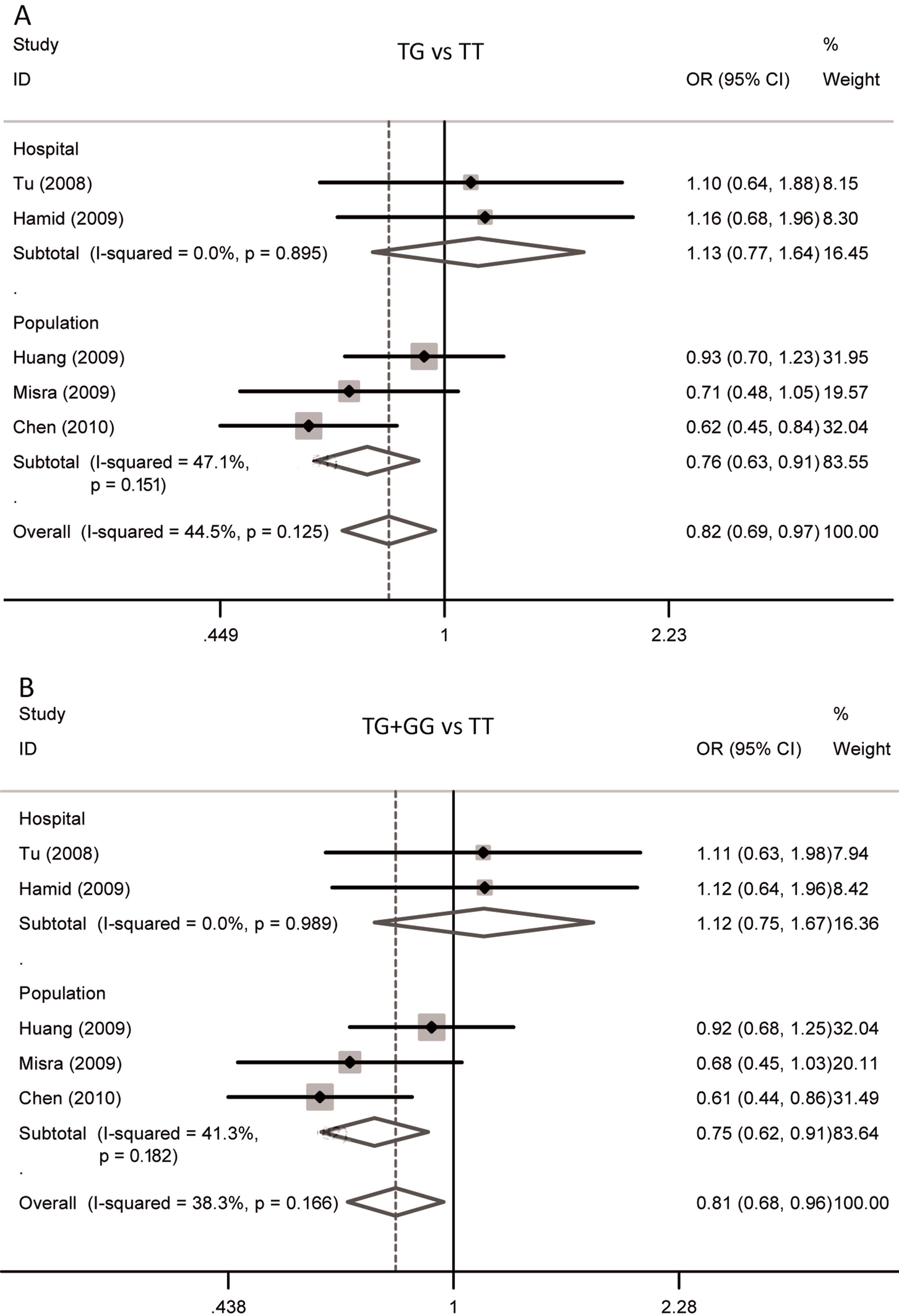
Meta-analysis results
The associations between MDM2 SNP309 variants and
OSCC risk are summarized in Table
II. In the overall analysis, the heterozygote (TG vs. TT:
OR=0.81; 95% CI: 0.68–0.96; P=0.02) and dominant genetic models
(TG+GG vs. TT: OR=0.82; 95% CI: 0.69–0.97; P=0.02) of MDM2 SNP309
were significantly associated with OSCC risk, while the results of
homozygote and recessive models and allele comparison were not
significant. Forest plots for the meta-analysis of the two
significant genetic models are shown in Fig. 2. In the subgroup analysis based on the
source of the controls, significant associations were observed in
the population-based controls among the heterozygote (TG vs. TT:
OR=0.75; 95% CI: 0.62–0.91; P=0.004) and dominant models (TG+GG vs.
TT: OR=0.76; 95% CI: 0.63–0.91; P=0.003), and allele comparison (G
vs. T: OR=0.89; 95% CI: 0.79–0.99; P=0.04). In the ethnicity
subgroup analysis, no significant association in Asians was found
among any genetic model. As there was only one Caucasian study, the
data of the Caucasian stratified analysis are not shown (Table II).
 | Table II.Summary of pooled odds ratios with 95%
confidence intervals in the meta-analysis. |
Table II.
Summary of pooled odds ratios with 95%
confidence intervals in the meta-analysis.
|
| Test of
association | Test of
heterogeneity |
|---|
|
|
|
|
|---|
| Group (no. of
cases/controls) | Genotype | OR (95% CI) | P-value | I2
(%) | P-value |
|---|
| Overall | TG vs. TT | 0.81
(0.68–0.96) | 0.02 | 38 | 0.17 |
| (1,369/2,167) | GG vs. TT | 0.86
(0.70–1.06) | 0.17 | 2 | 0.39 |
|
| TG+GG vs. TT | 0.82
(0.69–0.97) | 0.02 | 45 | 0.13 |
|
| GG vs. TG+TT | 0.98
(0.83–1.16) | 0.80 | 0 | 0.88 |
|
| G vs. T | 0.92
(0.83–1.02) | 0.11 | 34 | 0.20 |
| Subgroup |
|
|
Population-based controls | TG vs. TT | 0.75
(0.62–0.91) | 0.004 | 41 | 0.18 |
|
(973/1,935) | GG vs. TT | 0.80
(0.63–1.01) | 0.06 | 6 | 0.35 |
|
| TG+GG vs. TT | 0.76
(0.63–0.91) | 0.003 | 47 | 0.15 |
|
| GG vs. TG+TT | 0.96
(0.79–1.16) | 0.65 | 0 | 0.67 |
|
| G vs. T | 0.89
(0.79–0.99) | 0.04 | 48 | 0.15 |
|
Asian | TG vs. TT | 0.90
(0.73–1.11) | 0.32 | 8 | 0.41 |
|
(1,044/1,832) | GG vs. TT | 0.94
(0.75–1.19) | 0.61 | 2 | 0.66 |
|
| TG+GG vs. TT | 0.94
(0.75–1.19) | 0.37 | 0 | 0.42 |
|
| GG vs. TG+TT | 1.02
(0.85–1.22) | 0.87 | 0 | 0.98 |
|
| G vs. T | 0.98
(0.87–1.09) | 0.67 | 0 | 0.73 |
Test of heterogeneity
As seen in Table II,
there was no evidence of heterogeneity in any of the genetic models
(all P>0.1 for heterogeneity, Q-test). Therefore, a summary of
ORs was analyzed with the fixed-effects model
(Mantel-Haenszel).
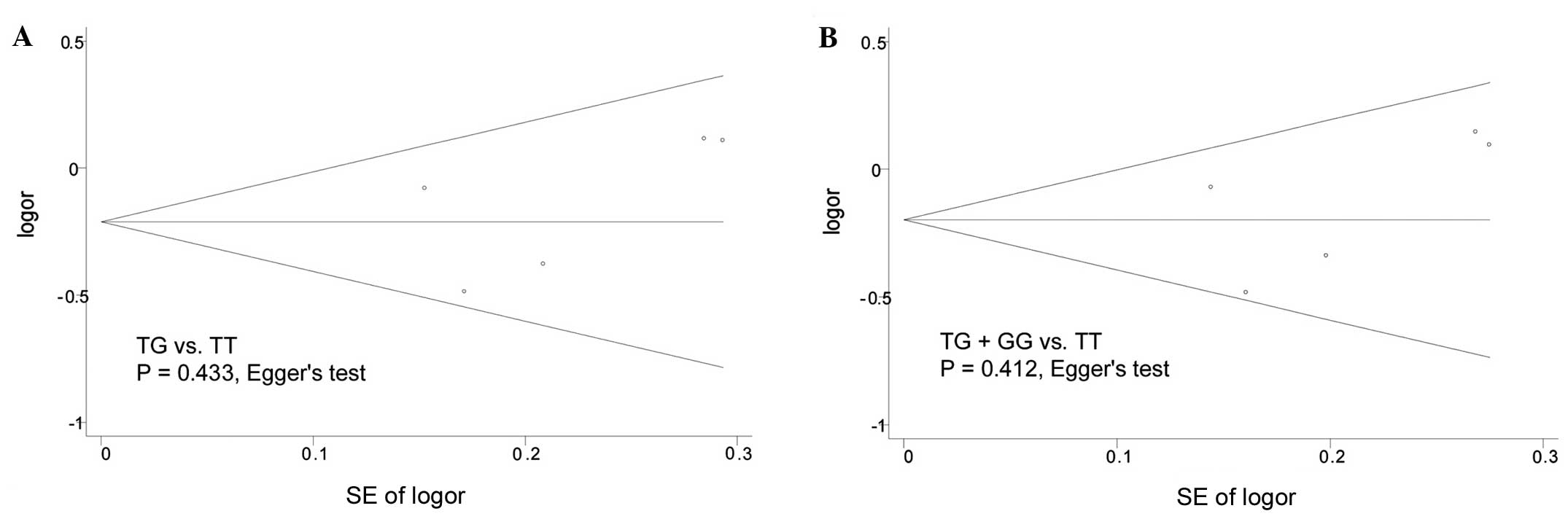
Publication bias
Publication bias was evaluated using the Begg's
funnel plot and Egger's test. As shown in Fig. 3, the shape of Begg's funnel plots
appeared to be largely symmetrical in both the heterozygote and
dominant models; in addition, the Egger's test revealed no
significant publication bias (P=0.433 and P=0.412,
respectively).
Discussion
MDM2 is an important regulator of the p53 suppressor
gene, and the associations between the MDM2 SNP309 mutation and
cancer susceptibility have been evaluated in a number of cancer
types (7,8). Previously published studies on the
association of this SNP with OSCC risk have generated variable
results (9–13). The aim of the present study was to
obtain a more precise estimation of the association between MDM2
SNP309 and OSCC susceptibility by performing a meta-analysis that
included all eligible case-control studies published to date.
Five independent studies investigating the genetic
effects of MDM2 SNP309T>G on OSCC risk (9–13) were
included in this meta-analysis. Among these, 1 study demonstrated
that MDM2 SNP309T>G was associated with a reduced risk of OSCC
(13), while the remaining 4 studies
provided no evidence of an association between this SNP and OSCC
risk (9–12). A meta-analysis of the 5 studies,
including 1,369 cases and 2,167 controls, was performed to
investigate the overall effects of MDM2 SNP309 on OSCC risk. The
overall analysis revealed that the MDM2 SNP309 heterozygote and
dominant models were inversely associated with OSCC risk, with no
significant heterogeneity. These results suggest that the MDM2
SNP309T>G may be a protective factor for OSCC, which is
consistent with previous analyses performed on prostate and ovarian
cancers (19–21).
Although a number of previous meta-analyses have
demonstrated that MDM2 SNP309T>G may be a risk factor for
several cancer types, including cervical, gastric and
hepatocellular cancers (7,8), there are also analyses that provide
evidence regarding its protective role in prostate and ovarian
cancers (19–21), in addition to the present analysis for
oral cancer. Cancer development is an extremely complex process,
and different cancer types exhibit genetic heterogeneity. Although
MDM2 SNP309 is a plausible carcinogenic hereditary factor in the
MDM2-p53 pathway, MDM2 is activated in response to a variety of
oncogenic pathways that are independent of p53 (22). Furthermore, Francis et al
(23) reported that MDM2 has no
significant role in stabilizing the expression of p53, and remarked
on the presence of unknown interactors that sequester p53 and lead
to its aberrant accumulation. Additionally, there is evidence that
decreased MDM2 expression is associated with a worse prognosis of
head and neck carcinomas (24). Taken
together, these findings suggest that MDM2 may have tumor
suppressor functions under certain conditions. Therefore, gene-gene
and gene-environment interactions regulate carcinogenesis, and the
presence of other causal factors as yet unidentified demonstrate an
involvement of MDM2 SNP309 in OSCC development.
Consistent with the results of the overall analysis,
the subgroup analysis revealed that MDM2 SNP309 significantly
reduced OSCC risk in the population-based controls, while no
significant association was found in hospital-based controls. A
possible explanation is that the hospital-based controls may
represent a sample of an ill-defined reference population rather
than the general population, and inherent selection bias may
therefore not be completely excluded. Thus, use of more appropriate
population-recruited control subjects may be crucial for reducing
study bias. Additionally, the ethnicity subgroup analysis
demonstrated a significant association between MDM2 SNP309 and OSCC
risk among Caucasians, but not Asians, suggesting that SNP309 may
be an ethnicity-dependent factor associated with OSCC risk.
Due to several limitations of the present
meta-analysis, the results should be interpreted with caution.
First, the number of the published studies eligible for inclusion
and the pooled sample size of the independent studies were
relatively small in the overall and subgroup analyses. In
particular, only one relevant study with Caucasian patients was
identified, and it is possible that some relevant unpublished
studies were overlooked. Second, the effect of the confounding
factors, resulting from gene-gene (such as the p53 pathway) and
gene-environment (such as tobacco and alcohol consumption)
interactions were not evaluated in the present study due to data
limitations. Thus, in order to obtain a more precise analysis of
the effect of MDM2 SNP309 mutation on OSCC risk, additional,
improved studies with larger sample sizes and diverse ethnicities,
particularly Caucasians and Africans, are required. However,
despite these limitations, the present meta-analysis has some
strengths. First, a systematic review of the association between
MDM2 SNP309 and OSCC risk is statistically more powerful compared
with any independent study. Second, the quality of the eligible
studies included in the present meta-analysis was acceptable, and
there was no evidence of publication bias or obvious outcome
heterogeneity.
In conclusion, the present meta-analysis achieved a
more precise evaluation of the association between MDM2 SNP309 and
OSCC risk compared with independent studies. The results of the
meta-analysis indicate that the MDM2 SNP309 mutation may serve as a
protective factor in OSCC; however, this is only a preliminary
analysis and the results presented herein should be interpreted
with caution. Further well-designed and larger studies are required
to elucidate the association between MDM2 SNP309 and OSCC risk.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (81472517, 81102049,
81302358 and 81202132), the Shanghai Pujiang Talent Program
(15PJD024), the Shanghai Education Commission Project of Young
Teachers (ZZjdyx13104) and the Shanghai Natural Science Foundation
(13ZR1457100).
References
|
1
|
Rao Krishna SV, Mejia G, Roberts-Thomson K
and Logan R: Epidemiology of oral cancer in Asia in the past
decade-an update (2000–2012). Asian Pac J Cancer Prev.
14:5567–5577. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Petersen PE: Oral cancer prevention and
control-the approach of the World Health Organization. Oral Oncol.
45:454–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zini A, Czerninski R and Sgan-Cohen HD:
Oral cancer over four decades: Epidemiology, trends, histology, and
survival by anatomical sites. J Oral Pathol Med. 39:299–305. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liao G, Wang Y, Zhou YQ, Li TW, Zeng DQ,
Zeng X, Li J, Dan HX and Chen QM: Host genetic susceptibility to
oral cancer: Evidence from meta-analyses and pooled analyses. Oral
Dis. 20:644–649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bond GL, Hu W, Bond EE, Robins H, Lutzker
SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al: A
single nucleotide polymorphism in the MDM2 promoter attenuates the
p53 tumor suppressor pathway and accelerates tumor formation in
humans. Cell. 119:591–602. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bond GL, Hu W and Levine A: A single
nucleotide polymorphism in the MDM2 gene: From a molecular and
cellular explanation to clinical effect. Cancer Res. 65:5481–5484.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wo X, Han D, Sun H, Liu Y, Meng X, Bai J,
Chen F, Yu Y, Jin Y and Fu S: MDM2 SNP309 contributes to tumor
susceptibility: A meta-analysis. J Genet Genomics. 38:341–350.
2011.PubMed/NCBI
|
|
8
|
Chen B, Cao L, Hu KW, Zhang JW, Meng XL
and Xiong MM: MDM2 SNP309 is an ethnicity-dependent risk factor for
digestive tract cancers. Tumour Biol. 35:3431–3438. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tu HF, Chen HW, Kao SY, Lin SC, Liu CJ and
Chang KW: MDM2 SNP 309 and p53 codon 72 polymorphisms are
associated with the outcome of oral carcinoma patients receiving
postoperative irradiation. Radiother Oncol. 87:243–252. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang SF, Chen IH, Liao CT, Wang HM, Liou
SH and Hsieh LL: Combined effects of MDM2 SNP 309 and p53 mutation
on oral squamous cell carcinomas associated with areca quid
chewing. Oral Oncol. 45:16–22. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Misra C, Majumder M, Bajaj S, Ghosh S, Roy
B and Roychoudhury S: Polymorphisms at p53, p73, and MDM2 loci
modulate the risk of tobacco associated leukoplakia and oral
cancer. Mol Carcinog. 48:790–800. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hamid S, Yang YH, Peng KN, Ismail SM, Zain
RB, Lim KP, Wan Mustafa WM, Abraham MT, Teo SH and Cheong SC: MDM2
SNP309 does not confer an increased risk to oral squamous cell
carcinoma but may modulate the age of disease onset. Oral Oncol.
45:496–500. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen X, Sturgis EM, Lei D, Dahlstrom K,
Wei Q and Li G: Human papillomavirus seropositivity synergizes with
MDM2 variants to increase the risk of oral squamous cell carcinoma.
Cancer Res. 70:7199–7208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Canova C, Hashibe M, Simonato L, Nelis M,
Metspalu A, Lagiou P, Trichopoulos D, Ahrens W, Pigeot I, Merletti
F, et al: Genetic associations of 115 polymorphisms with cancers of
the upper aerodigestive tract across 10 European countries: The
ARCAGE project. Cancer Res. 69:2956–2965. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Sturgis EM, Zhang Y, Huang Z, Zhou
Q, Wei Q and Li G: Combined p53-related genetic variants together
with HPV infection increase oral cancer risk. Int J Cancer.
131:E251–E258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Li S, Xiao HQ, Hu ZX, Xu YC and
Huang Q: Vascular endothelial growth factor gene polymorphisms and
renal cell carcinoma: A systematic review and meta-analysis. Oncol
Lett. 6:1068–1078. 2013.PubMed/NCBI
|
|
17
|
Wang JG, Zhang Y and Xiao TL: Quantitative
analysis of the association between CRP rs2808630 and rs1417938
polymorphisms and cancer risk. Oncol Lett. 9:994–998.
2015.PubMed/NCBI
|
|
18
|
Jiang LL and Ruan LW: Association between
FOXP3 promoter polymorphisms and cancer risk: A meta-analysis.
Oncol Lett. 8:2795–2799. 2014.PubMed/NCBI
|
|
19
|
Chen T, Yi SH, Liu XY and Liu ZG:
Meta-analysis of associations between the MDM2-T309G polymorphism
and prostate cancer risk. Asian Pac J Cancer Prev. 13:4327–4330.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu G, Jiang D, Shen S and Yu L: Murine
double minute 2 promoter SNP309 polymorphism and prostate cancer
risk: A meta-analysis. Int J Urol. 19:914–920. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma YY, Guan TP, Yao HB, Yu S, Chen LG, Xia
YJ, He XJ, Wang HJ, Jiang XT and Tao HQ: The MDM2 309T>G
polymorphism and ovarian cancer risk: A meta-analysis of 1534 cases
and 2211 controls. PLoS One. 8:e550192013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Manfredi JJ: The Mdm2-p53 relationship
evolves: Mdm2 swings both ways as an oncogene and a tumor
suppressor. Genes Dev. 24:1580–1589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Francis G, Dileep Kumar U, Nalinakumari
KR, Jayasree K and Kannan S: Accumulation of inactive p53 protein
in oral squamous cell carcinoma: Stabilization by protein
interaction. Eur J Oral Sci. 121:21–28. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Millon R, Muller D, Schultz I, Salvi R,
Ghnassia JP, Frebourg T, Wasylyk B and Abecassis J: Loss of MDM2
expression in human head and neck squamous cell carcinomas and
clinical significance. Oral Oncol. 37:620–631. 2001. View Article : Google Scholar : PubMed/NCBI
|