Introduction
Lung cancer is one of the most common malignant
tumors worldwide. Non-small-cell lung cancer (NSCLC) constitutes
~80% of lung cancers, with a 5-year survival of only 15%. Further
study on the pathogenic mechanism of lung cancer is required to
establish novel diagnostic or treatment strategies for this lethal
disease.
Bone morphogenetic proteins (BMPs) belong to the
transforming growth factor-β (TGF-β) superfamily. Thus far, >20
BMPs have been identified in humans. Six of the type I and three of
the type II serine/threonine kinase receptors have been shown to
mediate BMP signaling (1).
Both type I and type II receptors consist of a N-terminal
extracellular ligand-binding domain and a C-terminal
serine/threonine kinase domain (2).
When the BMP ligand binds to preformed hetero-oligomeric complexes,
the SMAD-dependent pathway may be activated (3). The pathway-restricted SMADs (such as
SMAD1, 2, 3, 5 or 8) are recruited and translocated into the
nucleus with the assistance of SMAD4 and regulate the transcription
of target genes, referred to as the SMAD-dependent pathway. The
activated BMP signaling may then enhance invasion and bone
metastasis of cancer cells via the SMAD pathway (4). BMPs significantly affect embryonic and
postnatal development and maintenance of homeostasis in organs and
tissues; they are also associated with cell proliferation,
differentiation, motility and survival (5) and they are involved in the development
and progression of certain malignant tumors, such as lung, prostate
and breast cancer (6). Recently,
accumulating evidence demonstrated that BMPs also participate in
tumor-related angiogenesis (7,8).
It was hypothesized that the polymorphisms in the
BMP component and regulatory genes may contribute to the
genetic susceptibility to lung cancer. To test this hypothesis, a
two-stage case-control study was performed, including a total of
2,340 patients and 2,340 cancer-free controls, with the specific
aim of evaluating the effects of gene polymorphisms in the 18
selected genes related to the BMP pathway [BMP2,
4, 6, 7 and 9, SMAD1, 4,
5, 6, 7 and 8, SMURF1 and
2, ACTR2, ALK2 (ACVR1), ALK3
(BMPR-1A), ALK6 (BMPR-1B) and BMPR2] on
lung cancer. To the best of our knowledge, this is the first study
to evaluate the associations between a comprehensive panel of
genetic variants related to the BMP pathway and lung cancer,
and to identify certain patient subgroups that may be at high risk
of developing lung cancer.
Patients and methods
Study population and design
The study design and subject recruitment have been
previously described (9). Briefly,
two independent case-control projects were conducted. The ‘stage I’
setting (discovery) included 1,422 patients and 1,422 controls
mainly from Guangzhou, Guangdong, China (9). The ‘stage II’ setting (replication)
included 918 cases and 918 controls from Xianyang, Hubei, China, to
test the results of stage I. A total of 2,340 cases with
histopathologically diagnosed lung cancer were enrolled from
September, 2009 onwards. The 2,340 cancer-free controls were
recruited during the same time period. The characteristics of the
cases and controls are summarized in Table I. Structured questionnaires were
conducted by experienced interviewers using a standardized
protocol. The inclusive criteria and definition of cigarette
smoking were previously detailed (9). This project was approved by the
Institutional Ethics Committee of each participating center. All
the participants provided written informed consent prior to
enrolment.
 | Table I.Frequency distributions of selected
variables in lung cancer patients and cancer-free controls. |
Table I.
Frequency distributions of selected
variables in lung cancer patients and cancer-free controls.
|
| Discovery | Replication | Combined |
|---|
|
|
|
|
|
|---|
| Variables | Cases, n (%)
(n=1,422) | Controls, n (%)
(n=1,422) | P-valuea | Cases, n (%)
(n=918) | Controls, n (%)
(n=918) | P-valuea | P-valueb | Cases, n (%)
(n=2,340) | Controls, n (%)
(n=2,340) | P-valuea |
|---|
| Age, years |
|
| 0.9855 |
|
| 0.0108 | 0.0448 |
|
| 0.3494 |
| ≤60 | 688 (50.00) | 688 (50.00) |
| 459 (53.19) | 404 (46.81) |
|
| 1,146 (49.31) | 1,178 (50.69) |
|
|
>60 | 734 (50.00) | 734 (50.00) |
| 459 (47.22) | 514 (52.78) |
|
| 1,194 (50.68) | 1,162 (49.32) |
|
| Gender |
|
| 0.9904 |
|
| 0.8757 | 0.9277 |
|
| 0.9471 |
|
Male | 1,007 (50.00) | 1,007 (50.00) |
| 714 (49.93) | 716 (50.07) |
|
| 1,721 (49.97) | 1,723 (50.03) |
|
|
Female | 415 (50.00) | 415 (50.00) |
| 204 (50.37) | 202 (49.63) |
|
| 619 (50.08) | 617 (49.92) |
|
| Smoking status |
|
|
<0.0001 |
|
|
<0.0001 |
<0.0001 |
|
|
<0.0001 |
|
Smoker | 702 (54.49) | 519 (42.51) |
| 555 (59.36) | 380 (40.64) |
|
| 1,258 (57.63) | 925 (42.37) |
|
| Former
smoker | 168 (68.85) | 76 (31.15) |
| 86 (44.79) | 106 (55.21) |
|
| 254 (57.34) | 189 (42.66) |
|
|
Non-smoker | 551 (39.99) | 827 (60.01) |
| 277 (39.12) | 432 (60.88) |
|
| 828 (40.31) | 1,226 (59.69) |
|
| Pack years |
|
|
<0.0001 |
|
|
<0.0001 | 0.7312 |
|
|
<0.0001 |
|
≥25 | 650 (65.46) | 343 (34.54) |
| 483 (61.12) | 306 (38.88) |
|
| 1,274 (61.49) | 798 (38.51) |
|
|
<25 | 220 (46.61) | 252 (53.39) |
| 94 (40.52) | 138 (59.48) |
|
| 238 (42.96) | 316 (57.04) |
|
| 0 | 551 (39.99) | 827 (60.01) |
| 277 (39.12) | 432 (60.88) |
|
| 828 (40.31) | 1,226 (59.69) |
|
| Gender and
smoking |
|
|
<0.0001 |
|
|
<0.0001 | 0.6112 |
|
|
<0.0001 |
| Male
smokers | 832 (58.47) | 591 (41.53) |
| 610 (56.53) | 469 (43.47) |
|
| 1,443 (56.90) | 1,093 (43.10) |
|
| Male
non-smokers | 174 (29.49) | 416 (70.51) |
| 104 (29.63) | 247 (70.37) |
|
| 278 (30.62) | 630 (69.38) |
|
| Female
smokers | 38 (90.48) | 4 (9.52) |
| 31 (64.58) | 17 (35.42) |
|
| 69 (76.67) | 21 (23.33) |
|
| Female
non-smokers | 377 (47.84) | 411 (52.16) |
| 173 (48.46) | 185 (51.54) |
|
| 549 (47.99) | 595 (52.01) |
|
Blood sampling, SNP selection and
genotyping
Blood samples were provided by all the participants.
The genomic DNA was extracted with the Qiagen Blood DNA kit
(Qiagen, Valencia, CA, USA). A total of 18 genes associated with
the BMP pathway were selected. For each of those genes, the
tagSNPs were selected using Haploview 4.2 software (http://haploview.software.informer.com/4.2/). The
genotypes of 314 selected tagSNPs and their associations with lung
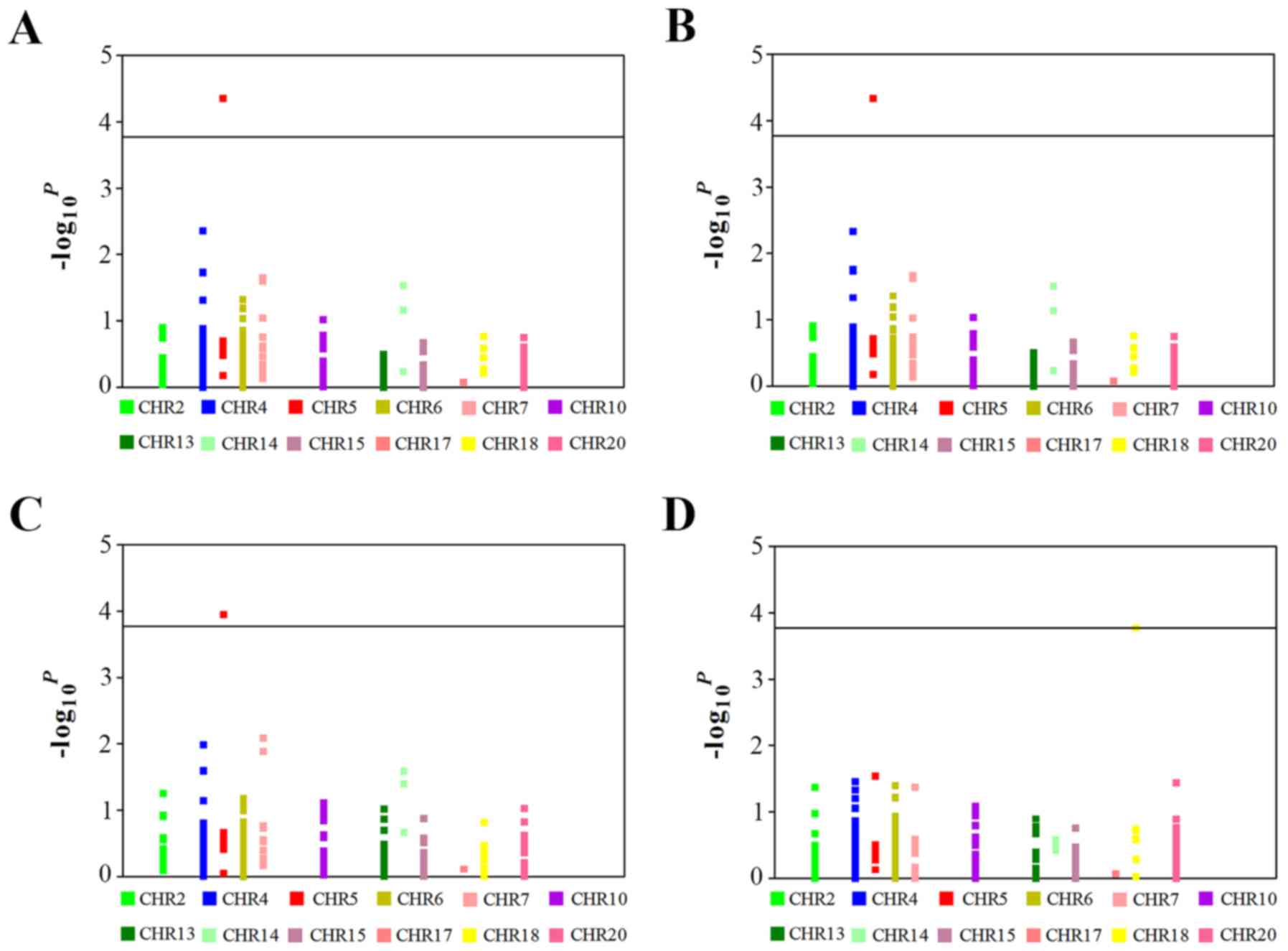
cancer are detailed in Fig. 1
(P<1.6×10−4) and Table
II (P<0.05). The Illumina high-throughput genotyping
platform and the Illumina Beadstudio software were used (Illumina,
San Diego, CA, USA). To ensure quality control, genotyping was
performed using blinded methods and the analysis was conducted
separately by two researchers. Over 15% of the samples were used
for confirmation, and the outcomes were 100% concordant. The
genotyping call rates were all >95%.
 | Table II.Genotyping of the 314 tagSNPs in the
BMP pathway and their associations with lung cancer
(P<0.05). |
Table II.
Genotyping of the 314 tagSNPs in the
BMP pathway and their associations with lung cancer
(P<0.05).
|
|
|
|
| Discovery | Replication | Combined |
|---|
|
|
|
|
|
|
|
|
|---|
|
|
|
|
| MAF, |
|
| MAF, |
|
| MAF, |
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| CHR | SNP | Gene | Allelea | Cases/controls
(n=1,422/1,422) |
Padd |
PHWE | Cases/controls
(n=918/918) |
Padd |
PHWE | Cases/controls
(n=2,340/2,340) |
Padd |
PHWE |
|---|
| 2 | rs7589059 | BMPR2 | A>G | 0.12/0.10 | 0.0216 | 0.6569 | 0.11/0.11 | 0.7100 | 0.1501 | 0.11/0.10 | 0.1237 | 0.5779 |
| 2 | rs6747756 | BMPR2 | A>G | 0.14/0.11 | 0.0101 | 0.8424 | 0.12/0.14 | 0.3826 | 0.081 | 0.13/0.12 | 0.1589 | 0.3288 |
| 4 | rs10516957 | ALK6
(BMPR-1B) | T>C | 0.84/0.94 | 0.0411 | 0.1689 | 0.87/0.82 | 0.3901 | 0.7904 | 0.85/0.89 | 0.2955 | 0.3682 |
| 5 | rs12719482 | SMAD5 | T>C | 0.14/0.11 | 0.0060 | 0.7201 | 0.14/0.10 | 0.0018 | 0.9762 | 0.14/0.11 | 4.39E-5 | 0.7971 |
| 10 | rs7095804 | ALK3
(BMPR-1A) | T>A | 0.46/0.41 | 0.0360 | 0.4681 | 0.48/0.47 | 0.9265 | 0.2704 | 0.47/0.43 | 0.0936 | 0.1967 |
| 20 | rs230194 | BMP7 | A>G | 0.24/0.27 | 0.0454 | 0.6498 | 0.28/0.26 | 0.4705 | 0.4835 | 0.25/0.27 | 0.2778 | 0.9208 |
| 20 | rs230198 | BMP7 | G>A | 0.23/0.27 | 0.0423 | 0.6011 | 0.28/0.26 | 0.4954 | 0.5107 | 0.25/0.27 | 0.2573 | 0.9831 |
| 20 | rs4811823 | BMP7 | G>A | 0.24/0.27 | 0.0492 | 0.6004 | 0.28/0.26 | 0.5102 | 0.5261 | 0.25/0.27 | 0.2713 | 0.9974 |
| 20 | rs6123677 | BMP7 | G>A | 0.24/0.28 | 0.0324 | 0.5969 | 0.27/0.26 | 0.6336 | 0.6184 | 0.25/0.27 | 0.1768 | 0.9296 |
Statistical analysis
The χ2 test was used to evaluate the
qualitative data and the Hardy-Weinberg equilibrium (HWE) applied.
The Akaike Information Criteria (AIC) (10) were employed to select a genetic model
for each SNP. Odds ratios (ORs) and corresponding 95% confidence
intervals (CIs) were calculated by an unconditional logistic
regression model adjusted for confounding factors. Stratification
analyses were performed with the variables of interest. The
pairwise linkage disequilibrium among the SNPs and haplotype
blocks/frequencies were measured accordingly (11–13).
Homogeneity between stage I and II populations was assessed by the
Breslow-Day test. Statistical power was assessed by Quanto 1.2.
software (http://biostats.usc.edu/Quanto.html) and corrected by
the Bonferroni test for each of the 314 SNPs at a significance
level of 0.05/314=1.6×10−4 (Fig. 1). All the analyses were performed
using SAS 9.2 software (SAS Institute Inc., NC, USA).
Results
Characteristics of the
populations
As summarized in Table
I, consistent results were found in stages I and II, with
significant differences in smoking status, pack-years and
gender-smoking (P<0.001 for all); there was no statistically
significant deviation in the distributions of gender and age
between the case and control groups (P>0.05 for all). The
frequency distribution of smoking status was heterogeneous
(Breslow-Day test P<0.0001). The two groups were then combined
to increase the study power, and almost identical change tendencies
were identified separately for stages I and II.
Genetic variants in SMAD5 and lung
cancer risk
As mentioned above, 314 tagSNPs were selected from
18 genes associated with the BMP pathway; those with
P<0.05 in stage I, stage II, or the combined populations, are
shown in Table II. Among these
tagSNPs, consistently significant associations were found between
SMAD5 rs12719482 and lung cancer risk in the three types of
sources of populations (P<0.05 for all). In particular, when
combining stages I and II, the differences were more significant
compared with any individual stage in the additive genetic model
(P=4.39×10−5). All observed genotype distributions among
these groups were in HWE (P≥0.05 for all). The frequency
distributions of the genotypes of rs12719482 are also summarized in
Table III. In the stage I set,
significant associations were found between rs12719482 T>C
genotypes, as well as alleles, and lung cancer risk. Compared with
zero-risk genotype carriers, the one- or two-risk genotype was
associated with increased lung cancer risk in a dose-dependent
manner (adjusted OR=1.27, 95% CI: 1.07–1.50, P=0.0060), while the
rs12719482 CT/CC genotype was associated with a significantly
increased risk in the dominant genetic model (OR=1.26; 95% CI:
1.05–1.52; P=0.0133). Allele C increased the risk of lung cancer by
23% compared with the wild-type T. Concordant with the results of
rs12719482 T>C and lung cancer risk analysis in stage I, the
variant genotype analyses revealed almost similar change tendencies
in the stage II setting. Importantly, the association remained
statistically significant when all the participants were
combined.
 | Table III.Distribution of genotypes in
SMAD5 and associations with risk of lung cancer. |
Table III.
Distribution of genotypes in
SMAD5 and associations with risk of lung cancer.
|
| Discovery | Replication | Combined |
|---|
|
|
|
|
|
|---|
|
Genotypes/alleles | Cases/controls | OR (95%
CI)a |
P-valuea | Case/control | OR (95%
CI)a |
P-valuea | Cases/controls | OR (95%
CI)a |
P-valuea |
|---|
| Total no. of
subjects | 1,422/1,422 |
| 0.0921 | 918/918 |
|
| 2,340/2,340 |
|
|
| Total no. of
alleles | 2,844/2,844 |
|
| 1,836/1,836 |
|
| 4,680/4,680 |
|
|
| rs12719482
T>C |
|
|
|
|
|
|
|
|
|
| Co-dominant
model |
|
|
|
|
|
|
|
|
|
| TT | 1,091/1,147 | 1.00 (ref.) |
| 711/762 | 1.00 (ref.) |
| 1,802/1,909 | 1.00 (ref.) |
|
| CT | 308/263 | 1.23
(1.02–1.48) | 0.0344 | 194/148 | 1.38
(1.08–1.76) | 0.0097 | 502/411 | 1.29
(1.11–1.50) | 0.0007 |
| CC | 22/12 | 2.10
(1.02–4.32) | 0.0437 | 13/7 | 1.88
(0.72–4.93) | 0.1989 | 35/19 | 2.08
(1.17–3.70) | 0.0128 |
| Trend test
P-value |
|
| 0.0060 |
|
| 0.0046 |
|
| 4.9e-5 |
| Additive model |
| 1.27
(1.07–1.50) | 0.0060 |
| 1.38
(1.10–1.72) | 0.0046 |
| 1.32
(1.16–1.51) | 4.9e-5 |
| Allele model |
|
|
|
|
|
|
|
|
|
| T | 2,490/2,557 | 1.00 (ref.) |
| 1616/1672 | 1.00 (ref.) |
| 4,106/4,229 | 1.00 (ref.) |
|
| C | 352/287 | 1.23
(1.06–1.49) | 0.0087 | 220/162 | 1.35
(1.08–1.70) | 0.0080 | 572/449 | 1.31
(1.14–1.50) | 1.1e-4 |
| Dominant model |
|
|
|
|
|
|
|
|
|
|
CT+CC | 330/275 | 1.26
(1.05–1.52) | 0.0133 | 207/155 | 1.40
(1.10–1.78) | 0.0056 | 537/430 | 1.33
(1.15–1.53) | 1.5e-4 |
| Recessive
model |
|
|
|
|
|
|
|
|
|
|
TT+CT | 1,399/1,410 | 1.00 (ref.) |
| 905/910 | 1.00 (ref.) |
| 2,304/2,320 | 1.00 (ref.) |
|
| CC | 22/12 | 2.02
(0.98–4.14) | 0.0565 | 13/7 | 1.77
(0.68–4.63) | 0.2467 | 35/19 | 1.98
(1.11–3.52) | 0.0203 |
Stratification analysis of rs12719482
T>C in the combined study
The associations of the rs12719482 (T>C) with
lung cancer risk stratified by selected variables using the
additive model was then assessed according to the AIC pattern. As
shown in Table IV, individuals
exhibited a significantly increased lung cancer risk as the number
of variant alleles increased, in the ages of ≤60 as well as >60
years (P=0.0039 and P=0.0087, respectively), in both genders
(P=0.0155 for men and P=0.0006 for women), in smokers as well as
non-smokers (P=0.0135 and P=0.0012, respectively), in ≥20 and 0
pack-years (P=0.0407 and P=0.0017, respectively) and in both male
smokers and female non-smokers (Pmax=0.0350)
following adjustment for confounding factors. There were also
statistically significant multiplicative interactions among gender,
smoking status, pack-years and allele genes (P<0.0001 for
all).
 | Table IV.Stratification analysis on rs12719482
T>C in BMP. |
Table IV.
Stratification analysis on rs12719482
T>C in BMP.
|
| Casesa |
Controlsa |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Variables | TT, n (%) | CT, n (%) | CC, n (%) | TT, n (%) | CT, n (%) | CC, n (%) | OR (95%
CI)b |
P-valueb |
P-valuec |
|---|
| Age, years |
|
|
|
|
|
|
|
| 0.3824 |
|
≤60 | 881 (76.88) | 245 (21.38) | 20 (1.75) | 961 (81.58) | 205 (17.40) | 12 (1.02) | 1.32
(1.09–1.59) | 0.0039 |
|
|
>60 | 921 (77.20) | 257 (21.54) | 15 (1.26) | 948 (81.65) | 206 (17.74) | 7 (0.60) | 1.30
(1.07–1.59) | 0.0087 |
|
| Gender |
|
|
|
|
|
|
|
|
<.0001 |
|
Male | 1,332 (77.40) | 362 (21.03) | 27 (1.57) | 1,392 (80.79) | 313 (18.17) | 18 (1.04) | 1.21
(1.04–1.42) | 0.0155 |
|
|
Female | 470 (76.05) | 140 (22.65) | 8 (1.29) | 517 (83.93) | 98 (15.91) | 1 (0.16) | 1.63
(1.23–2.15) | 0.0006 |
|
| Smoking status |
|
|
|
|
|
|
|
|
<.0001 |
|
Yes | 1,169 (77.31) | 319 (21.10) | 24 (1.59) | 900 (80.79) | 206 (18.49) | 8 (0.72) | 1.25
(1.05–1.50) | 0.0135 |
|
| No | 633 (76.54) | 183 (22.13) | 11 (1.33) | 1,009 (82.37) | 205 (16.73) | 11 (0.90) | 1.41
(1.14–1.73) | 0.0012 |
|
| Pack years |
|
|
|
|
|
|
|
|
<.0001 |
|
≥20 | 979 (76.84) | 275 (21.59) | 20 (1.57) | 645 (80.83) | 149 (18.67) | 4 (0.50) | 1.26
(1.01–1.58) | 0.0407 |
|
|
0-20 | 190 (79.83) | 44 (18.49) | 4 (1.68) | 255 (80.70) | 57 (18.04) | 4 (1.27) | 1.08
(0.70–1.65) | 0.7301 |
|
| 0 | 633 (76.54) | 183 (22.13) | 11 (1.33) | 1,009 (82.37) | 205 (16.73) | 11 (0.90) | 1.43
(1.14–1.79) | 0.0017 |
|
| Group by gender and
smoking |
|
|
|
|
|
|
|
|
|
| Male
smoker | 1,118 (77.48) | 302 (20.93) | 23 (1.59) | 881 (80.60) | 204 (18.66) | 8 (0.73) | 1.22
(1.01–1.46) | 0.0350 |
|
| Male
non-smoker | 214 (76.98) | 60 (21.58) | 4 (1.44) | 511 (81.11) | 109 (17.30) | 10 (1.59) | 1.21
(0.89–1.65) | 0.2176 |
|
| Female
smoker | 51 (73.91) | 17 (24.64) | 1 (1.45) | 19 (90.48) | 2 (9.52) | 0 (0) | 2.76
(0.58–13.1) | 0.2017 |
|
| Female
non-smoker | 419 (76.32) | 123 (22.40) | 7 (1.28) | 498 (83.70) | 96 (16.13) | 1 (0.17) | 1.62
(1.23–2.15) | 0.0007 |
|
| Source |
|
|
|
|
|
|
|
| 0.6009 |
|
Discovery | 1,091 (76.78) | 308 (21.67) | 22 (1.55) | 1,147 (80.66) | 263 (18.50) | 12 (0.84) | 1.26
(1.06–1.49) | 0.0086 |
|
|
Replication | 711 (77.45) | 194 (21.13) | 13 (1.42) | 762 (83.10) | 148 (16.14) | 7 (0.76) | 1.36
(1.08–1.70) | 0.0080 |
|
Discussion
Lung cancer remains a major health concern, due to
its 5-year survival of only 15% and associated high medical costs.
Genetic biomarkers may help identify susceptible subgroups for
screening, diagnosis and even therapy in earlier stages, and it may
also be beneficial for clinical outcomes. A total of 314 tagSNPs
were identified in the 18 pivotal genes from the BMP pathway
and evaluated for their association with lung cancer risk. In the
present study, a significant role was indicated for SMAD5 in
TGF-β-mediated lung cancer. The findings were concordant for a
significant association between the SMAD5 rs12719482 and
cigarette smoking. In addition, using Function Prediction websites
(https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html),
it was demonstrated that SMAD5 rs12719482 may be a
susceptibility marker by decreasing the expression of SMAD5
through binding of one of hsa-miR-1270, hsa-miR-571 or hsa-miR-920
to the polymorphic site in the Chinese population. To the best of
our knowledge, the present study is the first to estimate the
associations between a comprehensive panel of genetic polymorphism
of BMP genes and lung cancer risk, and to investigate the
potential susceptible subgroups. A series of functional experiences
are required in the future, as this is the first epidemiological
study supporting an association between SMAD5 rs12719482 and
lung cancer risk.
Identification of SMAD proteins has been helpful in
promoting the understanding of TGF-β signaling. BMPs may
signal through both canonical and non-canonical pathways. In
particular, in the canonical pathway, the BMP functions through BMP
ligand (BMPR1)-binding membrane-bound receptors, resulting in the
phosphorylation of intracellular mediators, known as the
receptor-regulated SMADs (R-SMADs). SMADs are crucial intracellular
signaling transmitters of the TGF-β superfamily of peptide growth
factors that regulate a series of biological processes (5). Controlling SMAD activity and
protein levels are crucial for proper signaling by TGF-β and its
related factors. Due to their ubiquitous expression and various
functions as regulators in various organs of the body, BMPs
were referred to as body morphogenetic proteins (14), as disruptions of BMP signaling
are associated with a wide variety of defects or severe
pathologies, such as vascular diseases, skeletal diseases and
cancer.
SMAD5, one of the MAD-homologues, is
situated on chromosome 5q31 in humans and acts downstream of the
TGF-β receptors. It is an important component of R-SMADs
(SMAD1/5/8) and the closest homolog of SMAD1, which
plays a role in the BMP-2 signaling pathway (15). Accumulating evidence indicates that
the SMAD5 gene functions in the signaling pathway involving
the inhibitory effect of TGF-β on cancer. For example, SMAD5
was considered to be a candidate tumor suppressor gene in
myelodysplastic syndrome and acute myeloid leukemia and is found in
human hematopoietic progenitor cells.
Animal studies have also revealed specific functions
of SMAD5. In the absence of SMAD5, mice die between
embryonic day (E)9.5 and E10.5 from impaired circulation.
Difficulties in angiogenesis and diminished number of vascular
smooth muscle cells (VSMCs) have been reported in
SMAD5-deficient mice (16,17).
Mice lacking SMAD5 exhibited increased apoptosis of cardiac
myocytes and craniofacial abnormalities. At the cellular level,
previous studies demonstrated that conditional knockout of
SMAD1 and SMAD5 in granulosa cells results in the
formation of vascularized granulosa cell tumors with complete
penetrance and with an elevated incidence of peritoneal metastases
and hemorrhagic ascites with increasing age (18). In addition, SMAD5-deficient
embryos exhibited defects in angiogenesis leading to decreased VSMC
numbers and enlarged dilated vessels. A disorganized allantois was
formed, primordial germ cells were largely diminished, with
left-right asymmetry and impaired embryo turning, all resulting
from SMAD5 deficiency (19,20).
Furthermore, the SMAD5−/− yolk sac may lead to
abnormalities in the vitelline network (17). However, the potential molecular
mechanism underlying SMAD5-dependent function remains to be
further elucidated.
The results of the present study also indicated that
the effect of SMAD5 rs12719482 (T>C) on humans was
detrimental, particularly among heavy smokers and when compared
between non-smokers and light smokers. In particular, the effect of
SMAD5 rs12719482 (T>C) was more prominent among light
smokers (0 pack-years) and non-smokers compared with the wild-types
TT. For example, the heavy smoking group (≥20 pack-years) exhibited
a 1.26-fold increased risk for rs12719482 (T>C), whereas its
effect was 1.43 among light smokers when wild-type TT was used as
the reference genotype in the additive model. When stratified by
smoking status, non-smokers showed a 1.41-fold increased risk for
rs12719482 (T>C) compared with the TT genotype. Therefore, there
may be a modified association between cigarette smoking and the
genotype variants of SMAD5 rs12719482 (T>C). The
potential mechanism underlying the interaction of SMAD5 and
smoking is not clear. However, the role of SMAD5 rs12719482
(T>C) was highlighted among non-smokers and light smokers.
Therefore, additional evidence and functional studies are required.
In addition, in the present study the frequency distribution of
smoking status was not homogeneous between cases and controls in
stage I as well as stage II, reflecting the differences in
lifestyle.
Several inherent biases of this study should be
elucidated. First, patients were mainly selected from hospitals and
the controls were selected from the center of health examination
during a routine health check; therefore, there was an inherent
selection bias. However, by matching the controls to the patients
according to age, gender and residential area, the underlying
confounding factors may be minimized. Second, risk factors other
than smoking status, such as occupational exposure and nutrition,
which may interact with BMP genotypes or function as
potential confounding factors, were not included in our study.
Possible interactions between those factors should be thoroughly
investigated in future studies. Finally, the functional relevance
of rs12719482 remains unknown, and further related investigations
should be performed.
In conclusion, we herein provided evidence
indicating that the SMAD5 rs12719482 polymorphism and its
interactions with smoking status may be a potential etiology of
lung cancer in Chinese patients. These findings remain to be tested
by larger scale studies in different ethnic groups.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (81520108001,
81220108001 and 81170052), the 973 Key Scheme of China
(2015CB553406), the National Key Research and Development Project
(2016YFC0903700), the Guangdong Province Universities and Colleges
Pearl River Scholar Funded Scheme (2014, to W. Lu), the Guangdong
Province Universities and Colleges Key Grant for Innovative
Research (cxzd1142), the Guangzhou Municipal Research Project
(201607020030), the Guangzhou Department of Education Innovative
Team (13C08), the Guangzhou Department of Education Yangcheng
Scholarship (12A001S), the Guangdong Natural Science Foundation
(2016A030313593) and the Guangzhou Municipal University Research
Projects (1201430298).
References
|
1
|
Babitt JL, Huang FW, Wrighting DM, Xia Y,
Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, et
al: Bone morphogenetic protein signaling by hemojuvelin regulates
hepcidin expression. Nat Genet. 38:531–539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu PB, Beppu H, Kawai N, Li E and Bloch
KD: Bone morphogenetic protein (BMP) type II receptor deletion
reveals BMP ligand-specific gain of signaling in pulmonary artery
smooth muscle cells. J Biol Chem. 280:24443–24450. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Daly AC, Randall RA and Hill CS:
Transforming growth factor beta-induced Smad1/5 phosphorylation in
epithelial cells is mediated by novel receptor complexes and is
essential for anchorage-independent growth. Mol Cell Biol.
28:6889–6902. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katsuno Y, Hanyu A, Kanda H, Ishikawa Y,
Akiyama F, Iwase T, Ogata E, Ehata S, Miyazono K and Imamura T:
Bone morphogenetic protein signaling enhances invasion and bone
metastasis of breast cancer cells through Smad pathway. Oncogene.
27:6322–6333. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kang JS, Saunier EF, Akhurst RJ and
Derynck R: The type I TGF-beta receptor is covalently modified and
regulated by sumoylation. Nat Cell Biol. 10:654–664. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng H, Jiang W, Phillips FM, Haydon RC,
Peng Y, Zhou L, Luu HH, An N, Breyer B, Vanichakarn P, et al:
Osteogenic activity of the fourteen types of human bone
morphogenetic proteins (BMPs). J Bone Joint Surg Am. 85-A:1–1552.
2003.PubMed/NCBI
|
|
7
|
Zhang L, Ye Y, Long X, Xiao P, Ren X and
Yu J: BMP signaling and its paradoxical effects in tumorigenesis
and dissemination. Oncotarget. 7:78206–78218. 2016.PubMed/NCBI
|
|
8
|
Ye L and Jiang WG: Bone morphogenetic
proteins in tumour associated angiogenesis and implication in
cancer therapies. Cancer Lett. 380:586–597. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Z, Wang J, He J, Zheng Z, Zeng X,
Zhang C, Ye J, Zhang Y, Zhong N and Lu W: Genetic variants in MUC4
gene are associated with lung cancer risk in a Chinese population.
PLoS One. 8:e777232013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Akaike H: A new look at the statistical
model identification. IEEE Trans Automat C ontr AC. 19:716–723.
1974. View Article : Google Scholar
|
|
11
|
Lewontin RC: On measures of gametic
disequilibrium. Genetics. 120:849–852. 1988.PubMed/NCBI
|
|
12
|
Gabriel SB, Schaffner SF, Nguyen H, Moore
JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart
M, et al: The structure of haplotype blocks in the human genome.
Science. 296:2225–2229. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stephens M and Donnelly P: A comparison of
bayesian methods for haplotype reconstruction from population
genotype data. Am J Hum Genet. 73:1162–1169. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wagner DO, Sieber C, Bhushan R, Börgermann
JH, Graf D and Knaus P: BMPs: From bone to body morphogenetic
proteins. Sci Signal. 3:mr12010.PubMed/NCBI
|
|
15
|
Hoodless PA, Haerry T, Abdollah S,
Stapleton M, O'Connor MB, Attisano L and Wrana JL: MADR1, a
MAD-related protein that functions in BMP2 signaling pathways.
Cell. 85:489–500. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang H, Huylebroeck D, Verschueren K, Guo
Q, Matzuk MM and Zwijsen A: Smad5 knockout mice die at
mid-gestation due to multiple embryonic and extraembryonic defects.
Development. 126:1631–1642. 1999.PubMed/NCBI
|
|
17
|
Yang X, Castilla LH, Xu X, Li C, Gotay J,
Weinstein M, Liu PP and Deng CX: Angiogenesis defects and
mesenchymal apoptosis in mice lacking SMAD5. Development.
126:1571–1580. 1999.PubMed/NCBI
|
|
18
|
Pangas SA, Li X, Umans L, Zwijsen A,
Huylebroeck D, Gutierrez C, Wang D, Martin JF, Jamin SP, Behringer
RR, et al: Conditional deletion of Smad1 and Smad5 in somatic cells
of male and female gonads leads to metastatic tumor development in
mice. Mol Cell Biol. 28:248–257. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang H and Matzuk MM: Smad5 is required
for mouse primordial germ cell development. Mech Dev. 104:61–67.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang H, Zwijsen A, Vogel H, Huylebroeck D
and Matzuk MM: Smad5 is essential for left-right asymmetry in mice.
Dev Biol. 219:71–78. 2000. View Article : Google Scholar : PubMed/NCBI
|