Introduction
Epidermal growth factor receptor (EGFR),
present on the cell surface, facilitates intercellular
communication. EGFR selectively targets growth signals and
permits the transfer of information (1,2). Cancer,
being a progressive disease, employs EGFR as a target,
which, when impaired, can successfully deregulate the downstream
cascade and favor tumorigenesis. As per mycancergenome (https://www.mycancergenome.org/content/disease/lung-cancer/),
~35% of patients with non-small cell lung cancer (NSCLC) in East
Asia have tumors associated with positive EGFR
expression.
Clinical diagnostics have evolved through multiple
facets, allowing difficult medical decisions to be accomplished
easily. Cancer is one field where opinions are widespread and
diverse, requiring extensive research and development. Cancer
diagnostics are complex and always changing, involving techniques
such as tissue-based immunohistochemistry (IHC), chromosome based
fluorescence in situ hybridization (FISH) and chromogenic
in situ hybridization (CISH), DNA-based gold-standard
sequencing techniques such as Sanger sequencing, and next
generation sequencing (NGS). These techniques not only support the
diagnosis of cancer, but also help to predict treatment success and
repercussions. NGS encompasses DNA-based genetic modifications,
such as single nucleotide variations (insertions, deletions and
rearrangements), in its therapeutic prediction, using massive
parallel sequencing operandi. Single nucleotide variations
constitute the substitution of one nucleotide base for another,
thus impairing the formation of hydrogen bonds between the strands.
These variations can be synonymous or non-synonymous; when they
occur at conserved domains they are able to impair normal function
of the translated protein. Insertions and deletions refer to the
introduction or removal of one or more nucleotides from the DNA
strand. Rearrangement variations occur because of chromosomal
breakage causing broken segments to adhere elsewhere on the
chromosome. These can include small or large segments of DNA, with
effects depending on the size and domain. NGS has been gradually
introduced into many clinical fields, especially oncology, for
which genetic modifications are a major factor (3). Lung cancer, one of the most common
types of cancer, has EGFR as a major therapeutic predictor.
The present study highlights the significance of other genomic
alterations in the therapeutic prediction of lung cancer, most of
which are frequently neglected.
Notable associations have been documented between
somatic mutations (deletions and single nucleotide variants) in
EGFR exons 19 and 21 and the corresponding tyrosine kinase
inhibitors (TKIs). Small molecule TKIs and monoclonal antibodies
are major treatment regimens pursued for cases of lung cancer
(4). EGFR TKIs have also been
approved by the Food and Drug Administration (FDA) as a first line
regimen for the treatment of NSCLC. Of all EGFR TK domain
mutations, 50% were exon 19 in-frame deletions, 42% were exon 21
missense mutations, 7% were exon 18 mutations, and 3% were in exon
20 (5,6). Noronha et al (6) showed that, the overall frequency of
EGFR mutations, including single nucleotide alterations,
deletions and insertions, in cases of Indian patients with
adenocarcinoma is 26%, compared with 3.8% in squamous cell
carcinoma. TKIs and monoclonal antibodies are useful in the
presence of somatic EGFR mutations, the absence of which
creates agitation in treating oncologists, as patients are unable
to have TKI treatment (7). Thus, new
methods and techniques for managing lung cancer, beyond
EGFR, are required.
Technology improved clinical diagnostic and
therapeutic methods at an enormous rate over recent years. Single
gene testing is used for the effective determination of mutations
in established genes and regions (8,9). Routine
diagnostic testing of functionally significant genes is recommended
by NCCN guidelines as it is beneficial in the treatment of lung
cancer, generally using formalin-fixed paraffin-embedded (FFPE)
tissue extracted from tumor samples. Despite its clinical
significance, the method also presents uncertainty with ‘tumor
heterogeneity’ (10–12). To overcome the limitations of hotspot
testing, comprehensive genomics-based testing panels have been
developed to provide a comprehensive picture of the genome, exome
or targeted regions to study various other rare or barely studied
oncogenic drivers (13–15). It is a technique with a great
capacity to identify alterations beyond EGFR, along with
other benefits such as early treatment response and resistance
evaluation, the assessment of molecular heterogeneity, early
detection of disease, and, most importantly, the identification of
genetic determinants for targeted therapy (16–18).
Herein, we present our data analysis and characterization of
pathogenic and targetable variants beyond EGFR for the
effective prediction of clinical outcomes in lung cancer.
Materials and methods
Reports were investigated pertaining to 137 patients
with lung cancer who had previously undergone commercially
available comprehensive hybrid capture NGS (PositiveSelect). This
included patients with different stages of the disease and a wide
therapeutic spectrum. All cases were predominantly diagnosed at
stage 4 with progressive disease condition and metastasis, as
presented in Table I. Few had
previously undergone molecular testing for NCCN recommended genes
like EGFR, ALK or ROS1. Molecular diagnostics mostly
comprised EGFR, with occasional testing for ALK and
other NCCN recommended genes. A few cases were previously treated
with targeted and chemotherapeutics based on hotspot molecular
diagnostics reports and NCCN guideline recommendations,
respectively. The PositiveSelect assay protocol for NGS (sequencing
and analysis) employed in the study is not yet been published. The
parameters utilized for NGS are briefly described below. Hybrid
capture libraries of 350 genes and 35 selected introns frequently
rearranged in cancer were sequenced in parallel with a high,
uniform coverage (×1,000). The resultant raw data were
demultiplexed using NextSeq reporter software (CASAVA 2.2;
Illumina, Inc., San Diego, CA, USA) for acquisition of individual
samples. The acquired data are often subjected to trimming to
eliminate primer artifacts, followed by alignment with a human
reference genome, variant calling and prioritization. Strict
cut-off parameters were subjected to variant calling algorithms
that segregate variants of allelic fractions greater than 1 as
clonal and those greater than 0.1, but less than 1 were regarded as
subclonal variations for further analysis. The detection of
subclonal alterations was accomplished effectively with the
utilization of liquid biopsy-based samples. The algorithm further
followed annotation according to databases with notable functional
evidence of genomic alterations, which includes dbsnp, ClinVar,
COSMIC, TCGA and ExAC, among others. Interpretation of the results
holds significance for the determination of clinically relevant
classes of alterations, such as base-pair substitutions,
insertions/deletions, copy number alterations and rearrangements.
Clinically relevant genomic alterations were thus defined as those
associated with responses to therapies currently available or in
target-driven clinical trials. In the present study the clinically
relevant alterations were dichotomized based on EGFR and
ALK positivity to understand the therapeutic utility of rare
and frequently occurring alterations beyond EGFR and
ALK. The analytical perspectives from the clinically
relevant classes of alterations characterized by PositiveSelect
were collated and assessed with international studies, constructing
a pooled meta-analysis executed from cBioPortal and proving
clinical concordance of our study with the international study
spectrum (http://www.cbioportal.org/).
 | Table I.Cohort characteristics included in
our analysis. |
Table I.
Cohort characteristics included in
our analysis.
|
| Total number of
cases (n=137) |
|
|---|
|
|
|
|
|---|
| Age (years) | Total (n) | Males (n=91) | Females (n=46) | Disease stage |
|---|
| 30–40 | 7 | 5 | 2 | 4 |
| 40–50 | 20 | 13 | 7 | 2, 3B and 4 |
| 50–60 | 47 | 29 | 18 | 1C, 2A, 3B and
4 |
| 60–70 | 37 | 26 | 11 | 2, 3B and 4 |
| 70–80 | 26 | 18 | 8 | 3 and 4 |
Results
Our analysis revealed a notable percentage of
genomic alterations beyond EGFR and ALK recommended
by NCCN guidelines in the study population, which are as follows:
Single nucleotide variations in KRAS, RET and BRAF
were detected at 6, 2 and 2%, respectively; single nucleotide
variation and amplification detected in MET and ERBB2
at 3 and 2%, respectively. Though the percentages do not appear
appreciable, these become significant when considered in light of
the clinical implications. This verifies the study objective of
moving beyond EGFR and ALK to achieve better clinical
outcomes.
EGFR positivity and beyond
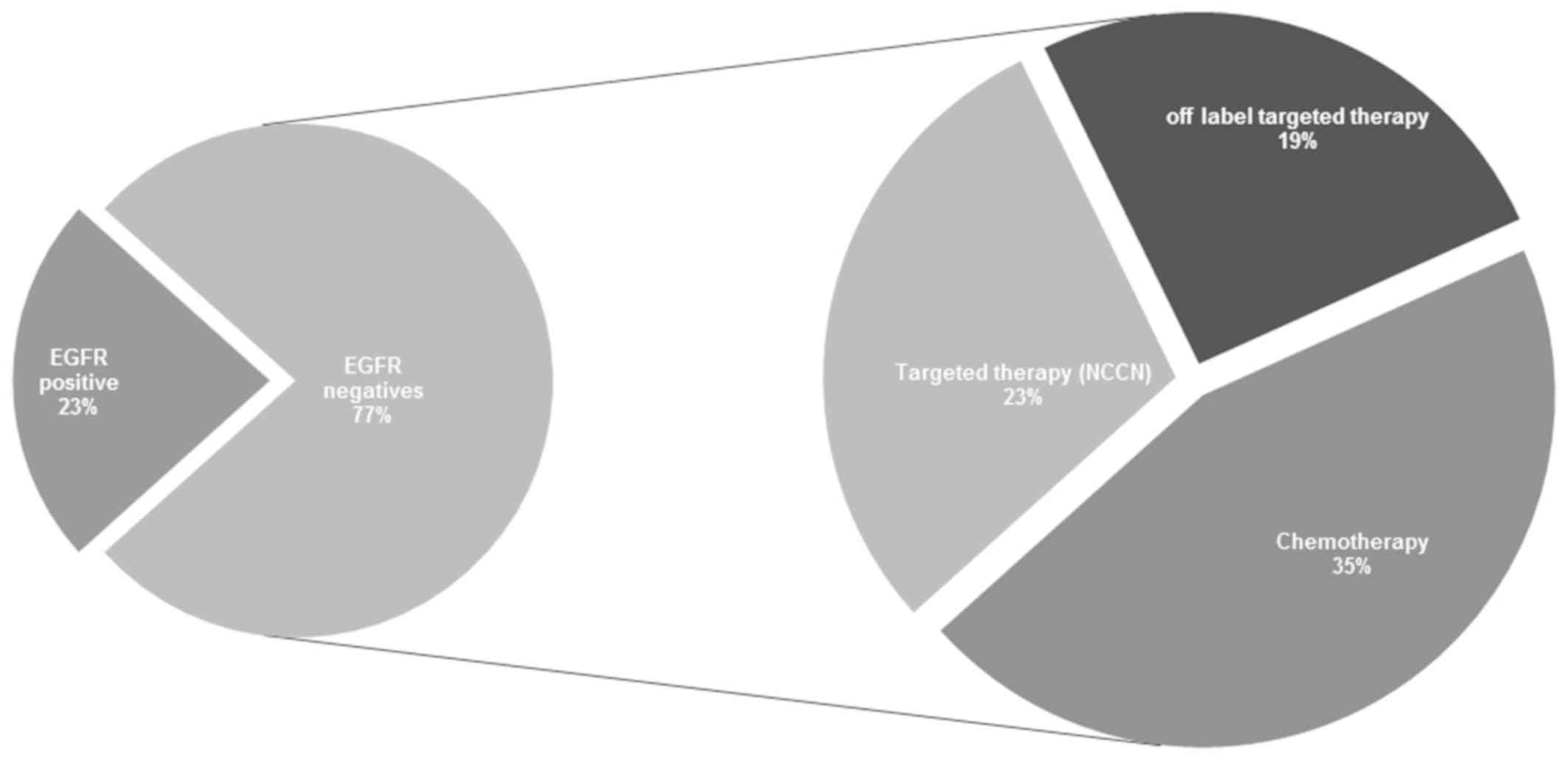
Of the 137 lung cancer cases in our cohort, 23%
(n=31) were identified to be EGFR-positive, indicating
benefit from the use of EGFR-TKI. However, the 77% (n=106)
of patients were EGFR-negative cases, which still pose a
therapeutic challenge. The present study provides new hope for the
treatment of patients who fall under the criteria of
EGFR-negative status. It was identified that 23% of
patients, who were EGFR-negative, may benefit from
NCCN-recommended targeted therapeutics as they possess
non-EGFR genomic alterations, while 19% of patients, who
were EGFR-negative, were identified to have other gene
alterations indicating that they may benefit from the use of
off-label targeted therapeutics. The remaining 35% of patients, who
were EGFR-negative, become eligible for NCCN-recommended,
and/or off-label chemotherapy, which would have otherwise been the
entire 77% as per current clinical practice. This finding has also
been summarized in Fig. 1. According
to our analysis, of the EGFR-negative population that may
benefit from NCCN guideline recommended targeted therapeutics, we
identified 7% (n=7) to bear a KRAS pathogenic single
nucleotide mutation, which allows the utilization of downstream
MEK, PIK3CA and mTOR inhibitors, such as Trametinib,
Buparlisib and Everolimus, to target the downstream
KRAS/MEK/ERK and
KRAS/PIK3CA/AKT/mTOR signaling pathways,
respectively. Apart from KRAS, other pathogenic alterations
detected include ALK rearrangements in 3% (n=3), and
ALK resistance mutations detected in 4% (n=4), BRAF
in 2% (n=2), ERBB2 amplifications and mutations detected in
1% (n=1) each, MET in 2% (n=2) and RET in 3% (n=3). A
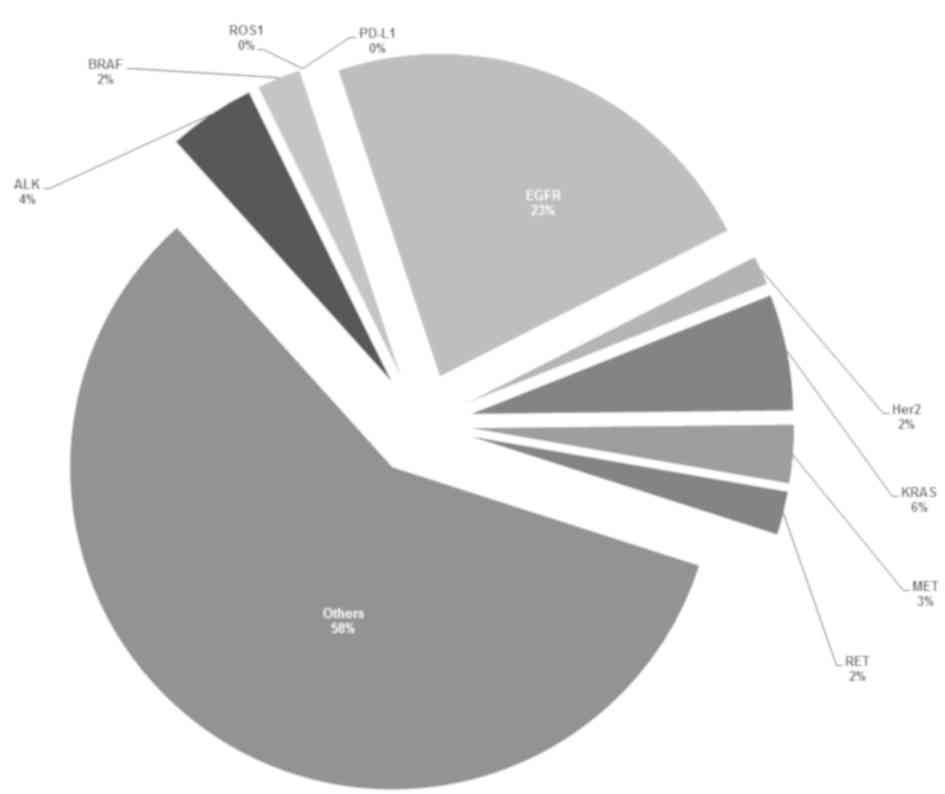
summary of the overall variant frequencies of NCCN recommended
genes in the 137 lung cancer cases is presented in Fig. 2.
EGFR and ALK dichotomy
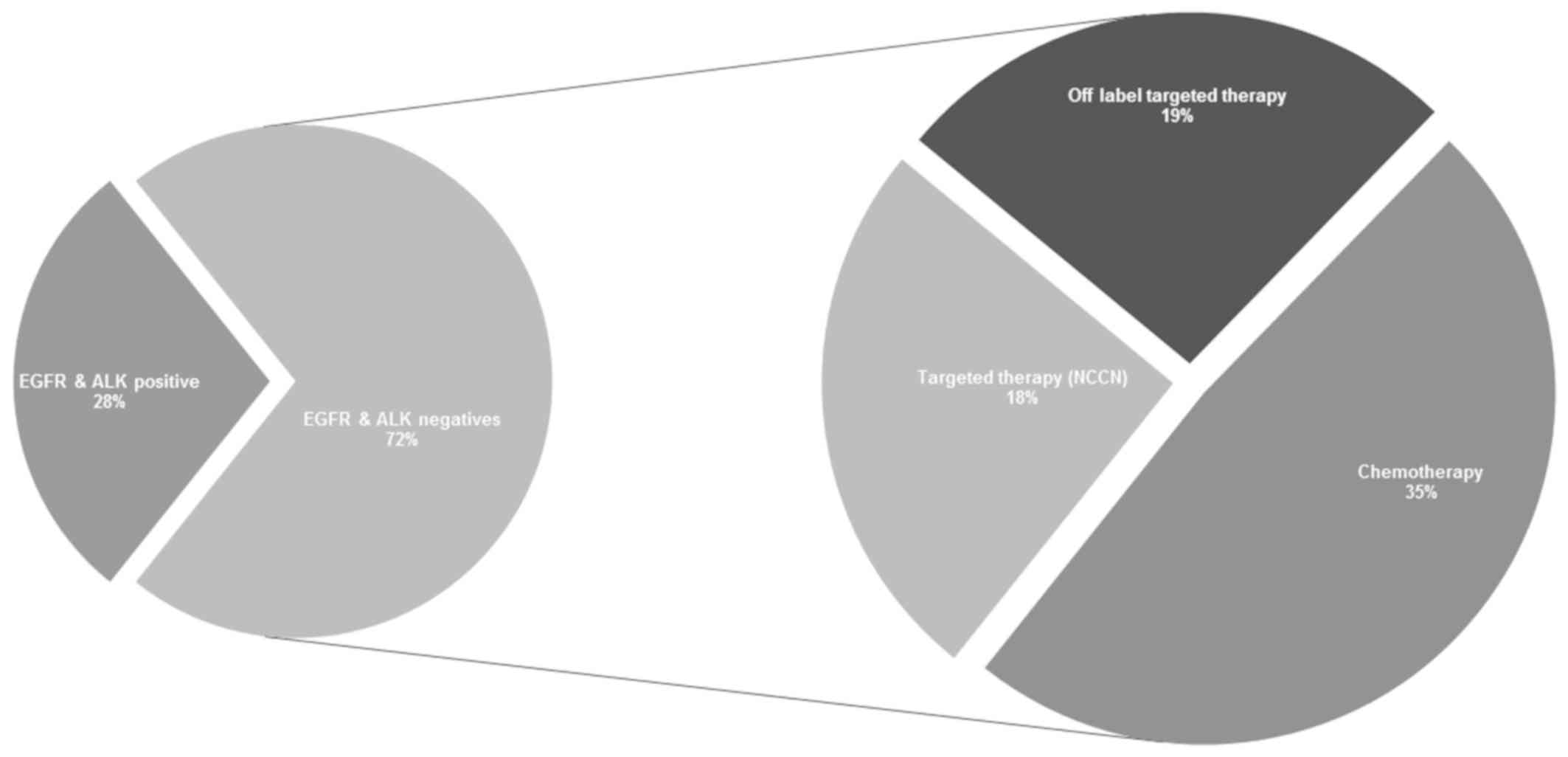
Based on EGFR and ALK dichotomization,
our study indicated that only 28% of the total cohort was eligible
for targeted therapy based on EGFR and ALK
positivity, taken as the two most frequently tested genes in lung
cancer therapeutics. The remaining major cohort (72% of patients),
who were negative for EGFR and ALK, received
conventional chemotherapy treatment rather than being subjected to
further molecular tests. Contrary to typical approaches, our
comprehensive NGS analysis identified other favorable treatment
options for the ALK- and EGFR-negative cohort, of
whom 18% harbored genomic alterations which made them eligible for
NCCN guideline recommended targeted therapy. A further 19% of the
cohort was identified to benefit from off-label targeted therapy
due to alterations detected in PTEN and TSC1/2.
Specifically, we detected KRAS pathogenic single nucleotide
variations in 6% (n=6) and amplifications in 2% (n=2) of the
negative population. Furthermore, 35% of the patients who were
ALK negative and EGFR negative, with no additional
genomic alterations, have been as potentially receiving benefiting
from chemotherapy, as no additional genomic alterations were
identified as highlighted in Fig.
3.
Meta-analysis and a pooled assessment
with International Lung Cancer studies
During a meta-analysis of three international
studies on the variant distribution frequency of the nine NCCN
recommended lung cancer genes, we observed concordance between our
data and the global published literature (Table II) (19–21). Of
the 137 lung cancer cases analyzed in this study, 23% (n=31) were
identified with targetable EGFR variants, marginally
concordant with the 19% reported in a relevant international lung
cancer study in 2012 by Imielinski et al (19) (Table
II). Additionally, ALK (4%), and RET (2%)
correlated with the statistics unveiled by the Broad Institute of
Harvard and MIT (Cambridge, MA, USA) in their study, which reported
frequencies at 4 and 2% for ALK and RET, respectively
(19). Additional similarities were
observed between our frequencies and those reported by
international studies, validating the detection of additional
genomic alterations in EGFR and ALK mutation-negative
cohorts (20,21). The results of the aforementioned
study demonstrated that 20% of the mutations harbored by the
EGFR-negative cohort occurred in other significant genes,
including PIK3CA, CDK4/6 or CCND, KDR, TSC1/2, mTOR,
TP53 and DNA repair genes recommended as drug targets by the
NCCN. This enhances the clinical benefits of the patients harboring
variants in these genes. It is evident that this 20% population
from the EGFR-negative cohort in the current study may also
benefit from the aforementioned gene-targeted therapy.
 | Table II.Comparison of frequencies with
published international literature. |
Table II.
Comparison of frequencies with
published international literature.
|
| Gene variant
distribution % (n) in all lung cancer cases | Gene variant
distribution % (n) in EGFR negative lung cancer cases |
|---|
|
|
|
|
|---|
|
| 1. Broad (19) | 2. Nature (20) | 3. MSKCC (21) | Meta analysis (1, 2
and 3) | Positive
Select | 1. Broad (19) | 2. Nature (20) | 3. MSKCC (21) | Meta analysis (1, 2
and 3) | Positive
Select |
|---|
| EGFR | 19 (34) | 3 (4) | 6
(2) | 12.3 (40) | 23 (31) | 81
(149) | 96 (106) | 94
(32) | 88
(287) | 77 (106) |
| ALK | 4
(10) | 5 (7) | 0 | 4
(17) | 4 (6) | 5 (8) | 7 (7) | 0 | 5
(15) | 3 (3) |
| BRAF | 6
(14) | 2 (2) | 3
(1) | 4
(17) | 2 (3) | 8
(12) | 2 (2) | 3
(1) | 5
(15) | 2 (2) |
| ERBB2 | 2 (3) | 1 (1) | 0 | 2 (4) | 2 (2) | 2 (3) | 1 (1) | 0 | 1 (4) | 1 (1) |
| KRAS | 19 (48) | 0 | 23 (8) | 14 (56) | 8 (6) | 32 (47) | 0 | 25 (8) | 19 (55) | 6 (7) |
| MET | 4 (8) | 2 (2) | 6
(2) | 4
(12) | 3 (4) | 4 (8) | 2 (2) | 6
(2) | 4 (12) | 2 (2) |
| RET | 2 (6) | 5 (6) | 0 | 3
(12) | 2 (3) | 3 (5) | 5 (5) | 0 | 4 (10) | 3 (3) |
However, 44% of the total cohort harbored ≥1 gene
mutation from the NCCN recommended panel for lung cancer. Ideally,
patients can be recommended agents with an improved therapeutic
benefit based on if they possess targetable and clinically
actionable gene mutations aside from the NCCN recommended genes
that may distinguish them from the conventional chemotherapy group.
A compilation of identified mutations, including single nucleotide
variations, insertions/deletions and copy number alterations, in
EGFR- and ALK+EGFR-negative cohorts, and their
corresponding recommended therapeutics, is listed in Table III.
 | Table III.Treatment approaches recommended by
our analysis in EGFR- and (EGFR+ALK)-negative
cases. |
Table III.
Treatment approaches recommended by
our analysis in EGFR- and (EGFR+ALK)-negative
cases.
| NCCN recommended
panel of genes | Frequency % (n)
EGFR negative | Frequency % (n)
(EGFR+ALK) negative | Potential
therapeutic recommendations |
|---|
| ALK | 3 (3) | 0 (0) | ALK
inhibitors (Crizotinib, Ceritinib) |
| BRAF | 2 (2) | 0 (0) | BRAF
inhibitors (Dabrafenib, Vemurafenib) |
| ERBB2 | 1 (1) | 1 (2) | ERBB2
inhibitors (Neratinib, Lapatinib) |
| KRAS | 7 (7) | 6 (8) | MEK
inhibitors (Trametinib), PI3K inhibitors (Buparlisib) |
| MET | 2 (2) | 1 (1) | MET
inhibitors (Crizotinib) |
| RET | 3 (3) | 3 (3) | RET
inhibitors (Cabozantinib, Lenvatinib, Sorafenib) |
Discussion
In lung cancer, EGFR is the most commonly
targeted gene and is regarded as a potential candidate for small
molecule therapeutics. The prevalence of EGFR mutations
across various countries was highlighted by a study conducted at
Tata Memorial Hospital, and has been tabulated in Table S1. Targeting EGFR has been a
hot research topic in the scientific community, and has modeled
current therapeutic interventions for lung cancer (22,23). The
present analysis is based on the essential functionality of every
gene in a cell, not just their statistics. Using this strategy,
distinguishing driver mutations beyond from EGFR becomes
straightforward. Thus, the aim was to determine the maximum number
of patients, who are currently over-prescribed chemotherapy, that
could potentially benefit from targeted therapeutics beyond
EGFR.
In current oncology, NCCN guidelines are a
repository of treatment protocols for patients with various cancer
types. As per the recently updated NCCN guidelines for NSCLC,
testing for 9 genes (EGFR, ALK, ROS1, PDL-1, RET, KRAS/NRAS,
BRAF, MET, ERBB2) is recommended as routine. Fig. 2 shows the distribution of alterations
of 9 NCCN recommended genes in the study population. Of these,
routine hotspot molecular tests using conventional methods
generally include EGFR, ALK and ROS, due to the
availability of approved targeted therapeutics (24). However, this type of limited testing
leaves the majority of patients diagnosed as EGFR- and
ALK-negative with no alternatives to chemotherapy. The
EGFR-negative population was identified to be 77% of the
total cohort, and accounts for the majority of patients with lung
cancer, who are then either subjected to genetic testing to detect
ALK status or recommended chemotherapy in current practice.
In our study, 42% of this negative population (23% NCCN recommended
targeted therapy + 19% off-label targeted therapy) was identified
to present other mutations in genes including KRAS, BRAF, ERBB2,
MET, RET, PTEN and TSC1/2, among others. Similarly, 72%
of the population did not present with pathogenic mutations in
EGFR and ALK, of which 37% (18% NCCN recommended
targeted therapy + 19% off-label targeted therapy) exhibited
mutations in other genes such as KRAS, MET, RET, PTEN and
TSC1/2. Only 35% of both the cohorts were classified to
benefit from chemotherapy. Though these therapeutic decisions are
challenging, the data could be beneficial to improving clinical
outcomes in the future.
The mutation frequencies listed in Table II assert that these genetic
alterations do have a significant impact on lung carcinogenesis. It
is understood that these alterations could develop as onco-drivers
under certain microenvironmental pressures. In this way, patients
would not have to deal with the genotoxic and cytotoxic effects on
the normal healthy cellular population caused by chemotherapy.
Though the current study approach may not boost therapeutic
practice, it will certainly enhance medical oncology clinical
outcomes in the near future.
There are a few targeted therapy options based on
genomic alterations beyond EGFR and ALK detected in
the present study population. In ALK and MET
pathogenic cases (ALK-rearrangements,
MET-amplifications and exon-skipping mutations), TKIs such
as crizotinib and ceritinib would deliver better treatment
responses. BRAF inhibitors such as dabrafenib and
vemurafenib could be effective against BRAF single
nucleotide variant pathogenicity (25). RET single nucleotide variant
pathogenicity could be efficiently targeted by inhibitors such as
cabozantinib, lenvatinib and sorafenib. A few inhibitors, such as
neratinib and lapatinib, have been recommended for ERBB2
mutations including, amplification and single nucleotide
variations.
Future cancer therapeutics can achieve better
clinical outcomes with drugs targeting additional cancer drivers
such as PTEN, PIK3CA and TSC1/2, among others, which
contribute to cancer growth and progression. The present study also
unveiled novel avenues for genomic biomarker discovery to assist
cancer diagnosis and therapy.
In summary, lung cancer is a rapidly progressive
class of cancer. The current state of oncology is such that the
treatment regimen is determined based on the mutation status of
specific guideline-recommended genes, tested for using routine
diagnostic procedures. The majority of routine diagnostic testing
focuses on established regional variants of EGFR and
ALK. Treatment strategies are straightforward if testing
indicates positivity for EGFR and ALK, or any of the
nine genes recommended by the guidelines. Unfortunately, the only
treatment option available for negative results is chemotherapy.
Additional parameters beyond EGFR and ALK were
examined to better deal with the toxicity by bringing in more
targeted therapeutic options in place of chemotherapy alone.
Additional genes beyond EGFR and ALK were examined to
increase the utilization of gene-specific-targeted drugs in place
of just chemotherapy. Thus, patients administered chemotherapy due
to their non-responsiveness to EGFR/ALK-based targeted
therapy could be minimized. The results of the present study will
allow reforms to the current cancer diagnostics framework, and
facilitate the development of better therapeutics.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GS designed the study concept and reviewed the
manuscript. SI performed study design, analyzed and interpreted the
data and contributed to manuscript preparation. SK contributed to
data acquisition, algorithms and performed statistical analysis and
figure generation. RP contributed to data acquisition, algorithms
and performed statistical analysis and figure generation. AR
contributed to data acquisition, statistical analysis, figure
generation and manuscript preparation. MB performed statistical
analysis and figure generation and contributed to manuscript
review. ST contributed to manuscript preparation and review. RV
contributed to data interpretation and analysis. RN contributed to
data interpretation. PK performed data analysis and interpretation.
AB contributed to manuscript preparation. VM contributed to
manuscript preparation and review. VM contributed to manuscript
review. SCT contributed to data interpretation and manuscript
review. MS contributed to manuscript preparation and review. AR
contributed to manuscript preparation and review.
Ethical approval and consent to
participate
All the experiments were carried out according to
the appropriate guidelines and are approved by the PositiveATGC's
(ACADEMY for TRAINING in GENOMICS and CLINICAL APPLICATION)
scientific review committee. Written informed consent was acquired
from all subjects as per the protocol of the review committee for
the utilization of clinical samples in this study.
Patient consent for publication
The patient, or parent, guardian or next of kin
provided written informed consent for the publication of any
associated data and accompanying images.
Competing interests
The authors declare no competing interests.
References
|
1
|
Bethune G, Bethune D, Ridgway N and Xu Z:
Epidermal growth factor receptor (EGFR) in lung cancer: An overview
and update. J Thorac Dis. 2:48–51. 2010.PubMed/NCBI
|
|
2
|
Siegelin MD and Borczuk AC: Epidermal
growth factor receptor mutations in lung adenocarcinoma. Lab
Invest. 94:129–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shyr D and Liu Q: Next generation
sequencing in cancer research and clinical application. Biol Proced
Online. 15:42013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu TC, Jin X, Wang Y and Wang K: Role of
epidermal growth factor receptor in lung cancer and targeted
therapies. Am J Cancer Res. 7:187–202. 2017.PubMed/NCBI
|
|
5
|
Zhang Z, Stiegler AL, Boggon TJ, Kobayashi
S and Halmos B: EGFR-mutated lung cancer: A paradigm of molecular
oncology. Oncotarget. 1:497–514. 2010.PubMed/NCBI
|
|
6
|
Noronha V, Pinninti R, Patil VM, Joshi A
and Prabhash K: Lung cancer in the Indian subcontinent. South Asian
J Cancer. 5:95–103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Linardou H, Dahabreh IJ, Bafaloukos D,
Kosmidis P and Murray S: Somatic EGFR mutations and efficacy of
tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 6:352–366.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Van Assche K, Ferdinande L, Lievens Y,
Vandecasteele K and Surmont V: EGFR mutation positive stage IV
non-small-cell lung cancer: Treatment beyond progression. Front
Oncol. 4:350–358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Capelozzi VL: Role of immunohistochemistry
in the diagnosis of lung cancer. J Bras Pneumol. 35:375–382. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mafficini A, Amato E, Fassan M, Simbolo M,
Antonello D, Vicentini C, Scardoni M, Bersani S, Gottardi M, Rusev
B, et al: Reporting tumor molecular heterogeneity in
histopathological diagnosis. PLoS One. 9:e1049792014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Janku F: Tumor heterogeneity in the
clinic: Is it a real problem? Ther Adv Med Oncol. 6:43–51. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han HS and Magliocco AM: Molecular testing
and the pathologist's role in clinical trials of breast cancer.
Clin Breast Cancer. 16:166–179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McCoach CE and Doebele RC: The minority
report: Targeting the rare oncogenes in NSCLC. Curr Treat Options
Oncol. 15:644–657. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Levy MA, Lovly CM and Pao W: Translating
genomic information into clinical medicine: Lung cancer as a
paradigm. Genome Res. 22:2101–2108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meldrum C, Doyle MA and Tothill RW:
Next-generation sequencing for cancer diagnostics: A practical
perspective. Clin Biochem Rev. 32:177–195. 2011.PubMed/NCBI
|
|
16
|
Khoo C, Rogers TM, Fellowes A, Bell A and
Fox S: Molecular methods for somatic mutation testing in lung
adenocarcinoma: EGFR and beyond. Transl Lung Cancer Res. 4:126–141.
2015.PubMed/NCBI
|
|
17
|
Gagan J and Van Allen EM: Next-generation
sequencing to guide cancer therapy. Genome Med. 7:802015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rothschild SI: Targeted therapies in
non-small cell lung cancer - Beyond EGFR and ALK. Cancers (Basel).
7:930–949. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
George J, Lim JS, Jang SJ, Cun Y, Ozretić
L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al:
Comprehensive genomic profiles of small cell lung cancer. Nature.
524:47–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 348:124–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang YL, Yuan JQ, Wang KF, Fu XH, Han XR,
Threapleton D, Yang ZY, Mao C and Tang JL: The prevalence of EGFR
mutation in patients with non-small cell lung cancer: A systematic
review and meta-analysis. Oncotarget. 7:78985–78993.
2016.PubMed/NCBI
|
|
23
|
Choughule A, Noronha V, Joshi A, Desai S,
Jambhekar N, Utture S, Thavamanni A, Prabhash K and Dutt A:
Epidermal growth factor receptor mutation subtypes and geographical
distribution among Indian non-small cell lung cancer patients.
Indian J Cancer. 50:107–111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Korpanty GJ, Graham DM, Vincent MD and
Leighl NB: Biomarkers that currently affect clinical practice in
lung cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front Oncol.
4:2042014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng L, Lopez-Beltran A, Massari F,
MacLennan GT and Montironi R: Molecular testing for BRAF mutations
to inform melanoma treatment decisions: A move toward precision
medicine. Mod Pathol. 31:24–38. 2018. View Article : Google Scholar : PubMed/NCBI
|