Introduction
Multiple myeloma (MM) is a common neoplastic
disorder (1). It is the second most
diagnosed hematological malignancy in China with the incidence rate
of ~1.1/100,000 that accounts for 2.1% of all new cancer cases in
2016(2). Typically, MM is an
incurable cancer of the plasma cells, which is identified by the
infiltration and clonal proliferation of antibody-secreting
post-germinal center plasma cells in the bone marrow that lead to
renal insufficiency, bone disease and anemia (3,4).
Recent articles have indicated that age, male sex,
obesity and ionizing radiation exposure are the most common risk
factors for MM (5,6). Nevertheless, the exact cause of MM is
still unknown (6). Epidemiological
studies have shown an increasing incidence rate of MM in Caucasian
countries. Although African populations have the highest incidence
rate of MM, while the lowest ones belong to the Asian and
Mediterranean populations (7,8).
The pathology of MM is speculated to involve
multiple factors including genetic, immune system and environmental
causes. Since MM is a heterogeneous disease, genetic factors play
an important role in its etiology (9). Gene polymorphisms of inflammatory
factors may be involved in the progression of MM by unbalancing
pro- and anti-inflammatory cytokines profile (9,10).
Interleukin (IL) family that are involved in the
immune response and inflammatory processes of MM, consist of a
group of lymphatic factors with a multiplicity of biological
activities (11,12). Many experimental evidences have
revealed specific associations between the metastatic risk of MM
and activation of interleukin (12).
In light of this, among the interleukin family, IL-6 with its
receptor IL-6R is the major interleukin for the T helper (Th)
mediated inflammation and it is important in maintaining the
Th1/Th2 balance in the inflammatory stage of MM (13,14).
These observations indicate that human IL-6 and IL-10 polymorphisms
might act as a biomarker for monitoring the clinical course of MM
(15,16). Consequently, the prognostic value of
genetic factors are potentially limited in genotype-related
interleukin, specific interleukin loci and individual phenotype
(14).
Recently, a multivariable analysis was used to
determine if interleukin family polymorphisms are a prognostic
factor for survival in MM patients (17-19).
It has been widely reported that many different polymorphisms in
different loci are related to poor survival and prognosis of MM
(19,20). The polymorphism of interleukin family
occurs in MM patients, and it is associated with statistically
increased of MM risk (20-22).
Interleukin production is regulated by the polymorphisms of
cytokine genes in promoter regions (20,23). The
high level of pro-inflammatory interleukin level is a significant
predictor of both progression-free survival (PFS) and overall
survival (OS) in Caucasian MM patients (24,25).
Although, the results of these studies are
controversial or inconclusive due to their limited genuine
heterogeneity, stage of MM and sample size. Therefore, we conducted
a quantitative systematic review along with a comprehensive
meta-analysis investigation to resolve inconsistent and often
ambiguous findings. Hence, this paper attempted to investigate the
associations between the interleukin family gene polymorphisms and
MM patients.
Materials and methods
Ethics statement
The present meta-analysis was approved by an
independent Ethics Committee/institutional review board at
Southwest Medical University, Department of Hematology in Luzhou,
China. This investigation was carried out by following
recommendations of the Preferred Reporting Items for Systematic
Reviews and Meta-Analysis (PRISMA) (26).
Search strategy and study
identification
A comprehensive literature search was conducted in
MEDLINE electronic databases of PubMed, Embase, Wiley Online
Library, Web of Science, Science Direct and VIP-Google Scholar
Database. All databases were searched without using language
restrictions to assess the prognostic value of interleukin family
polymorphism in MM patients prior to July 05, 2018, with no lower
date limit. The search string was conducted by using MeSH terms and
following the main heading term or word (both the US and UK
variants). Based on the research question, the combinations of the
keywords or main headings were determined as follows: ‘IL’ OR
‘interleukin’, OR ‘IL-’ AND ‘polymorphism’ OR ‘polymorphisms’ OR
‘interleukin polymorphisms’ OR ‘single nucleotide polymorphism’ OR
‘SNP’, AND ‘myeloma’ OR ‘multiple myeloma’. We have repeated these
terms for each of the investigated interleukins, including:
IL-1α/β, -4, -6, -10, -17, -21, -23, -26 and their receptors.
Alternative synonyms and spellings were incorporated using Boolean
‘OR’ and the main terms were linked with Boolean ‘AND’.
Inclusion/exclusion criteria
The following inclusion standards were used to
select potential studies for this meta-analysis: i) Evaluated
association between the individual IL and/or the IL receptor
polymorphism with MM risk; ii) Human genotypes involvement iii) Use
of a case-control design; iv) Contained raw genotype frequencies
for at least one IL promoter and/or IL receptor polymorphism; v)
Publication in English. All case reports, retrospective studies,
editorials and review articles, family-based studies, unrelated
articles, studies without available genotype frequencies, studies
that only investigated the impact of IL polymorphisms in response
to therapy or drug resistance and genome-wide association studies
were excluded. When an author had published several articles with
data obtained from the same patient population, only the newest or
the most informative article was selected.
Data collection
The titles and abstracts of all selected articles
were analyzed according to the PICO principle by two independent
investigators (MNS and II) (27).
Any disagreement or discrepancy was adjudicated through debate or
consultation; if a consensus was not reached then a third
investigator (SH) was consulted. The electronic study was
supplemented by a hand-search of relevant articles from reference
lists to ensure that all relevant research were identified.
Synchronously, references of review articles were checked for any
relevant articles and bibliographies. The following key components
of all qualified studies were recorded: First author's name, year
of publication, ethnicity, country of origin, genotyping methods,
sample size, characteristics of controls and matching criteria,
study design, TNM stage, clerk stage, reference control, total
number of cases and controls that were stratified by genotype
frequencies (homozygous wild-type, heterozygous and homozygous
mutant), P-value for Hardy-Weinberg equilibrium (HWE) of controls
and true and false positives and negatives. We also e-mailed the
corresponding authors of the selected articles to obtain additional
and/or any missing information, as well as the copies of the
original data required for the meta-analysis. Finally, if the above
information was not mentioned in the original study or we did not
receive any response via email, the item was preserved as ‘not
reported (NR)’.
Quality assessment
The quality of the included studies were assessed by
two authors (CY and CH), independently according to the
Newcastle-Ottawa Scale (NOS) (28)
and Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2)
(29). Low-quality studies with
scores of 4 or lower were excluded. Additionally, the risk of bias
was calculated according to criteria from the Cochrane
Collaboration's tool (Cochrane Handbook for Systematic Reviews of
Interventions Version 5.1.0.).
Statistical analysis
The systematic search was performed using Review
Manager Software version 5.2 (The Nordic Cochrane Centre,
Copenhagen, Denmark). The RevMan version 5.2 was used to combine
data (free software was downloaded from http://www.cochrane.org, The Cochrane Collaboration,
The Nordic Cochrane Centre, Copenhagen, Denmark, 2012). Data were
presented as means ± standard deviation (SD) or median (range), and
a description of qualitative variables as number and percentage.
HWE was checked by χ² test (30).
The results of the meta-analysis were reported as odds ratios (ORs)
with 95% confidence intervals (CIs). The ORs were pooled for each
allele comparison in the three models, the dominant model, the
recessive model and heterozygote model. The Chi-square-based Q-test
was applied to testify between-study heterogeneity. Hazard ratio
(HR) was calculated by the fixed effects model when P
(heterogeneity) >0.05, as a secondary analysis. Otherwise, the
random-effects model was used. According to our hypothesis and
inclusion criteria, HR >1 implies poor prognosis for any IL
polymorphisms. We used pooled HRs with 95% CIs to find the
relationship between IL single nucleotide polymorphisms (SNPs) and
MM susceptibility. The Hazard ratios and 95% CIs were calculated by
Tierney's method if the data were not reported in the original
paper. Furthermore, subgroup analysis was conducted for the type of
each allele polymorphisms and ethnicity, genotyping methods, IL
type and reference control. Publication bias was evaluated by
Begg's funnel plots and Egger's regression test (31). A value of ‘Pr>|z|’ less than 0.05
was considered as potential publication bias (32). Moreover, we also conducted a
sensitivity analysis by precluding a single study to observe
whether the pooled HRs changed. Statistical analyses were conducted
using Meta-DiSc version 1.4 and R software Packages (version
3.3.1), included ‘mada’ (for sensitivity and specificity analysis).
P<0.05 and I2 >50% was considered as statistically
significant.
Results
Study selection
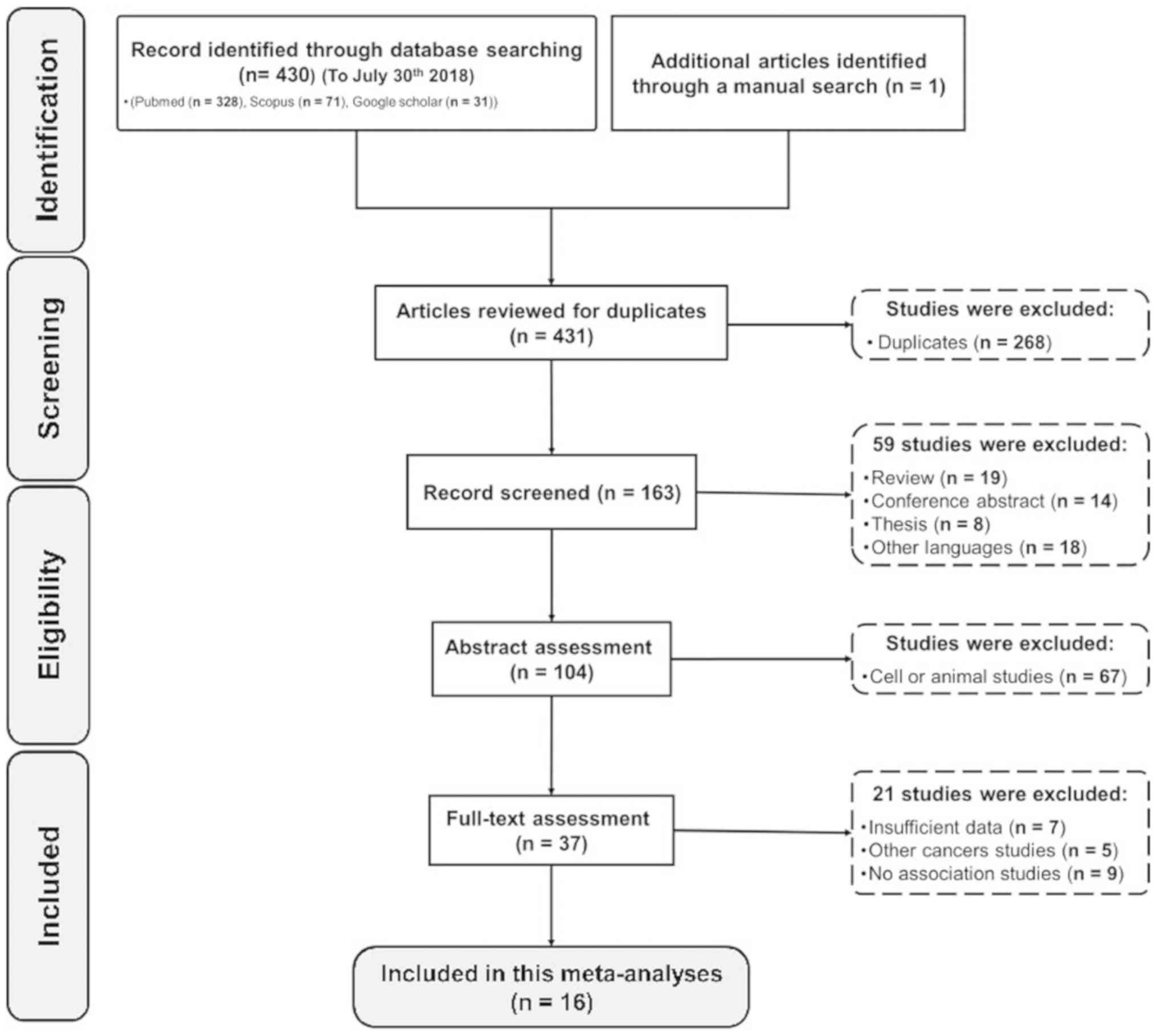
A detailed PRISMA flowchart of the study
identification, screening and exclusion process are shown in
Fig. 1. A total of 431 studies were
retrieved by database searching, with 340 potentially eligible
studies that met the inclusion and exclusion criteria and 1 record
attained through manual search. By screening the titles, 268
studies were excluded due to being duplicated. After carefully
reviewing abstracts, 126 studies were excluded including conference
studies, review articles, thesis compilations, those that were not
either in English or Chinese, cell culture and animal studies data.
Of the remaining 37 full-text candidate articles, 21 potential
studies were excluded involving insufficient data, other cancer
studies and unrelated studies. Finally, 16 studies were selected
and presented in the current meta-analysis that attempts to find a
relationship between interleukin polymorphism and risk of MM.
Study and SNP characteristics
The main characteristics of the included studies are
shown in Table I. The published
dates of all research studies varied from 2000 to 2018. Most of the
research was conducted within Caucasian communities: 10 studies in
Caucasian populations (62.5%) (20,22,23,25,33-38), 3 within Asian
populations (18.75%) (17,39,40) and
3 in Mixed Afro-American populations (18.75%) (24,41,42). Of
note, the quantitative TaqMan method (TaqMan PCR) was often used to
detect the SNP state with control references (11 studies, 68.75%)
(20,22-25,36-39,41,42).
Also, polymerase chain reaction-restriction fragment length
polymorphism (PCR-RFLP) method was used in 5 studies (31.25%)
(17,33-35,40).
The variables from sixteen relevant case-control studies were
contained 2,597 cases and 3,851 controls. Source controls were
mainly selected by population-based method with age/sex-matched (12
studies, 75%) (17,20,23-25,33,34,36,38,40-42).
Regarding the NOS methodological quality, all included studies were
of high quality with ≥7 out of 10 (mean 8.02 point).
 | Table ISummary of included articles. |
Table I
Summary of included articles.
| First author | Year | Population
(ethnicity) | Genotyping
method | Sample size
(case/control) | Source of
control | Sex | NOS score | (Refs.) |
|---|
| Zheng | 2000 | Sweden (C) | PCR-RFLP | 73/129 | PB | Not matched | 7 | (33) |
| Zheng | 2001 | Sweden (C) | PCR-RFLP | 73/109 | PB | Not matched | 7 | (34) |
| Mazur | 2005 | Poland (C) | Taq-PCR | 54/50 | PB | Matched | 8 | (23) |
| Cozen | 2006 | USA (C, M) | Taq-PCR | 146/125 | PB | Matched | 8 | (24) |
| Duch | 2007 | Brazil (C, M) | Taq-PCR | 52/60 | PB | Matched | 9 | (41) |
|
Abazis-Stamboulieh | 2007 | Greece (C) | Taq-PCR | 74/160 | HB | Not matched | 8 | (22) |
| Aladzsity | 2009 | Hungary (C) | Taq-PCR and
PCR-RFLP | 100/99 | | | 8 | (35) |
| Birmann | 2009 | USA (C) | Taq-PCR | 82/164 | PB | Not matched | 8 | (20) |
| Martino | 2012 | Italy and Germany
(C) | Taq-PCR | 201/234 | PB | Matched | 9 | (38) |
| Vangsted | 2012 | Denmark (C) | Taq-PCR | 348/1,700 | PB | Matched | 8 | (36) |
| Stephens | 2012 | USA (C, M) | Taq-PCR | 626/44 | PB | Not matched | 8 | (42) |
| Iakupova | 2003 | Russia (C) | Taq-PCR | 69/102 | PB | Not matched | 9 | (25) |
| Chakraborty | 2017 | India (A) | Taq-PCR | 103/117 | HB | Matched | 8 | (39) |
| Kasamatsu | 2017 | Japan (A) | PCR-RFLP | 128/202 | PB | Matched | 9 | (40) |
| Nielsen | 2017 | Denmark (C) | Taq-PCR | 348/355 | HB | Not matched | 7 | (37) |
| Kasamatsu | 2018 | Japan (A) | PCR-RFLP | 120/201 | PB | Matched | 8 | (17) |
The genotype susceptibility of MM and interleukin
family gene polymorphisms are shown in Table SI. In Total, 16
published studies had detected interleukin polymorphism of IL-1β,
IL-1α, IL-4, IL-6, IL-6R, IL-10, IL-10R, IL-17 and IL-23 in the
prediction of MM patients. In more details, 2 studies had
identified the relationship between the SNP of IL-1α/β (22,36), one
study of IL-4 37, ten studies of IL-6 and IL-6R (20,22-25,34,37-39,42),
four studies of IL-10 and IL-10R (17,23,33,37),
three studies of (IL-17 39-41) and one IL-23R polymorphisms 17. The
majority of SNP associations were reported by Birmann et al
(20). IL-6 promoter was the most
frequently reported (11 of 16 datasets; 68.75%) involving 1,854 MM
patients and 1,479 controls in 5 different SNPs. The IL-6 promoter
rs1800795 (174G>C) was the most frequently reported SNP of the
IL-6 promoter (9 of all SNPs; 81.82%). Furthermore, A:G (10 of 33
SNPs; 30.3%) and C:T (11 of 33 SNPS; 33.4%) substations are the
most reported SNP allies in this meta-analysis. The genotypic
frequencies of the controls in these 16 studies were all consistent
with the HWE (Table SI).
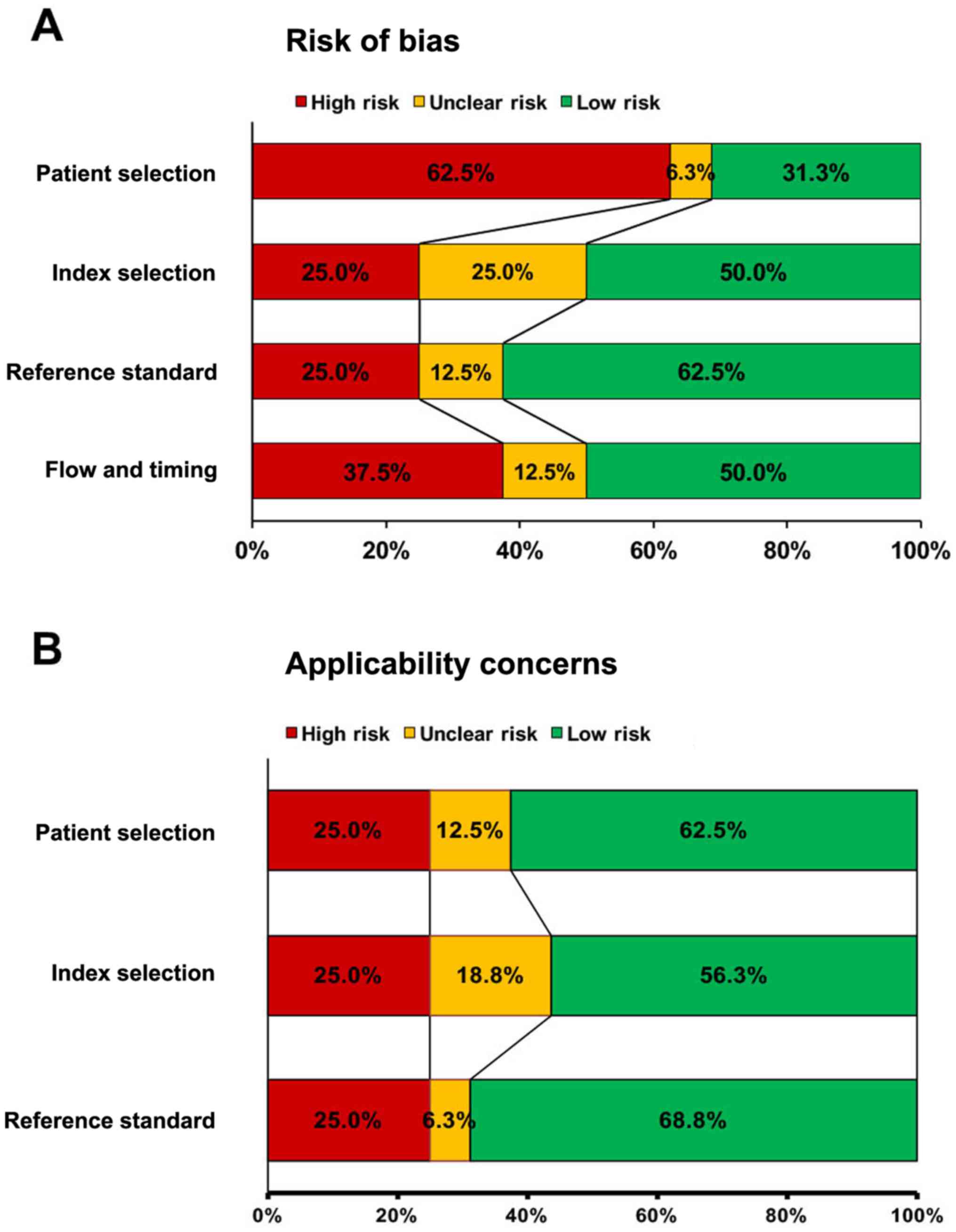
Quality assessment
All 16 selected papers were methodologically assayed
by NOS and QUADAS-2 quality evaluation standards of the Cochrane
Reviewers' Handbook. The detailed quality assessment of eligible
studies, according to the NOS score, was summarized in Table SII.
Overall, all studies included in the current meta-analysis were
judged to be at moderate to high risk of bias, with scores ≥7
points (Table SII). The average NOS score was 8.01 out of 10, that
was relatively classified in the high quality. Many studies
provided sufficient information about study design and execution.
Also, QUADAS-2 results confirmed that significant bias were not
present in this meta-analysis. Fig.
2 shows all parameters of QUADAS-2 assessment, regarding bias
risk and applicability concerns. Most studies had an acceptable
range with regard to completeness of outcome data (attrition bias)
and other sources of bias. More than half of the included studies
were rated as low risk for most parameters of the bias risk
(48.84%) and applicability concerns (62.5%). As shown in Fig. 2, no signification bias (Fig. 2A) and applicability concerns
(Fig. 2B) were found in any of the
selected studies.
The outcome of the meta-analysis
The present meta-analysis was performed in the both
homozygous and heterozygous allele genetic model. Based on our
systematic approach, we tried to find the associations between the
MM risk and SNP of G:A, G:C and T:C in IL-1β, IL-1α, IL-4, IL-6,
IL-6R, IL-10, IL-10R, IL-17 and IL-23 polymorphisms. Also, the
association between the type of each allele polymorphisms of G:A,
G:C, T:C and ethnicity, genotyping methods, IL type and control
reference were measured as subgroup analysis.
G:A polymorphisms and MM
susceptibility
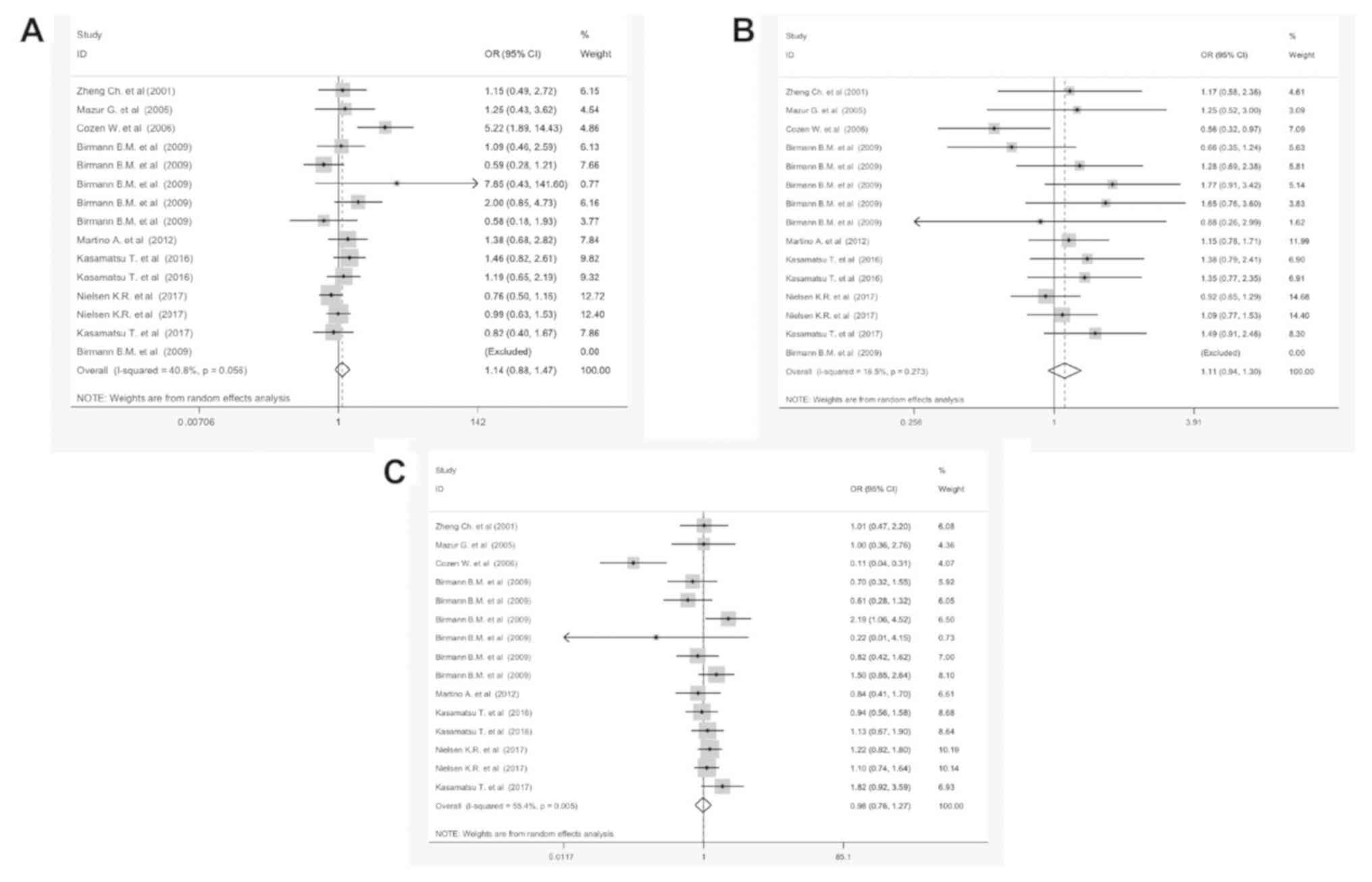
Table II shows the
results of the meta-analysis for G:A and MM in the three different
genotypes GG vs. AA, GG vs. AG and AA vs. GA. The combined analysis
of 14 studies indicated that GG/AA polymorphism was associated with
a statistically significant improvement of 40.8% in MM, when
compared with the control group (OR=1.14, 95% CI, 0.88-1.47,
P<0.05); suggesting that the over-expression of GG/AA
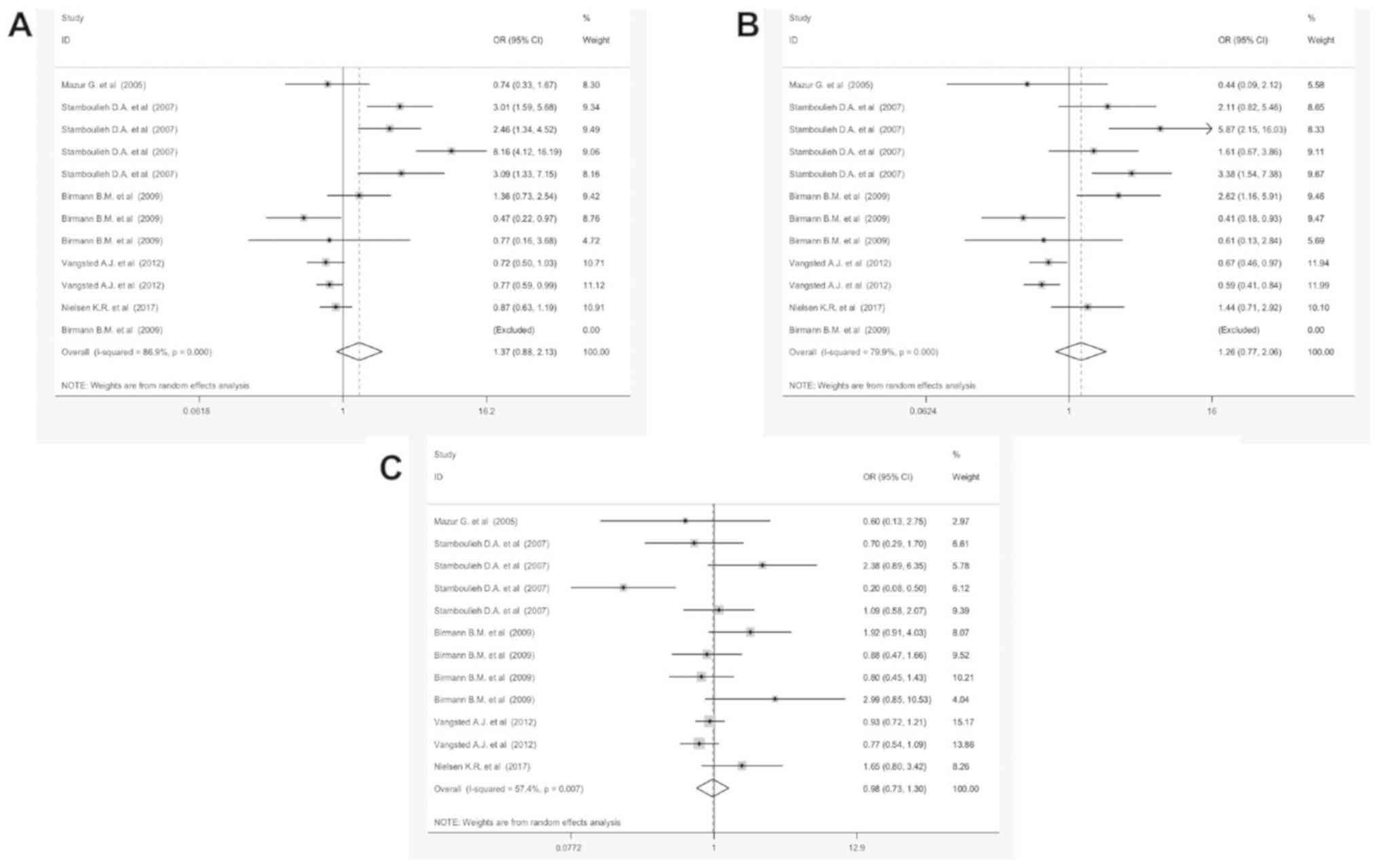
polymorphism is a prognostic factor for MM (Fig. 3A). Also, the subtotal OR of GG/AG and
AA/GA were 1.18 [95% CI, 0.94-1.3; P=0.27 (Fig. 3B)] and 0.98 [95% CI, 0.76-1.27;
P=0.005 (Fig. 3C)], respectively. No
significant coloration was found between IL-17Ars2275913 and
IL-10Rαrs2228055 polymorphism (OR=0.64, 95% CI, 0.48-1.33, P=0.26
and OR=0.72, 95% CI, 0.62-1.83, P=0.43, respectively). Strikingly,
the OR of GG/AA was notably different compared with other
polymorphisms. Subgroup analyses was conducted according to
ethnicity, genotyping methods, IL type and control reference (Fig.
S1). Furthermore, the G:A polymorphism detected in the IL-6
promoter (OR=1.05, 95% CI, 0.78-1.44) is more accurate in MM
samples of the Asian population (OR=1.24, 95% CI, 0.92-1.74). The
PCR-RFLP based methods are more valuable methods for the detection
of the G:A polymorphism in MM samples (OR=1.18, 95% CI, 0.94-1.62,
P=0.002).
 | Table IIMeta-analysis results for G:A gene
polymorphism. |
Table II
Meta-analysis results for G:A gene
polymorphism.
| Allele | Subgroup | Number study | OR | 95% CI | P-value |
|---|
| GG vs. AA | Ethnicity | (C) | 10 | 0.98 | (0.77-1.25) | 0.352 |
| | | (A) | 3 | 1.17 | (0.82-1.68) | 0.465 |
| | Method | Taq | 10 | 1.17 | (0.80-1.70) | 0.017 |
| | | PCR | 4 | 1.17 | (0.84-1.63) | 0.675 |
| | Source | PB | 12 | 1.25 | (0.92-1.71) | 0.083 |
| | | HB | 2 | 0.86 | (0.63-1.17) | 0.398 |
| | Gene | Interleukin-6
promoter | 4 | 1.43 | (0.69-2.97) | 0.006 |
| | | Interleukin-6
receptor | 3 | 1.37 | (0.42-4.50) | 0.038 |
| | | Interleukin-10 | 3 | 1.04 | (0.72-1.50) | 0.893 |
| | | Interleukin-10
receptor | 2 | 1.33 | (0.87-2.02) | 0.631 |
| | Overall | | 14 | 1.14 | (0.88-1.47) | 0.056 |
| GG vs. AG | Ethnicity | (C) | 10 | 1.09 | (0.92-1.29) | 0.626 |
| | | (A) | 3 | 1.41 | (1.03-1.92) | 0.960 |
| | Method | Taq | 10 | 1.03 | (0.84-1.26) | 0.202 |
| | | PCR | 4 | 1.37 | (1.03-1.81) | 0.956 |
| | Source | PB | 12 | 1.16 | (0.95-1.41) | 0.223 |
| | | HB | 2 | 1.00 | (0.78-1.27) | 0.504 |
| | Gene | Interleukin-6
promoter | 4 | 0.84 | (0.62-1.14) | 0.150 |
| | | Interleukin-6
receptor | 3 | 1.53 | (1.03-2.26) | 0.767 |
| | | Interleukin-10 | 3 | 1.12 | (0.83-1.50) | 0.949 |
| | | Interleukin-10
receptor | 2 | 1.36 | (0.92-2.02) | 0.949 |
| | Overall | | 14 | 1.11 | (0.94-1.30) | 0.273 |
| GA vs. AA | Ethnicity | (C) | 11 | 1.08 | (0.88-1.32) | 0.373 |
| | | (A) | 3 | 1.18 | (0.83-1.67) | 0.318 |
| | Method | Taq | 11 | 0.89 | (0.62-1.26) | 0.002 |
| | | PCR | 4 | 1.15 | (0.85-1.55) | 0.492 |
| | Source | PB | 13 | 0.92 | (0.66-1.29) | 0.002 |
| | | HB | 2 | 1.16 | (0.88-1.53) | 0.732 |
| | Gene | Interleukin-6
promoter | 5 | 0.60 | (0.30-1.180) | 0.001 |
| | | Interleukin-6
receptor | 3 | 1.14 | (0.50-2.90) | 0.077 |
| | | Interleukin-10 | 3 | 1.07 | (0.77-1.50) | 0.972 |
| | | Interleukin-10
receptor | 2 | 1.03 | (0.71-1.49) | 0.631 |
| | Overall | | 15 | 0.98 | (0.76-1.27) | 0.005 |
G:C polymorphisms and MM
susceptibility
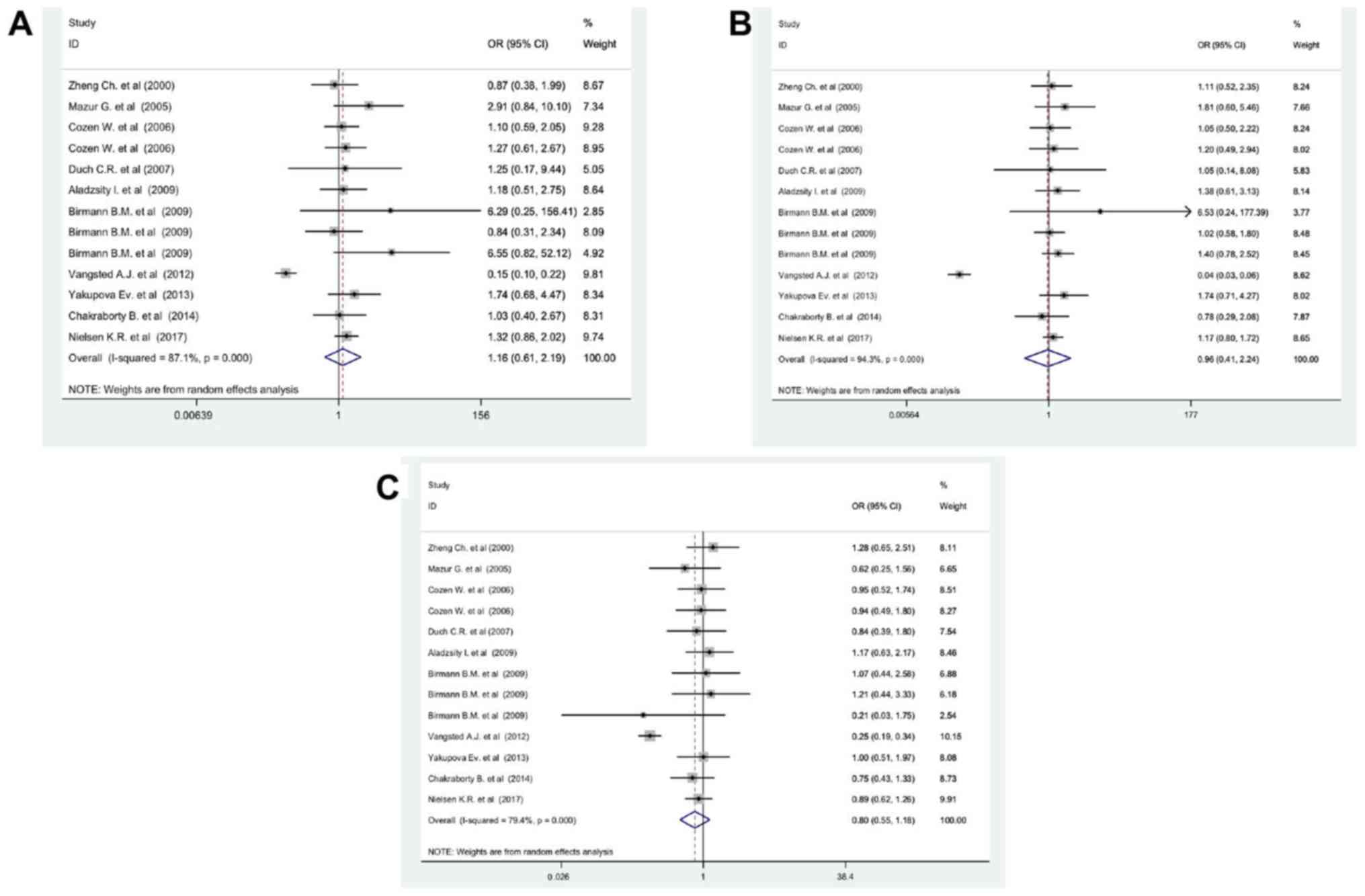
As shown in Fig. 4,
based on heterogeneity, the dominant model GG/CC has an appropriate
effect model in the detection of the MM. The results in Fig. 4 clearly show that the GG genotype for
the IL-6 promoter and IL-1β would increase MM risk by approximately
81% compared with the CC genotype. Correspondingly, G:C
polymorphism for IL-1β1464G>C and IL-6572G>C would increase
MM risk (Table III). Meanwhile,
the sub-analysis findings suggest that hospital-based samples were
more accurate in the detection of the G:C polymorphism in MM
patients (OR=1.02, 95% CI, 0.62-1.42, P=0.001; Fig. S2). Although,
other subgroup analyses such as ethnicity, genotyping methods and
IL type did not have any significant effect on MM prognosis and
therefore they were ignored (Fig. S2).
 | Table IIIMeta-analysis results for G:C gene
polymorphism. |
Table III
Meta-analysis results for G:C gene
polymorphism.
| Allele | Subgroup | Number study | OR | 95% CI | P-value |
|---|
| GG vs. CC | Ethnicity | (C) | 9 | 1.21 | (0.50-2.93) | 0.001 |
| | | (C, M) | 3 | 1.17 | (0.74-1.86) | 0.96 |
| | Method | Taq | 11 | 1.21 | (0.57-2.57) | 0.001 |
| | Source | PB | 10 | 1.19 | (0.51-2.80) | 0.001 |
| | | HB | 3 | 1.25 | (0.88-1.78) | 0.89 |
| | Gene | Interleukin-6
promoter | 11 | 1.24 | (0.97-1.58) | 0.9 |
| | Overall | | 13 | 1.16 | (0.61-2.19) | >0.001 |
| GC vs. CC | Ethnicity | (C) | 9 | 0.96 | (0.31-2.92) | 0.001 |
| | | (C, M) | 3 | 1.11 | (0.63-1.92) | 0.97 |
| | Method | Taq | 11 | 0.92 | (0.34-2.47) | 0.001 |
| | Source | PB | 10 | 0.94 | (0.30-2.93) | 0.001 |
| | | HB | 3 | 1.15 | (0.83-1.59) | 0.67 |
| | Gene | Interleukin-6
promoter | 11 | 1.18 | (0.94-1.48) | 0.97 |
| | Overall | | 13 | 0.96 | (0.41-2.24) | >0.001 |
| GG vs. GC | Ethnicity | (C) | 9 | 0.77 | (0.45-1.33) | 0.001 |
| | | (C, M) | 3 | 0.92 | (0.63-1.34) | 0.96 |
| | Method | Taq | 11 | 0.74 | (0.48-1.13) | 0.001 |
| | Source | PB | 10 | 0.76 | (0.46-1.28) | 0.001 |
| | | HB | 3 | 0.90 | (0.69-1.18) | 0.58 |
| | Gene | Interleukin-6
promoter | 11 | 0.94 | (0.78-1.13) | 0.98 |
| | Overall | | 13 | 0.81 | (0.55-1.18) | >0.001 |
T:C polymorphisms and MM
susceptibility
Data from 12 studies on IL-1, -6 and -10
polymorphisms were pooled and analyzed for the detection of the T:C
polymorphisms and MM susceptibility (Table IV). A high risk of MM was found in
the heterozygous model (Fig. 5A) and
dominant model (Fig. 5B) of the T:C
polymorphisms (TC/CC: OR=1.37, 95% CI, 0.88-2.16, P=0.001 and
TT/CC: OR=1.26, 95% CI, 0.77-2.06, P=0.007). Moreover, sub-analysis
results of the T:C polymorphisms revealed a significant correlation
between IL-1β polymorphism (OR=1.18, 95% CI, 0.66-1.89, P=0.005) in
hospital-based source samples (OR=1.82, 95% CI, 0.54-2.06, P=0.073)
and MM risk (Fig. S3). There was no significant correlation between
other subgroup analysis and MM risk in the T:C polymorphisms (data
not shown).
 | Table IVMeta-analysis results for G:C gene
polymorphism. |
Table IV
Meta-analysis results for G:C gene
polymorphism.
| Allele | Subgroup | Number study | OR | 95% CI | P-value |
|---|
| TT vs. CC | Source | PB | 7 | 0.95 | (0.74-1.23) | 0.19 |
| | | HB | 5 | 0.92 | (0.43-1.99) | 0.002 |
| | Gene | Interleukin-1β | 4 | 0.91 | (0.67-1.25) | 0.174 |
| | | Interleukin-1α | 2 | 0.49 | (0.09-2.58) | 0.003 |
| | | Interleukin-6
receptor | 3 | 1.06 | (0.64-1.75) | 0.161 |
| | | Interleukin-10 | 2 | 1.26 | (0.52-3.04) | 0.280 |
| | Overall | | 12 | 0.98 | (0.73-1.30) | 0.007 |
| TT vs. TC | Source | PB | 6 | 0.72 | (0.46-1.13) | 0.025 |
| | | HB | 5 | 2.39 | (1.46-3.90) | 0.157 |
| | Gene | Interleukin-1β | 4 | 1.28 | (0.59-2.75) | 0.001 |
| | | Interleukin-1α | 2 | 2.39 | (1.16-4.95) | 0.214 |
| | | Interleukin-6
receptor | 3 | 0.90 | (0.24-3.33) | 0.006 |
| | | Interleukin-10 | 2 | 0.99 | (0.34-2.89) | 0.180 |
| | Overall | | 12 | 1.26 | (0.77-2.06) | 0.001 |
| TC vs. CC | Source | PB | 6 | 0.768 | (0.63-0.93) | 0.391 |
| | | HB | 5 | 2.693 | (1.15-6.33) | 0.001 |
| | Gene | Interleukin-1β | 4 | 1.342 | (0.71-2.54) | 0.001 |
| | | Interleukin-1α | 2 | 5.181 | (1.99-13.43) | 0.078 |
| | | Interleukin-6
receptor | 3 | 0.81 | (0.37-1.78) | 0.090 |
| | | Interleukin-10 | 2 | 0.851 | (0.634-1.14) | 0.001 |
| | Overall | | 11 | 1.37 | (0.88-2.13) | 0.001 |
C:A polymorphisms and MM
susceptibility
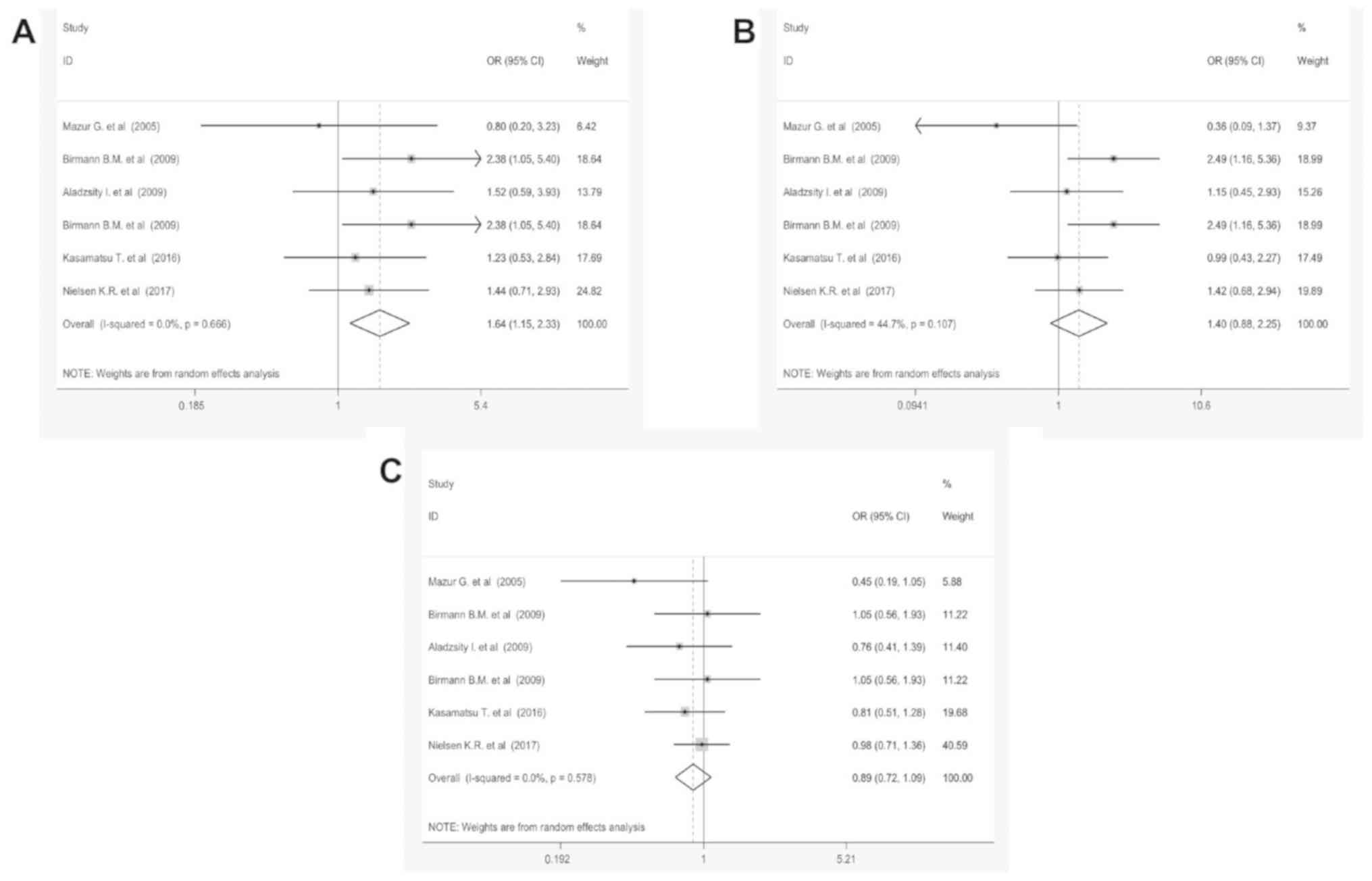
Table V and Fig. 6 demonstrate the meta-analysis results
of C:A polymorphisms. No significant association was identified
between the overall MM risk and three polymorphism models, CC/AA,
CC/AC and AC/AA in IL-6 (rs8192284) and IL-10 (rs1800872) receptors
(P>0.05). In the stratified analyses by ethnicity, genotyping
methods, IL type and control reference no significant association
were detected between C/A polymorphism and MM (Fig. S4). In the
overall analysis, no significant association was detected between
T:A and T:G polymorphism and MM under all three genetic models
(data not shown).
 | Table VMeta-analysis results for C:A gene
polymorphism. |
Table V
Meta-analysis results for C:A gene
polymorphism.
| Allele | Subgroup | Number study | OR | 95% CI | P-value |
|---|
| CC vs. AA | Source | PB | 4 | 1.79 | (1.17-2.75) | 0.462 |
| | | HB | 2 | 1.35 | (0.72-2.53) | 0.743 |
| | Gene | Interleukin-6
receptor | 3 | 2.11 | (1.29-3.46) | 0.730 |
| | | Interleukin-10 | 3 | 1.26 | (0.76-2.10) | 0.759 |
| | Ethnicity | (C) | 5 | 1.74 | (1.18-2.57) | 0.614 |
| | Method | Taq | 4 | 1.80 | (1.17-2.75) | 0.462 |
| | Overall | | 6 | 1.64 | (1.15-2.33) | 0.666 |
| CC vs. AC | Source | PB | 4 | 1.56 | (0.80-3.04) | 0.062 |
| | | HB | 2 | 1.06 | (0.57-1.97) | 0.816 |
| | Gene | Interleukin-6
receptor | 3 | 2.05 | (1.28-3.28) | 0.373 |
| | | Interleukin-10 | 3 | 0.95 | (0.49-1.84) | 0.211 |
| | Ethnicity | (C) | 5 | 1.50 | (0.86-2.59) | 0.094 |
| | Method | Taq | 4 | 1.56s | (0.80-3.04) | 0.062 |
| | Overall | | 6 | 1.40 | (0.87-2.25) | 0.107 |
| AC vs. AA | Source | PB | 4 | 0.93 | (0.72-1.21) | 0.362 |
| | | HB | 2 | 0.79 | (0.54-1.14) | 0.869 |
| | Gene | Interleukin-6
receptor | 3 | 0.94 | (0.66-1.34) | 0.7 |
| | | Interleukin-10 | 3 | 0.82 | (0.59-1.16) | 0.228 |
| | Ethnicity | (C) | 5 | 0.91 | (0.72-1.14) | 0.463 |
| | Method | Taq | 4 | 0.93 | (0.72-1.21) | 0.362 |
| | Overall | | 6 | 0.89 | (0.72-1.09) | 0.578 |
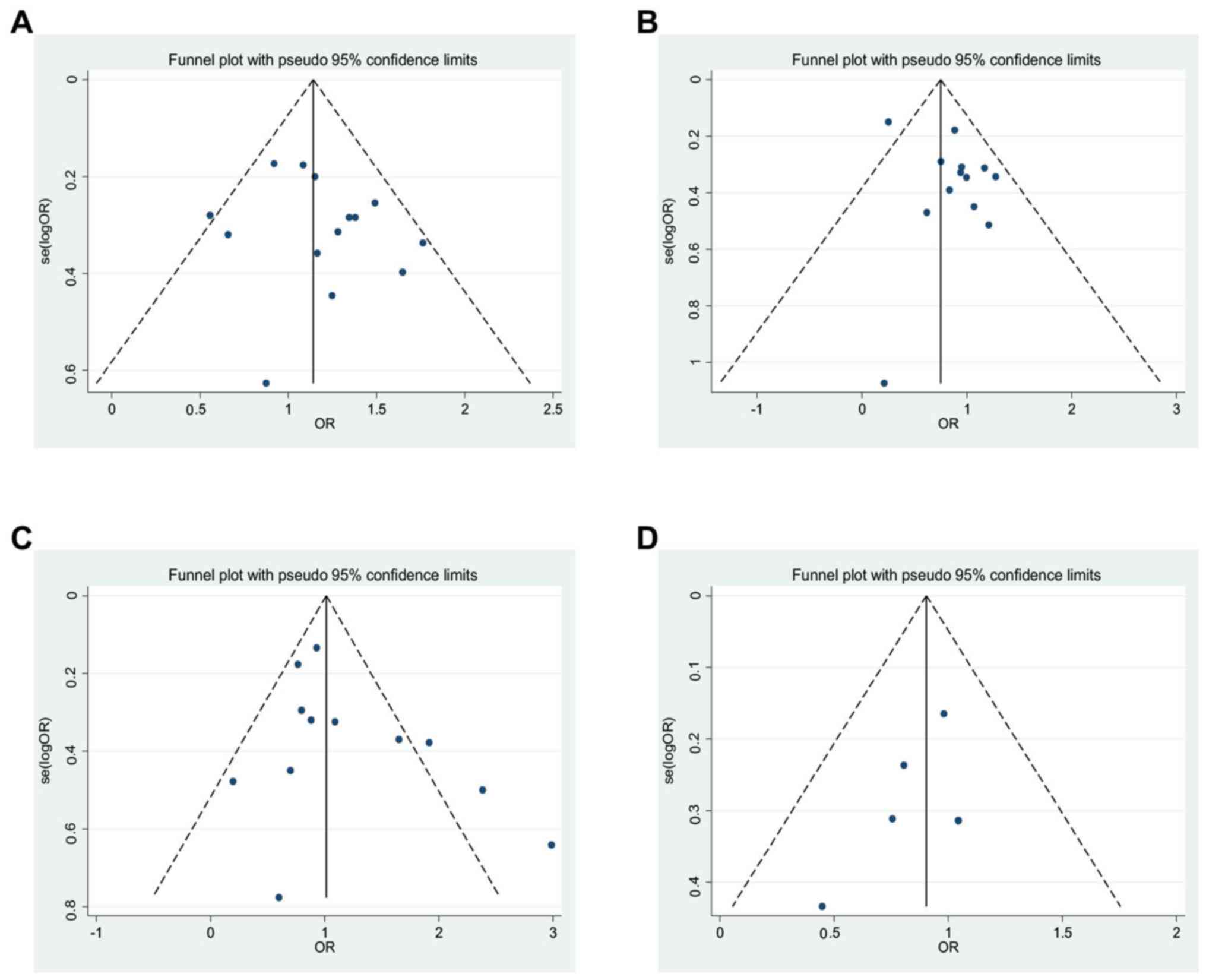
Publication bias
Begg's and Egger's tests were used to estimate the
publication bias of each allele polymorphism. The analysis was
carried out by precluding a single study at a time (Fig. 7) (43). The shape of funnel plot and Egger's
test provided no statistical evidence for publication bias of the
G:A [t=-0.92, P=0.38, 13 study (Fig.
7A)], G:C [t=-2.02, P=0.0.069, 14 study (Fig. 7B)], T:C [t=-1.51, P=0.162, 12 study
(Fig. 7C)] and G:A [t=-1.23, P=0.31,
5 study (Fig. 7D)]. Likewise, Table
SIII shows detailed publication bias of all investigated allele in
three different models. P-values were revealed in Table SIII and no
publication bias was found. Hence, there is no noticeable evidence
for significant publication bias in our meta-analysis, which
signifies that our meta-analysis results were stable and
credible.
Discussion
To the best of our knowledge, this is the first
comprehensive systematic review and meta-analysis study that has
been conducted to identify the prognostics accuracy of all
interleukin family gene polymorphisms in advanced MM patients. Data
were extracted from a total of 16 publications that contained
individuals carrying the GG genotype for IL-6 and IL-1 with an
increased risk of MM by approximately 40.8 and 80.2% compared with
the AA and CC genotypes. The results implied that a high MM risk
was similar for IL-1 as well as IL-6; and revealed a significant
association between their T:C polymorphism and MM risk.
Furthermore, this study showed that patients with the heterozygous
and dominant genotype of the T:C polymorphism have the highest MM
risk. Notably, no significant association was found between the MM
risk and C:A polymorphism in the IL-6 (rs8192284) and IL-10
(rs1800872) receptors (Fig. 6).
Furthermore, hospital-based samples detected by PCR-RFLP methods
are more attributable in identifying the interleukin family gene
polymorphisms in MM patients. Keeping in mind the mentioned
background, we tried to find significant correlations between
ethnicity, IL type and MM sensitivity. We examined an overall
sub-analysis of 2,597 patients and ORs for MM. Our results
indicated that there was no significant association between IL gene
polymorphism of the myeloma samples, ordered by ethnicity and type
of IL family; which adversely affect cancer survival. Remarkably,
we could not find any evidence about MM samples associated with sex
and IL type. Our results clearly show that GG genotype for IL-6
promoter and IL-1β would increase MM risk by approximately 81%
compared with the CC genotype (R=1.02, 95% CI, 0.62-1.42,
P=0.001).
It is well established that interleukin family
polymorphisms may be a prognostic factor for survival in MM
patients (20-23). Single-nucleotide polymorphisms in the promoter
region of IL-1, -6 and -10 are associated with multiple
inflammatory and immune-mediated diseases, including cancer,
Crohn's disease, ulcerative colitis and type 1 diabetes (44,45). As
a major pro-inflammatory cytokine, IL-6 and IL-1 could mediate
chemokine, adhesion molecule expression, recruit monocytes and
macrophages to release a large number of growth factors and
cytokines (20). Many studies show
that overexpression of the pro-inflammatory interleukin is a
significant prognostic biomarker in patients with MM (25). In the present meta-analysis, the G:C
polymorphisms (mainly GG genotype) in IL-6-572G>C and
IL-6-174G>C are associated with an increased susceptibility to
MM. Therefore, both tagging SNPs of IL-6rs1800795 and IL-6rs1800797
are most reliable and could be used for the prediction of MM
progression (23,24). These SNPs located in the promoter
region of the IL-6 gene may regulate IL-6 production and correlate
with the susceptibility of some other cancers (23,25).
Recently, Li et al, (2017) reported that IL-6 may be a
potential biomarker for MM diagnosis (18). It suggested that IL-6 SNPs are very
useful prognostic biomarkers in clinical usage and could be truly
specific in prognosis of a particular cancer type (18). These results are completely
consistent with other investigations (46-48).
IL-1α and IL-1β, two members of IL-1, have a common
receptor with shared SNPs (IL-1α-889C>T and IL-1β-3737C>T)
(49). Our results show that C:T
polymorphisms of the IL-1α-889C>T and IL-1β-3737C>T were
associated with MM risk. IL-10 is one of the well-investigated
pro-inflammatory cytokines (34). It
is documented that IL-10-592G/A and IL-10-1,082G>A SNPs are
located in the negative and positive regulatory sequence of the
promoter region, respectively (34).
The IL-10 gene with -592 (C) or -1,082 (G) is associated with a
high expression of IL-10, and it has a low expression with -592 (A)
or -1,082 (A). Our results indicate that the C allele of IL-10-592
has no significant association with MM susceptibility under the
three allele model, CC/AA, CC/AC and AC/AA18. In
contrast, our cumulative results indicate that IL-10-1082G>A is
found to be associated with MM risk (R=1.18, 95% CI, 0.94-1.3).
Recently, Sultana et al, (2018) (48) indicate that the C allele of IL-1α-889
is higher in MM patients than in controls. Similarly, Ziakas et
al, (2013) (50) show that the G
allele of IL-1β-1464G>C was significantly associated with MM in
an Asian population. Of course, the discrepancies between the
mentioned studies may also be due to small sample size and ethnic
differences.
Despite these competent studies, the conventional
formation and balancing of interleukin family gene polymorphisms in
the interconnecting angiogenesis of the human MM neoplasms are not
well-defined yet (45). Location
variety and heterogenic morphology of MM tumors that have a close
relationship with the interleukin family SNPs, represented
noteworthy challenge for oncologist. With these presuppositions,
the present investigation allows us to acquire a better
understanding of the prognostic role of interleukin family gene
SNPs in MM patients by using statistical approaches. Conversely,
the correlation between the interleukin gene SNPs and the survival
of cancer patients are controversial or inconclusive. On the other
hand, several published meta-analyses have been concerned with the
evaluation of the dissimilarity in interleukin family gene
polymorphisms related to the prognosis of cancer (20-25,51).
The overall findings help us describe the possible prognostic role
of the interleukin polymorphism in MM progress, and identify a
novel therapeutic target of MM progression. We tried to include all
the possible literature on the relationship between interleukin
polymorphism and MM risk and to obtain the maximum sample size and
comprehensive results. Although, it should be noted that many
future studies are needed for the description of the molecular
mechanisms, because at the moment the published proof is scant.
Unavoidably, there are still some limitations: i) A few studies and
a small sample size have demonstrated correlations between
interleukin gene family polymorphism and MM prognosis; ii) we only
included the papers in English language, while published papers in
other languages, especially Chinese and Russian were ignored; iii)
poor homogeneous distribution and population based on subgroup
parameters existed in some studies; iv) the occurrence and
development of MM are on the basis of many hereditary and
environmental factors as well as the interaction between them, but
these factors and the pathological states of MM were not considered
in this meta-analysis; and v) the low quality of some included
studies led to meta-analysis results that were based on unadjusted
estimates, because some studies did not provide detailed
information to calculate the adjusted estimates. Besides, a lot of
confounding factors were not controlled or reported in biased
statistical results. We like to point out that future well-accepted
clinical studies with larger samples sizes, systematized protocols
and more homogenized populations would be needed to fully
investigate the prognostic potential of each SNP in interleukin
gene family of MM patients.
In conclusion, the present meta-analysis shows that
G:C polymorphism of the IL-1β1464G>C and IL-6572G>C would
increase MM risk. However, it has been determined that there is a
significant association between MM risk and C:T polymorphism in
IL-1α-889C>T and IL-1β-3737C>T. Additionally, hospital-based
source samples are more accurate and attributable in the detection
of interleukin family gene polymorphisms in Asian patients with MM,
by using the PCR-RFLP method. These SNP loci may be appropriate
biomarkers for gene screening and an effective adjuvant therapeutic
strategy in prognostics of MM patients.
Acknowledgements
Not applicable.
Funding
This study was supported in part by National Natural
Science and Technology Fund Project (grant no. 81450030) and
Sichuan Provincial Department of Education major training project
(grant no. 14CZ0017).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CH and MNS were in charge of the idea and study
designation. II, SN, and CY searched and collected data. FL and SL
performed data analysis. MNS and II wrote the manuscript and were
in charge of language revision. SN edited the paper. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present meta-analysis was approved by an
independent Ethics Committee/institutional review board at
Southwest Medical University, Department of Hematology (Luzhou,
China). This investigation was carried out by following
recommendations of the Preferred Reporting Items for Systematic
Reviews and Meta-Analysis (PRISMA).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Shi H, Chen Z, Xie J and Chen N: The
prevalence and management of multiple myeloma-induced kidney
disease in China. Kidney Dis (Basel). 1:235–40. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Holstein SA and McCarthy PL:
Immunomodulatory drugs in multiple myeloma: Mechanisms of action
and clinical experience. Drugs. 77:505–520. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Huang QC and Ding JY: Research advance in
light Chain escape of multiple myeloma-Review. Zhongguo Shi Yan Xue
Ye Xue Za Zhi. 25:1833–1836. 2017.(In Chinese). PubMed/NCBI View Article : Google Scholar
|
|
5
|
T V, V G and A ND: Multiple myeloma index
for risk of infection. J Cancer. 9:2211–2214. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen YF and Lu YL: Survival and prognosis
analysis of 57 patients with multiple myeloma. Zhongguo Shi Yan Xue
Ye Xue Za Zhi. 25:1436–1443. 2017.(In Chinese). PubMed/NCBI View Article : Google Scholar
|
|
7
|
Curado MP, Oliveira MM, Silva DRM and
Souza DLB: Epidemiology of multiple myeloma in 17 Latin American
countries: An update. Cancer Med. 7:2101–2108. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tang CH, Liu HY, Hou HA, Qiu H, Huang KC,
Siggins S, Rothwell LA and Liu Y: Epidemiology of multiple myeloma
in Taiwan, a population based study. Cancer Epidemiol. 55:136–141.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Soekojo CY, de Mel S, Ooi M, Yan B and
Chng WJ: Potential clinical application of genomics in multiple
myeloma. Int J Mol Sci. 9(pii: E1721)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Willenbacher W, Seeber A, Steiner N,
Willenbacher E, Gatalica Z, Swensen J, Kimbrough J and Vranic S:
Towards Molecular profiling in multiple myeloma: A literature
review and early indications of its efficacy for informing
treatment strategies. Int J Mol Sci. 19(pii: E2087)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jupe S, Ray K, Roca CD, Varusai T,
Shamovsky V, Stein L, D'Eustachio P and Hermjakob H: Interleukins
and their signaling pathways in the Reactome biological pathway
database. J Allergy Clin Immunol. 141:1411–1416. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yasui H, Hideshima T, Richardson PG and
Anderson KC: Novel therapeutic strategies targeting growth factor
signalling cascades in multiple myeloma. Br J Haematol.
132:385–397. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Naka T, Nishimoto N and Kishimoto T: The
paradigm of IL-6: From basic science to medicine. Arthritis Res. 4
(Suppl 3):S233–S242. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Ishikawa H, Tsuyama N and Kawano MM:
Interleukin-6-induced proliferation of human myeloma cells
associated with CD45 molecules. Int J Hematol. 78:95–105.
2003.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liu FT, Zhu PQ, Ou YX, Liu WW, Xia GF and
Luo HL: Positive association between IL-16 rs1131445 polymorphism
and cancer risk: A meta-analysis. Minerva Med. 107:84–91.
2016.PubMed/NCBI
|
|
16
|
Hong JB, Zuo W, Wang AJ and Lu NH:
Helicobacter pylori infection synergistic with IL-1β gene
polymorphisms potentially contributes to the carcinogenesis of
gastric cancer. Int J Med Sci. 13:298–303. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kasamatsu T, Kimoto M, Takahashi N, Minato
Y, Gotoh N, Takizawa M, Matsumoto M, Sawamura M, Yokohama A, Handa
H, et al: IL17A and IL23R gene polymorphisms affect the clinical
features and prognosis of patients with multiple myeloma. Hematol
Oncol. 36:196–201. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Li Y, Du Z, Wang X, Wang G and Li W:
Association of IL-6 promoter and receptor polymorphisms with
multiple myeloma risk: A systematic review and meta-analysis. Genet
Test Mol Biomarkers. 20:587–596. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Banu C, Moise A, Arion CV, Coriu D, Tănase
A and Constantinescu I: Cytokine gene polymorphisms support
diagnostic monitoring of Romanian multiple myeloma patients. J Med
Life. 4:264–268. 2011.PubMed/NCBI
|
|
20
|
Birmann BM, Tamimi RM, Giovannucci E,
Rosner B, Hunter DJ, Kraft P, Mitsiades C, Anderson KC and Colditz
GA: Insulin-like growth factor-1- and interleukin-6-related gene
variation and risk of multiple myeloma. Cancer Epidemiol Biomarkers
Prev. 18:282–288. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Vangsted AJ, Klausen TW, Ruminski W,
Gimsing P, Andersen NF, Gang AO, Abildgaard N, Knudsen LM, Nielsen
JL, Gregersen H and Vogel U: The polymorphism IL-1beta T-31C is
associated with a longer overall survival in patients with multiple
myeloma undergoing auto-SCT. Bone Marrow Transplant. 43:539–545.
2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Abazis-Stamboulieh D, Oikonomou P,
Papadoulis N, Panayiotidis P, Vrakidou E and Tsezou A: Association
of interleukin-1A, interleukin-1B and interleukin-1 receptor
antagonist gene polymorphisms with multiple myeloma. Leuk Lymphoma.
48:2196–2203. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mazur G, Bogunia-Kubik K, Wróbel T,
Karabon L, Polak M, Kuliczkowski K and Lange A: IL-6 and IL-10
promoter gene polymorphisms do not associate with the
susceptibility for multiple myeloma. Immunol Lett. 96:241–246.
2005.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cozen W, Gebregziabher M, Conti DV, Van
Den Berg DJ, Coetzee GA, Wang SS, Rothman N, Bernstein L, Hartge P,
Morhbacher A, et al: Interleukin-6-related genotypes, body mass
index, and risk of multiple myeloma and plasmacytoma. Cancer
Epidemiol Biomarkers Prev. 15:2285–2291. 2006.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Iakupova EV, Grinchuk OV, Kalimullina DKh,
Bakirov BA, Galimova RR, Makarova OV, Khusnutdinova EK and
Viktorova TV: Molecular genetic analysis of the interleukin 6 and
tumor necrosis factor alpha gene polymorphisms in multiple myeloma.
Mol Biol (Mosk). 37:420–424. 2003.(In Russian). PubMed/NCBI
|
|
26
|
Shamseer L, Moher D, Clarke M, Ghersi D,
Liberati A, Petticrew M, Shekelle P and Stewart LA: PRISMA-P Group:
Preferred reporting items for systematic review and meta-analysis
protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ.
350(g7647)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Richardson WS, Wilson MC, Nishikawa J and
Hayward RS: The well-built clinical question: A key to
evidence-based decisions. ACP J Club. 123:A12–A13. 1995.PubMed/NCBI
|
|
28
|
Stang A: Critical evaluation of the
Newcastle-Ottawa scale for the assessment of the quality of
nonrandomized studies in meta-analyses. Eur J Epidemiol.
25:603–605. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Whiting PF, Rutjes AW, Westwood ME,
Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA and Bossuyt
PM: QUADAS-2 Group: QUADAS-2: A revised tool for the quality
assessment of diagnostic accuracy studies. Ann Intern Med.
155:529–536. 2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Heavener T and Vassar M: A review of
publication bias in the gastroenterology literature. Indian J
Gastroenterol. 37:58–62. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zheng C, Huang DR, Bergenbrant S, Sundblad
A, Osterborg A, Björkholm M, Holm G and Yi Q: Interleukin 6, tumour
necrosis factor alpha, interleukin 1beta and interleukin 1 receptor
antagonist promoter or coding gene polymorphisms in multiple
myeloma. Br J Haematol. 109:39–45. 2000.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zheng C, Huang D, Liu L, Wu R, Bergenbrant
Glas S, Osterborg A, Bjorkholm M, Holm G, Yi Q and Sundblad A:
Interleukin-10 gene promoter polymorphisms in multiple myeloma. Int
J Cancer. 95:184–188. 2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Aladzsity I, Kovács M, Semsei A, Falus A,
Szilágyi A, Karádi I, Varga G, Füst G and Várkonyi J: Comparative
analysis of IL6 promoter and receptor polymorphisms in
myelodysplasia and multiple myeloma. Leuk Res. 33:1570–1573.
2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Vangsted AJ, Nielsen KR, Klausen TW,
Haukaas E, Tjønneland A and Vogel U: A functional polymorphism in
the promoter region of the IL1B gene is associated with risk of
multiple myeloma. Br J Haematol. 158:515–518. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Nielsen KR, Rodrigo-Domingo M, Steffensen
R, Baech J, Bergkvist KS, Oosterhof L, Schmitz A, Bødker JS,
Johansen P, Vogel U, et al: Interactions between SNPs affecting
inflammatory response genes are associated with multiple myeloma
disease risk and survival. Leuk Lymphoma. 58:2695–2704.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Martino A, Buda G, Maggini V, Lapi F,
Lupia A, Di Bello D, Orciuolo E, Galimberti S, Barale R, Petrini M
and Rossi AM: Could age modify the effect of genetic variants in
IL6 and TNF-α genes in multiple myeloma? Leuk Res. 36:594–597.
2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chakraborty B, Vishnoi G, Gowda SH and
Goswami B: Interleukin-6 gene-174 G/C promoter polymorphism and its
association with clinical profile of patients with multiple
myeloma. Asia Pac J Clin Oncol. 13:e402–e407. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kasamatsu T, Saitoh T, Ino R, Gotoh N,
Mitsui T, Shimizu H, Matsumoto M, Sawamura M, Yokohama A, Handa H,
et al: Polymorphism of IL-10 receptor β affects the prognosis of
multiple myeloma patients treated with thalidomide and/or
bortezomib. Hematol Oncol. 35:711–718. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Duch CR, Figueiredo MS, Ribas C, Almeida
MS, Colleoni GW and Bordin JO: Analysis of polymorphism at site-174
G/C of interleukin-6 promoter region in multiple myeloma. Braz J
Med Biol Res. 40:265–267. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Stephens OW, Zhang Q, Qu P, Zhou Y, Chavan
S, Tian E, Williams DR, Epstein J, Barlogie B and Shaughnessy JD
Jr: An intermediate-risk multiple myeloma subgroup is defined by
sIL-6r: Levels synergistically increase with incidence of SNP
rs2228145 and 1q21 amplification. Blood. 119:503–512.
2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tobias A: Assessing the influence of a
single study in the meta-analysis estimate. Stata Techn Bull.
47:15–17. 1999.
|
|
44
|
Dukat-Mazurek A, Bieniaszewska M, Hellmann
A, Moszkowska G and Trzonkowski P: Association of cytokine gene
polymorphisms with the complications of allogeneic haematopoietic
stem cell transplantation. Hum Immunol. 78:672–683. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lu Y, Gu J, Lu H, Zhu Q, Zhang F, Wang X,
Lu L and Zhang C: Association between IL-17A +197 G/A polymorphism
and cancer risk: A meta-analysis. Genet Test Mol Biomarkers.
20:24–30. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Galicia JC, Tai H, Komatsu Y, Shimada Y,
Akazawa K and Yoshie H: Polymorphisms in the IL-6 receptor (IL-6R)
gene: Strong evidence that serum levels of soluble IL-6R are
genetically influenced. Genes Immun. 5:513–516. 2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Fishman D, Faulds G, Jeffery R,
Mohamed-Ali V, Yudkin JS, Humphries S and Woo P: The effect of
novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6
transcription and plasma IL-6 levels, and an association with
systemic-onset juvenile chronic arthritis. J Clin Invest.
102:1369–1376. 1998.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sultana Z, Bankura B, Pattanayak AK,
Sengupta D, Sengupta M, Saha ML, Panda CK and Das M: Association of
Interleukin-1 beta and tumor necrosis factor-alpha genetic
polymorphisms with gastric cancer in India. Environ Mol Mutagen.
59:653–667. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Demeter J, Messer G, Ramisch S, Mee JB, di
Giovine FS, Schmid M, Herrmann F and Porzsolt F: Polymorphism
within the second intron of the IL-1 receptor antagonist gene in
patients with hematopoietic malignancies. Cytokines Mol Ther.
2:239–242. 1996.PubMed/NCBI
|
|
50
|
Ziakas PD, Karsaliakos P, Prodromou ML and
Mylonakis E: Interleukin-6 polymorphisms and hematologic
malignancy: A re-appraisal of evidence from genetic association
studies. Biomarkers. 18:625–631. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Singh PK, Chandra G, Bogra J, Gupta R,
Kumar V, Jain A, Hussain SR, Mahdi AA and Ahmad MK: Association of
interleukin-6 genetic polymorphisms with risk of OSCC in Indian
population. Meta Gene. 4:142–51. 2015.PubMed/NCBI View Article : Google Scholar
|