Introduction
Cancer is a leading cause of human death. Esophageal
cancer is the nine most common cancer worldwide, with an estimated
572,034 new cases and 508,585 deaths in 2018. Men have a
substantially higher incidence than women (1). The cancers arise from the esophageal
mucosa. There are two main histological types: Esophageal squamous
cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). ESCC is
the predominant histological type in southeast Asian countries,
including Thailand (2). Major risk
factors for ESCC are smoking and excessive alcohol consumption
(3,4). These risk factors may lead to
esophageal cancer through multiple genetic alterations, such as
activated oncogenes and inhibited tumor suppressor genes (5).
Despite recent advances in surgical techniques and
perioperative management, the prognosis of patients who undergo
surgery alone for esophageal cancer remains poor. Neoadjuvant
chemotherapy and chemoradiotherapy followed by surgery have emerged
as a promising strategy for advanced esophageal cancer, and, in
fact, good responders to such preoperative therapy show improved
survival (6-9).
Cisplatin/5-fluorouracil (5-FU) has been accepted as a standard
treatment in for ESCC (10).
However, following Cisplatin/5-FU based chemotherapy,
non-responders are likely to receive no survival benefit (11,12). The
ability to predict the response to chemotherapy before treatment
should limit the application of chemotherapy to selected ESCC
patients who are likely to show benefits. However, the prognosis of
patients who are resistant to 5-FU treatment is poor. Resistance to
treatment with anticancer drugs results from a variety of factors,
including individual variations in patients.
miRNAs are noncoding RNAs that are approximately 22
nucleotides in length. They act through repressing the translation
of target mRNAs by binding to the 30-nucleotide untranslated region
of those mRNAs (13). miRNAs exist
stably in various tissues and play pivotal roles in differentiation
and development (14,15). The role of miRNAs has been well
established in various human cancers. The evidence has shown that
miRNA mutations or misexpression correlates with various human
cancers, indicating that miRNAs can function as tumor suppressors
or oncogenes. In addition, aberrant expression of miRNAs has been
reported in various types of cancers (16,17).
Recent studies of ESCC reported the oncogenic microRNAs: miR-21,
miR-10b, miR-31, and miR-373; the oncosuppressor microRNAs: let-7,
miR-34a, miR-133a, miR-150, miR-375, miR-205, miR-145, miR-29c, and
miR-210(18). The mi-R-25, miR-99a,
miR-133a and miR-133b showed good potential as diagnostic markers
and interestingly the mi-R-21, miR-27b, miR-126, miR-143 and
miR-145 appeared to be useful both as diagnostic and
prognostic/predictive markers (19).
A recent publication showed the involvement of several miRNAs in
resistance to 5-FU treatment as follows: The miRNA profiles of
neoadjuvant radiochemotherapy non-responders showed upregulation of
has-miR-1323, has-miR-3678-3p, hsv2-miR-H7-3p, has-miR-194,
has-miR-3152, kshv-miR-K12-4-3p, has-miR-665 and has-miR-3659, and
downregulation of has-miR-126, has-miR-484, has-miR-330-3p and
has-miR-3653(20). The aim of this
study was to investigate the expression profile of ESCC, revealing
differential expression between ESCC and 5-FU resistant ESCC.
Materials and methods
Cell lines and cell culture
Human ESCC cell lines (TE4, TE10, TE11 and TE15)
were obtained from Tohoku University. All cells were cultured in
Dulbecco's Modified Eagle's Medium (DMEM) (Nacalai Tesque, Inc.)
containing 10% fetal bovine serum (Life Technologies Inc.), 10%
penicillin/streptomycin (100 U/ml penicillin and 100 µg/ml
streptomycin) (Nacalai Tesque, Inc.) in a humidified atmosphere
under 5% CO2 at 37̊C.
Establishment of 5-FU resistant cell
lines
5-FU resistant (5-FUR) cell lines were cultured
through gradual increases in 5-FU concentration. The cultured cells
were exposed to 5-FU at an initial concentration of 1 nM/ml. After
24 h, the cells were cultured in 5-FU free medium until confluence.
Next, 5-FU concentrations were increased by 2- to 3-fold and the
cycle was repeated.
Proliferation assay
The WST-8
(2-(2-methoxy-4-nitrphenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,
monosodium salt) assay was conducted as described by the
manufacturer (Nacalai Tesque Inc.) and was used to determine the
IC50 (50% growth inhibition concentration) value of
5-FU. Cells were plated in 96-well microplates and cultured for 12
h before exposure to various concentrations of 5-FU (0, 0.5, 1, 5,
10, 50 µg/ml) for 48 h. The optical density (OD) value was detected
by RAINBO SUNRISE (Wako Pure Chemical Industries Ltd.) at 450 nm
test wavelength and 650 nm reference wavelength. The
IC50 value of 5-FU was calculated from the dose-response
curve.
Isolation of miRNA and miRNA
microarray
The miRNA was isolated from the cell lines using the
mirVana™ miRNA Isolation kit according to the
manufacturer's protocol (Ambion; Thermo Fisher Scientific, Inc.).
The concentration of RNA was quantified using the NanoDrop ND-1000
spectrophotometer (Thermo Fisher Scientific, Inc.). The miRNA
expression profiling of human ESCC cell lines (TE10, TE11) and
their corresponding 5-FU resistant (5-FUR) daughter lines (TE10
5-FUresistant cells: TE10-5-FUR; TE11 5-FU resistant cell:
TE11-5-FUR) were examined by TaqMan® Human MicroRNA
Array (Applied Biosystems; Thermo Fisher Scientific, Inc.). It
contained 384 miRNA targets (and 7 control miRNAs) and was
performed using Megaplex™ RT Primers. The miRNA
microarray analysis was performed with Applied Biosystems 7900HT
fast real-time PCR System and RT-PCR StatMiner™
software. The expression of each miRNA in 5-FU resistant cell lines
was compared with that in the control parental cell line, and the
ratio of miRNA expression in 5-FU resistant cells to control cells
was calculated for all 384 miRNAs.
Statistical analysis
The significance validation data of miRNA expression
are expressed as mean ± standard error of the mean. The cell
viability is computed and differences between viability curves are
compared. The parameters are compared using the χ2 test
for categorical data and continuous variables are compared using
Student's t-test. All data were analyzed with SPSS 22.0 data (IBM
Corp.). A P-value <0.05 was considered to be statistically
significant.
Results
The study first established 5-FU resistant ESCC cell
lines (TE4, TE10, TE11 and TE15) by gradually increasing 5-FU
concentration (starting from 0.1 µg/ml) and evaluating the cultures
by WST-8 assay every 4 weeks. After 8 weeks, we determined the
IC50 values. The results showed a significant
fold-increase in the concentration of 5-FU that inhibited TE10 and
TE11 cell growth by 50%. TE10-5-FUR cells were relatively resistant
to 5-FU, with an IC50 of 42.66±2.38 µg compared to a
value of 4.08±2.06 µg in the parent cells, a 10.5-fold increase in
concentration (P<0.01). TE11-5-FUR cells were also
relatively resistant to 5-FU with an IC50 of 21.62±11.91
µg compared to 2.73±0.81 µg in TE11 parent cells, a 7.91-fold
increase (P<0.01) (Fig.
1).
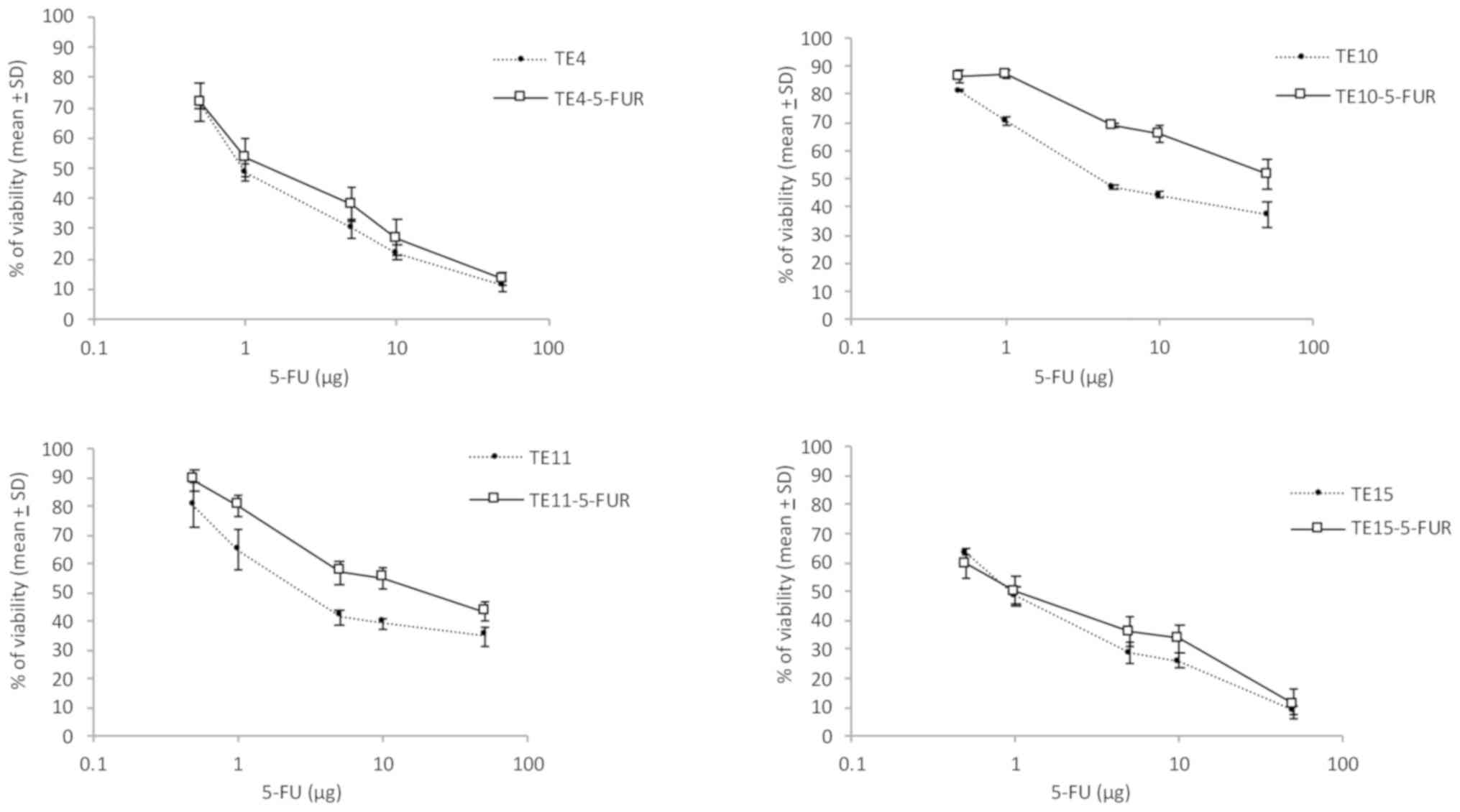 | Figure 1Quantitation of 5-FU resistance of
esophageal squamous cell lines. TE4, TE4-5-FUR, TE10, TE10-5-FUR,
TE11, TE11-5-FUR, TE15 and TE15-5-FUR cells were seeded into
96-well microplates (5x103 per well) 12 h before
treatment and were then exposed to different concentrations (0,
0.5, 1, 5, 10 and 50 µg) of 5-FU for 48 h. The percentage of
cellular proliferation was evaluated with WST-8. The 5-FU resistant
TE10-5-FUR cells showed resistance to 5-FU with an IC50
value of 42.66±2.38 µg and TE11-5-FUR achieved an IC50
value of 21.62±11.91 µg, both P<0.01 compared to their parental
cells. For 5-FU resistant TE4-5-FUR cells and TE15-5-FUR, the
results were not difference to their parental cells. Data are
presented as means ± SD and evaluated using Student's t-test.
WST-8,
2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,
monosodium salt; SD, standard deviation. |
To assess miRNA expression levels, we used miRNA
microarray to evaluate both 5-FU resistant and wild-type ESCC cell
lines. The results of the miRNA expression study were subjected to
a differential expression analysis and visualized using Expression
Suite Software (Applied Biosystems; Thermo Fisher Scientific,
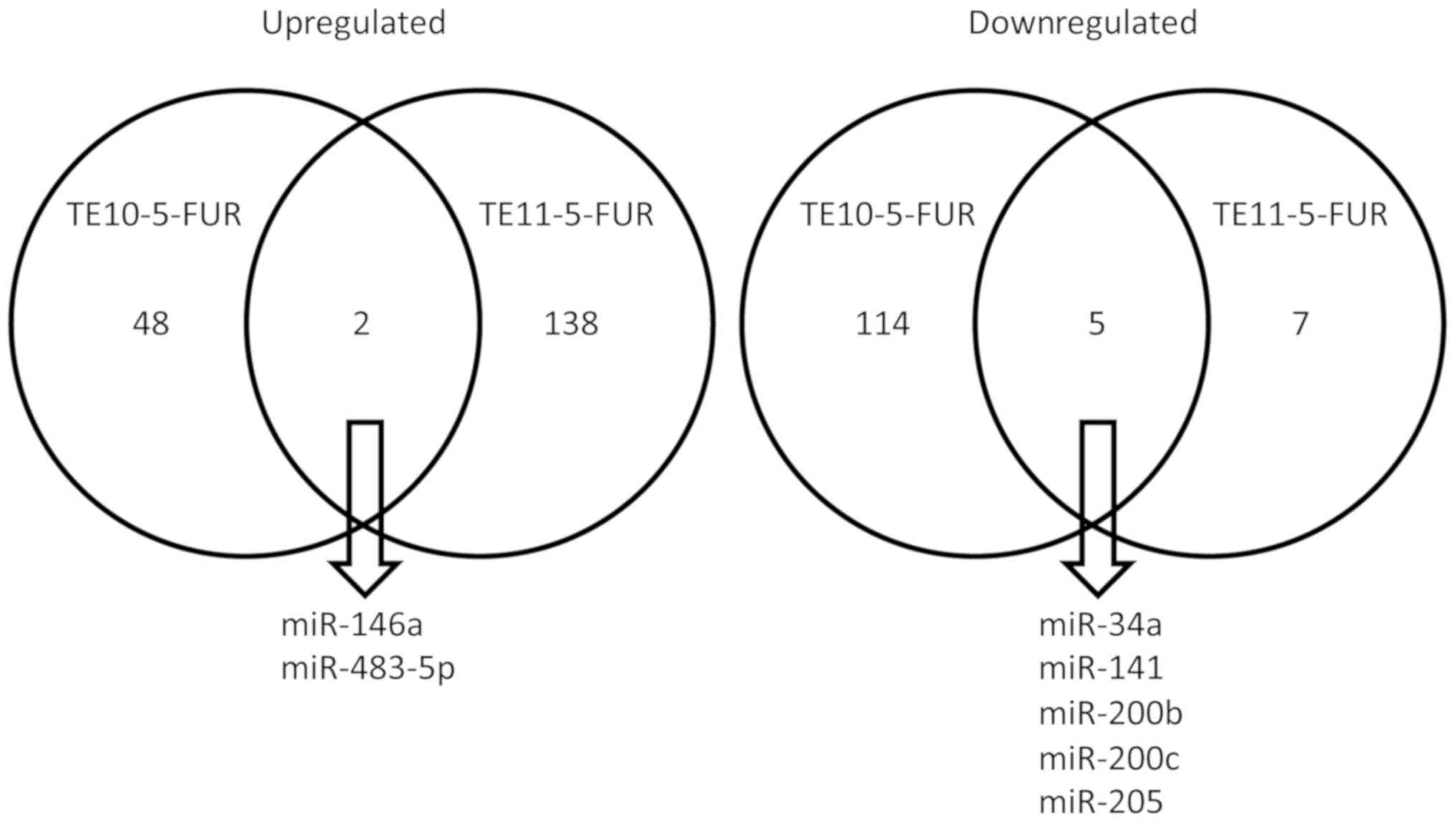
Inc.). The analysis showed 50 miRNAs upregulated in TE10-5-FUR
compared to TE10, while 119 miRNAs were downregulated. The
TE11-5-FUR demonstrated 140 miRNAs upregulated compared to TE11
with 12 miRNAs were downregulated. Among the most significantly
upregulated miRNAs of TE10-5-FUR were has-miR-99a-4373008,
has-miR-100-4373160, has-miR-125b-4373148, has-miR-140-5p-437374,
has-miR-146a-4373132, has-miR-155-4375459, has-miR-196b-4395326,
has-miR-302b-4378071, has-miR-499a-4373207, has-miR-483-5p-4395449
while the most downregulated were has-miR-34a-4395168,
has-miR-130a-4373145, has-miR-141-4373137, has-miR-152-4395170,
has-miR-183-4395380, has-miR-200a-4378069, has-miR-200b-4395362,
has-miR-200c-4395411, has-miR-205-4373093, has-miR-429-4373203
(Table I). For TE11-5-FUR, the
prominent upregulated miRNAs were has-miR-let7b-4395446,
has-miR-let7c-4373167, has-miR-10b-4395329, has-miR-22-4373079,
has-miR-137-4373301, has-miR-146a-4373132, has-miR-296-5p-4373066,
has-miRs-449b-4381011, has-miR-483-5p-4395449, has-miR-522-4395524,
while all those downregulated were has-miR-18a-4395533,
has-miR-34a-4395168, has-miR-141-4373137, has-miR-200b-4395362,
has-miR-200c-4395411, has-miR-203-4373095, has-miR-205-4373093,
has-miR-331-5p-4395344, has-miR-429-4373203, has-miR-708-4395452
(Table II). The result in both cell
lines observed at intersection of 2 miRNAs upregulated (miR-146a
and miR-483-5p) and 5 miRNAs downregulated (miR-34a, miR-141,
miR-200b, miR-200c and miR-205) (Fig.
2). Identification of potential target genes of miRNAs
associated with 5-FU resistant ESCC cell lines was essential to
investigate their biological functions. Candidate miRNAs of both
cell lines were reviewed using the database of miRNA.org site (http://www.microrna.org) (21).
 | Table IThe list of differentially expressed
microRNAs in TE10-5-FUR vs. TE10. |
Table I
The list of differentially expressed
microRNAs in TE10-5-FUR vs. TE10.
| miRNAs | Fold-change | Regulation | microRNA family |
|---|
|
has-miR-99a-4373008 | 5.25707 | Up | miR-99a |
|
has-miR-100-4373160 | 3.79643 | Up | miR-100 |
|
has-miR-125b-4373148 | 4.09845 | Up | miR-125b |
|
has-miR-140-5p-437374 | 3.76036 | Up | miR-140-5p |
|
has-miR-146a-4373132 | 5.46298 | Up | miR-146a |
|
has-miR-155-4375459 | 4.67046 | Up | miR-155 |
|
has-miR-196b-4395326 | 6.09578 | Up | miR-196b |
|
has-miR-302b-4378071 | 4.67347 | Up | miR-302b |
|
has-miR-499a-4373207 | 5.78943 | Up | miR-499a |
|
has-miR-483-5p-4395449 | 4.54962 | Up | miR-483-5p |
|
has-miR-34a-4395168 | 3.08635 | Down | miR-34a |
|
has-miR-130a-4373145 | 1.87463 | Down | miR-130a |
|
has-miR-141-4373137 | 2.98572 | Down | miR-141 |
|
has-miR-152-4395170 | 0.56493 | Down | miR-152 |
|
has-miR-183-4395380 | 1.59869 | Down | miR-183 |
|
has-miR-200a-4378069 | 2.78942 | Down | miR-200a |
|
has-miR-200b-4395362 | 0.89423 | Down | miR-200b |
|
has-miR-200c-4395411 | 2.05483 | Down | miR-200c |
|
has-miR-205-4373093 | 1.68473 | Down | miR-205 |
|
has-miR-429-4373203 | 1.87439 | Down | miR-429 |
 | Table IIThe list of differentially expressed
microRNAs in TE11-5-FUR vs. TE11. |
Table II
The list of differentially expressed
microRNAs in TE11-5-FUR vs. TE11.
| miRNAs | Fold-change | Regulation | microRNA
family |
|---|
|
has-miR-let7b-4395446 | 6.43275 | Up | let7b |
|
has-miR-let7c-4373167 | 7.59883 | Up | let7c |
|
has-miR-10b-4395329 | 5.58848 | Up | miR-10b |
|
has-miR-22-4373079 | 6.15343 | Up | miR-22 |
|
has-miR-137-4373301 | 5.42084 | Up | miR-137 |
|
has-miR-146a-4373132 | 5.58873 | Up | miR-146a |
|
has-miR-296-5p-4373066 | 5.02579 | Up | miR-296-5p |
|
has-miR-449b-4381011 | 4.96571 | Up | miR-449b |
|
has-miR-483-5p-4395449 | 6.19015 | Up | miR-483-5p |
|
has-miR-522-4395524 | 7.06593 | Up | miR-522 |
|
has-miR-18a-4395533 | 0.47938 | Down | miR-18a |
|
has-miR-34a-4395168 | 0.30295 | Down | miR-34a |
|
has-miR-141-4373137 | 3.15697 | Down | miR-141 |
|
has-miR-200b-4395362 | 3.18372 | Down | miR-200b |
|
has-miR-200c-4395411 | 4.10116 | Down | miR-200c |
|
has-miR-203-4373095 | 0.73426 | Down | miR-203 |
|
has-miR-205-4373093 | 1.49047 | Down | miR-205 |
|
has-miR-331-5p-4395344 | 0.80691 | Down | miR-331-5p |
|
has-miR-429-4373203 | 3.88948 | Down | miR-429 |
|
has-miR-708-4395452 | 0.56551 | Down | miR-708 |
Discussion
Esophageal cancer is a major global health problem.
Squamous cell carcinoma is the main histological type. Current
management of ESCC depends on the stage of the disease and includes
surgery, chemotherapy and radiation therapy. In multimodal
treatment of esophageal cancer, chemotherapy has an important role
in combination with radiation therapy and/or surgery.
Fluoropyrimidine plus platinum (FP) are the chemotherapeutic drugs
most frequently used to treat ESCC. This regimen has been reported
to be effective, with improved overall survival (22,23).
However, drug resistance often occurs, and the mechanisms of
resistance to 5-FU are still not clear.
The establishment of 5-FU resistant (5-FUR) cell
lines model provides an approach to analyze the mechanism of drug
resistance. The resistant cells were created from their parental
lines by exposing them to gradually increasing 5-FU concentrations
for 2 months. The TE10-5-FUR and TE11-5-FUR lines were partially
resistant to 5-FU with IC50 values of 42.66±2.38 µg and
21.62±11.91 µg.
miRNAs are short noncoding RNAs that regulate gene
expression and play an important role in human cancers. They can
also modulate the sensitivity and resistance to anticancer drugs.
This study demonstrated 50 miRNAs upregulated and 119 miRNAs
downregulated in TE10-5-FUR, 140 miRNAs upregulated and 12 miRNAs
downregulated in TE11-5-FUR, compared to their wild type. The
result in both of cell lines found 2 candidate miRNAs upregulated
(miR-146a and miR-483-5p) and 5 miRNAs were downregulation
(miR-34a, miR-141, miR-200b, miR-200c and miR-205). Recent studies
also showed the involvement of several miRNAs in resistance to
anticancer treatment and roles in esophageal cancer. miR-146a has
been reported as perhaps being associated with the cisplatin-base
susceptibility to lung cancer by downregulating cyclin J (23,24) and
as a potential therapeutic target for multidrug-resistant lung
cancer by targeting DNA damage inducible transcript 3(25). Polymorphism in miR-146a could be
associated with the lymph node metastasis and prognosis of gastric
cancer patients treated with oxaliplatin and fluoropyrimidines
(26). miR-483-5p has been described
upregulation with might be a tumor promoter of ESCC that correlated
with TNM stage and survival (27).
miR-483-5p could inhibit mitochondrial fission protein FIS1 with
significant association with cisplatin sensitivity and with overall
survival (28). miR-34a showed
significantly expressed reduction in ESCC tissues and exerted its
anticancer function by suppressing PLCE1(29). miR-34a has shown upregulation in
cisplatin sensitivity for lung cancer treatment via
p53/miR-34a/MYCN axis (30),
mediates oxaliplatin resistance of colorectal cancer cells by
inhibiting macroautophagy via the TGF-β/Smad4 pathway (31) and the patients with high levels of
expression were found to benefit more from 5-FU based chemotherapy
than patients with low levels of expression with the potential
targets including CREB1, Bcl-2, Notch 1, Sirt1, and E2F3(32). For miR-141, the overexpression could
abolish the self-renewal ability and carcinogenicity of esophageal
cancer stem-like cells and decrease cell invasion and migration by
suppressing TM4SF1(33). It enhanced
the effected of 5-FU and suppressed the malignant biological
behaviors of colorectal cancer by MAP4K4 signaling pathway
(34). miR-141 was significantly
decreased and correlated with advanced TNM stage and lymph node
metastasis with predicted possible target MACC1 in gastric cardia
adenocarcinoma (35). miR-200b has
been reported down-regulated in the multi-drug resistance of small
cell lung cancer via ZEB2(36). For
miR-200c, the serum levels in advanced ESCC patients were
significantly increased and associated with poor outcome of
platinum-based chemotherapy (37).
miR-200c also related to 5-FU chemotherapy with the potential
targets PTEN and E-cadherin in colorectal cancer (38-40).
miR-205 has been published as a tumor suppressor in adenocarcinoma
and an oncogene in squamous cell carcinoma of esophagus through
regulation of epithelial-mesenchymal transition (EMT) (41) and the Sp1-mediated transcriptional
activation of miR-205 promotes radioresistance through PTEN via
PI3K/AKT pathway in ESCC (42). For
TE11-5-FUR, the let-7b and let-7c demonstrated upregulation with
related to the previous publication that reported the let7 play the
role of oncosuppressor microRNAs (18).
This study acknowledges its own limitations-the
study, the sample size is too small to make any reasonable
conclusion. The cell viability and miRNA microarray experiments are
not performed on non-cancer cell lines as a control. This study is
not verified the function analysis of miRNAs that have related to
resistance to 5-FU. Their function should be testing by gene
transfer or knockdown with in vitro studies. Alternatively, the
association of these miRNAs with clinical efficacy of chemotherapy
be examining in a cohort of patients with esophageal cancer. The
current study revealed differentially regulated miRNAs that are
involved in 5-FU resistant ESCC. The identification of miRNA
expression profiles and candidates in 5-FU resistant ESCC could
provide a better understanding of the mechanisms involved in
chemo-sensitivity or resistance. By predicting the response to
chemotherapy, one could offer another treatment option for patients
who would otherwise be resistant. Further study is needed to select
the potential targets and explore the pathways that are upregulated
or downregulated after induction of 5-FU therapy. Moreover, these
findings suggest that it may be helpful to develop novel strategies
for targeted therapies in ESCC patients.
Acknowledgements
Not applicable.
Funding
This research supported in part by Thammasat
University.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
PM and PT contributed to the conception and design
of the study. PM contributed to data collection, conduction of the
study, performed the experiments, analysis and interpretation of
the data. PM and PT reviewed the manuscript, designed the figures
and tables. All the authors have read and approved the final
version of this manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nun-Anan P and Vilaichone RK: Late stage
and grave prognosis of esophageal cancer in Thailand. Asian Pac J
Cancer Prev. 16:1747–1749. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Morita M, Kumashiro R, Kubo N, Nakashima
Y, Yoshida R, Yoshinaga K, Saeki H, Emi Y, Kakeji Y, Sakaguchi Y,
et al: Alcohol drinking, cigarette smoking, and the development of
squamous cell carcinoma of the esophagus: Epidemiology, clinical
findings, and prevention. Int J Clin Oncol. 15:126–134.
2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Toh Y, Oki E, Ohgaki K, Sakamoto Y, Ito S,
Egashira A, Saeki H, Kakeji Y, Morita M, Sakaguchi Y, et al:
Alcohol drinking, cigarette smoking, and the development of
squamous cell carcinoma of the esophagus: Molecular mechanisms of
carcinogenesis. Int J Clin Oncol. 15:135–144. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Huang FL and Yu SJ: Esophageal cancer:
Risk factors, genetic association, and treatment. Asian J Surg.
41:210–215. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hsu PK, Chen HS, Huang CS, Liu CC, Hsieh
CC, Hsu HS, Wu YC and Wu SC: Patterns of recurrence after
oesophagectomy and postoperative chemoradiotherapy versus surgery
alone for oesophageal squamous cell carcinoma. Br J Surg.
104:90–97. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pasquali S, Yim G, Vohra RS, Mocellin S,
Nyanhongo D, Marriott P, Geh JI and Griffiths EA: Survival after
neoadjuvant and adjuvant treatments compared to surgery alone for
resectable esophageal carcinoma: A network meta-analysis. Ann Surg.
265:481–491. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ando N, Iizuka T, Ide H, Ishida K, Shinoda
M, Nishimaki T, Takiyama W, Watanabe H, Isono K, Aoyama N, et al:
Surgery plus chemotherapy compared with surgery alone for localized
squamous cell carcinoma of the thoracic esophagus: A Japan clinical
oncology group study-JCOG9204. J Clin Oncol. 21:4592–4596.
2003.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ando N, Kato H, Igaki H, Shinoda M, Ozawa
S, Shimizu H, Nakamura T, Yabusaki H, Aoyama N, Kurita A, et al: A
randomized trial comparing postoperative adjuvant chemotherapy with
cisplatin and 5-fluorouracil versus preoperative chemotherapy for
localized advanced squamous cell carcinoma of the thoracic
esophagus (JCOG9907). Ann Surg Oncol. 19:68–74. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Haisley KR, Hart KD, Nabavizadeh N, Bensch
KG, Vaccaro GM, Thomas CR Jr, Schipper PH, Hunter JG and Dolan JP:
Neoadjuvant chemoradiotherapy with concurrent
cisplatin/5-fluorouracil is associated with increased pathologic
complete response and improved survival compared to
carboplatin/paclitaxel in patients with locally advanced esophageal
cancer. Dis Esophagus. 30:1–7. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Rumiato E, Cavallin F, Boldrin E, Cagol M,
Alfieri R, Basso D, Castoro C, Ancona E, Amadori A, Ruol A and
Saggioro D: ERCC1 C8092A (rs3212986) polymorphism as a predictive
marker in esophageal cancer patients treated with
cisplatin/5-FU-based neoadjuvant therapy. Pharmacogenet Genomics.
23:597–604. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Miyata H, Yoshioka A, Yamasaki M,
Nushijima Y, Takiguchi S, Fujiwara Y, Nishida T, Mano M, Mori M and
Doki Y: Tumor budding in tumor invasive front predicts prognosis
and survival of patients with esophageal squamous cell carcinomas
receiving neoadjuvant chemotherapy. Cancer. 115:3324–3334.
2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chakraborty C, Chin KY and Das S:
miRNA-regulated cancer stem cells: Understanding the property and
the role of miRNA in carcinogenesis. Tumour Biol. 37:13039–13048.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Barbato S, Solaini G and Fabbri M:
MicroRNAs in oncogenesis and tumor suppression. Int Rev Cell Mol
Biol. 333:229–268. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of microRNAs in cancer.
Cancer Res. 76:3666–3670. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hemmatzadeh M, Mohammadi H, Karimi M,
Musavishenas MH and Baradaran B: Differential role of microRNAs in
the pathogenesis and treatment of esophageal cancer. Biomed
Pharmacother. 82:509–519. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sakai NS, Samia-Aly E, Barbera M and
Fitzgerald RC: A review of the current understanding and clinical
utility of miRNAs in esophageal cancer. Semin Cancer Biol.
23:512–521. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hummel R, Sie C, Watson DI, Wang T, Ansar
A, Michael MZ, Van der Hoek M, Haier J and Hussey DJ: MicroRNA
signatures in chemotherapy resistant esophageal cancer cell lines.
World J Gastroenterol. 20:14904–14912. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Slotta-Huspenina J, Drecoll E, Feith M,
Habermehl D, Combs S, Weichert W, Bettstetter M, Becker K and
Langer R: MicroRNA expression profiling for the prediction of
resistance to neoadjuvant radiochemotherapy in squamous cell
carcinoma of the esophagus. J Transl Med. 16(109)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36 (Database issue):D149–D153. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Vellayappan BA, Soon YY, Ku GY, Leong CN,
Lu JJ and Tey JC: Chemoradiotherapy versus chemoradiotherapy plus
surgery for esophageal cancer. Cochrane Database Syst Rev.
8(CD010511)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kidane B, Coughlin S, Vogt K and Malthaner
R: Preoperative chemotherapy for resectable thoracic esophageal
cancer. Cochrane Database Syst Rev. (CD001556)2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hao X, Xia L, Qu R, Yang X, Jiang M and
Zhou B: Association between miR-146a rs2910164 polymorphism and
specific cancer susceptibility: An updated meta-analysis. Fam
Cancer. 17:459–468. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shi L, Xu Z, Wu G, Chen X, Huang Y, Wang
Y, Jiang W and Ke B: Up-regulation of miR-146a increases the
sensitivity of non-small cell lung cancer to DDP by downregulating
cyclin J. BMC Cancer. 17(138)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tan W, Liao Y, Qiu Y, Liu H, Tan D, Wu T,
Tang M, Zhang S and Wang H: miRNA 146a promotes chemotherapy
resistance in lung cancer cells by targeting DNA damage inducible
transcript 3 (CHOP). Cancer Lett. 428:55–68. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liao YQ, Liao YL, Li J, Peng LX, Wan YY
and Zhong R: Polymorphism in miR-146a associated with clinical
characteristics and outcomes in gastric cancer patients treated
with adjuvant oxaliplatin and fluoropyrimidines. Onco Targets Ther.
8:2627–2633. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xue L, Nan J, Dong L, Zhang C, Li H, Na R,
He H and Wang Y: Upregulated miR-483-5p expression as a prognostic
biomarker for esophageal squamous cell carcinoma. Cancer Biomark.
19:193–197. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fan S, Chen WX, Lv XB, Tang QL, Sun LJ,
Liu BD, Zhong JL, Lin ZY, Wang YY, Li QX, et al: miR-483-5p
determines mitochondrial fission and cisplatin sensitivity in
tongue squamous cell carcinoma by targeting FIS1. Cancer Lett.
362:183–191. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cui XB, Peng H, Li RR, Mu JQ, Yang L, Li
N, Liu CX, Hu JM, Li SG, Wei Y, et al: MicroRNA-34a functions as a
tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh
esophageal squamous cell carcinoma. Oncotarget. 8:92454–92469.
2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Song C, Lu P, Sun G, Yang L and Wang Z and
Wang Z: miR-34a sensitizes lung cancer cells to cisplatin via
p53/miR-34a/MYCN axis. Biochem Biophys Res Commun. 482:22–27.
2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC,
Yao L and Qiao PF: miR-34a mediates oxaliplatin resistance of
colorectal cancer cells by inhibiting macroautophagy via
transforming growth factor-β/Smad4 pathway. World J Gastroenterol.
23:1816–1827. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhang Q, Wang J, Li N, Liu Z, Chen Z, Li
Z, Lai Y, Shen L and Gao J: miR-34a increases the sensitivity of
colorectal cancer cells to 5-fluorouracil in vitro and in vivo. Am
J Cancer Res. 8:280–290. 2018.PubMed/NCBI
|
|
35
|
Xue L, Yu X, Jiang X, Deng X, Mao L, Guo
L, Fan J, Fan Q, Wang L and Lu SH: TM4SF1 promotes the self-renewal
of esophageal cancer stem-like cells and is regulated by miR-141.
Oncotarget. 8:19274–19284. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang F, Zhao L, Zhang J, Meng Z, Zhou C
and Wang G, Liu Y, Li M, Xi J, Niu W and Wang G:
Chemotherapy-induced miR-141/MAP4K4 signaling suppresses
progression of colorectal cancer. Biosci Rep.
38(BSR20180978)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li S, Zhu J, Li J, Li S and Li B:
MicroRNA-141 inhibits proliferation of gastric cardia
adenocarcinoma by targeting MACC1. Arch Med Sci. 14:588–596.
2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fang S, Zeng X, Zhu W, Tang R, Chao Y and
Guo L: Zinc finger E-box-binding homeobox 2 (ZEB2) regulated by
miR-200b contributes to multi-drug resistance of small cell lung
cancer. Exp Mol Pathol. 96:438–444. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yu H, Duan B, Jiang L, Lin M, Sheng H,
Huang J and Gao H: Serum miR-200c and clinical outcome of patients
with advanced esophageal squamous cancer receiving platinum-based
chemotherapy. Am J Transl Res. 6:71–77. 2013.PubMed/NCBI
|
|
40
|
Heydari K, Saidijam M, Sharifi MR, Dermani
FK, Soleimani Asl S, Shabab N and Najafi R: The effect of miR-200c
inhibition on chemosensitivity (5-FluoroUracil) in colorectal
cancer. Pathol Oncol Res. 24:145–151. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326.
2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Diaz T, Tejero R, Moreno I, Ferrer G,
Cordeiro A, Artells R, Navarro A, Hernandez R, Tapia G and Monzo M:
Role of miR-200 family members in survival of colorectal cancer
patients treated with fluoropyrimidines. J Surg Oncol. 109:676–683.
2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Hezova R, Kovarikova A, Srovnal J,
Zemanova M, Harustiak T, Ehrmann J, Hajduch M, Sachlova M, Svoboda
M and Slaby O: MiR-205 functions as a tumor suppressor in
adenocarcinoma and an oncogene in squamous cell carcinoma of
esophagus. Tumour Biol. 37:8007–8018. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Pan F, Mao H, Bu F, Tong X, Li J, Zhang S,
Liu X, Wang L, Wu L, Chen R, et al: Sp1-mediated transcriptional
activation of miR-205 promotes radioresistance in esophageal
squamous cell carcinoma. Oncotarget. 8:5735–5752. 2017.PubMed/NCBI View Article : Google Scholar
|