Introduction
Kidney cancer accounts for 2-3% of all malignancies
in adults and is the 12th most common cancer type worldwide
(1,2). Overall, 80-90% of kidney cancer cases
develop in the renal parenchyma and are referred to as renal cell
carcinoma (RCC) (3). There are
three main RCC types: Clear cell RCC (ccRCC), papillary RCC (pRCC;
type I and II) and chromophobe RCC (chRCC) (3). Unfortunately, 25% of patients present
with metastatic disease, while up to 40% of patients with
locoregional RCC experience recurrence postoperatively, indicating
the necessity to optimize treatment strategies for such patients.
In addition to clinical and histological prognostic factors,
numerous molecular factors have also been identified. These factors
indicate which patients with localized RCC are at greater risk for
recurrence, and which patients with metastatic disease are at risk
of progression or death (2,4).
The tumour microenvironment is characterized by
decreased oxygen levels. Cancer cells, with their specific genetic
and epigenetic mechanisms, have the ability to adapt to these
hypoxic conditions (5,6). This is partly achieved by regulation
of gene products in response to hypoxia. A number of these
hypoxia-regulated genes require the mediation of hypoxia inducible
factors (HIFs), HIF-1α and HIF-2α (7). These two molecules heterodimerise with
the aryl hydrocarbon nuclear translocator protein (also known as
HIF-β), move into the nucleus and bind to DNA, leading to gene
transcription activation of the responsive genes, which plays a
pivotal role in angiogenesis (8).
Under normoxia condition, HIF-1α and HIF-2α are continuously
expressed and degraded. Degradation is mediated by the prolyl
hydroxylases (PHDs). PHD1, PHD2 and PHD3 hydroxylate two proline
residues (Pro-402/564) in the oxygen-dependent degradation domain
of the HIF-α subunits. This allows the binding of HIF-α subunits to
von Hippel-Lindau tumour suppressor (pVHL) to form a dimer that is
degraded by the proteasome (Fig. 1)
(9).
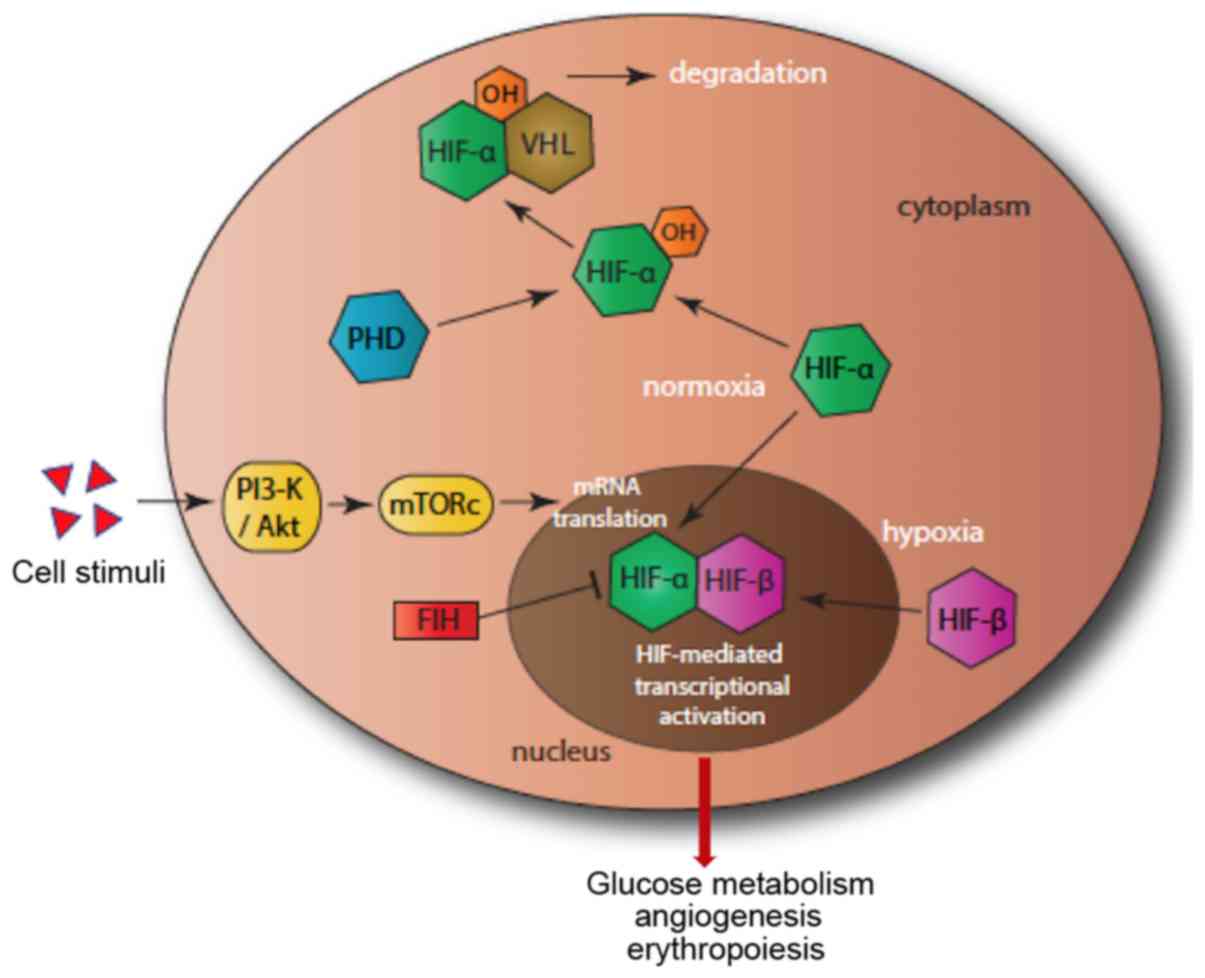 | Figure 1Hypoxia pathway. In normoxia, PHD
(blue) hydroxylates HIF-α (green), which then binds to VHL, leading
to proteosomal degradation. In hypoxia, HIF-α enters the nucleus,
forms a dimer with HIF-β, which then binds to DNA, leading to gene
transcription activation. A second pathway, that of PI3-K/Akt
(yellow), initiates the mTOR and enhances the translation of HIF.
FIH (red) adds a further level of control by reducing the
transcriptional activity of HIF-α. FIH, factor-inhibiting HIF;
HIF-α, hypoxia inducible factor α; PHD, prolyl hydroxylase; VHL,
von Hippel-Lindau tumor suppressor. |
The present study investigated the possible
involvement of the HIF/PHD pathway in RCC by evaluating HIF-1α,
HIF-2α, PHD1, PHD2 and PHD3 mRNA expression levels using reverse
transcription-quantitative PCR (RT-qPCR). The associations of these
expression levels with clinicopathological parameters and survival
outcomes were then analysed.
Patients and methods
Patients
The present study included 41 patients who underwent
radical or partial nephrectomy for histopathologically verified RCC
between December 2010 and September 2013. All patients were treated
by the same surgical team at the 1st Department of Urology,
Aristotle University of Thessaloniki, Greece. Written informed
consent was obtained from every patient and the study protocol was
approved by the Ethics Committee of Aristotle University of
Thessaloniki.
Patients' clinical and pathological data were
obtained prospectively, while follow-up and survival data were
obtained by clinical appointments and/or telephone communication.
Data included age, sex, tumour-node-metastasis (TNM)
classification, Fuhrman grade, ECOG performance status, primary
tumour size and presence of tumour necrosis. The RCC types were
assessed according to the Heidelberg classification system
(3). Tumour size was measured at
the maximum diameter of the surgical specimen. Tumour stage and
grade were determined by two independent pathologists, and
controversies were resolved by consensus. Patients were evaluated
from the time of diagnosis to the end of the study (June 2018).
Overall survival (OS) time was defined as the time from nephrectomy
to mortality from any cause.
Tissue processing and RNA template
preparation
Hematoxylin and eosin (H&E)-stained slides from
formalin-fixed paraffin-embedded (FFPE) tissue blocks were reviewed
for confirmation of diagnosis and abundance of tumour tissue.
Tumour and matched normal tissues were used for the construction of
low-density tissue microarrays (TMAs) with a manual arrayer (Model
I; Beecher Instruments Inc.). TMAs contained two 1.5-mm sections
per tumour (one from the front of the tumour and one from the
centre of the tumour) and two sections from adjacent normal kidney
tissue.
Total RNA was extracted from 8-µm TMA sections, one
from adjacent normal tissue and one from tumour tissue per patient.
The tumour section contained tissue from both the front and centre
of the tumour. Tissue samples were transferred into a lysis buffer
containing 500 µg/ml proteinase K for overnight lysis at 56̊C.
Tissue lysates were then processed for total RNA extraction with
TRIzol LS reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. Subsequently, 4-5 µg
total RNA was reverse transcribed into single-stranded
complementary DNA (cDNA) with random hexamers and
SuperScript® III Reverse Transcriptase (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions (10,11).
RT-qPCR
Relative mRNA expression was assessed by qPCR with
the ABI 7900HT system under default conditions. Β-glucuronidase
(GUSB) was used as the endogenous reference for quantifying the
relative mRNA expression levels (12). Reactions with 100 ng template cDNA
were performed in duplicate.
The following TaqMan-MGB probes (Thermo Fisher
Scientific, Inc.) were used for the interrogated gene targets:
HIF-1α (Hs00153153_m1; NM_001243084.1; exons 4-5; 76 bp), HIF-2α
(Hs01026149_m1; NM_001430.4; exons 7-8; 70 bp), PHD1
(Hs00363196_m1; NM_053046.3; exons 5-6; 77 bp), PHD2
(Hs00254392_m1; NM_022051.2; exons 1-2; 70 bp), PHD3
(Hs00222966_m1; NM_022073.3; exons 3-4; 62 bp) and GUSB
(Hs00939627_m1; NM_000181.3; exons 8-9; 96 bp). The data in
parentheses refer to assay ID; Genbank reference; amplicon
location; and size.
Relative mRNA expression of the target
genes
Following amplification of all cDNA samples, the
standard curves and the cycle threshold values of the samples (Cq)
were recorded. Then, the relative gene expression was calculated
using the 2-ΔΔCq method. The corresponding values in
matched normal samples were used a reference (Fig. S1) (13). Samples with an endogenous control Cq
≥36 were excluded. A 2-fold increase (≥2) or decrease (≤0.5) in
expression was considered biologically significant (for
overexpression or downregulation, respectively), as previously
published (14). Two samples for
PHD1 and PHD3 and three samples for PHD2 were not eligible for
analysis as the cDNA sample was inadequate or they had an
endogenous control Cq ≥36.
Statistical analysis
Continuous variables are presented as the median and
interquartile range (IQR), while certain categorical variables are
presented as frequencies and percentages [n (%)]. Fisher's exact
test was used to analyse the associations between categorical
variables. The non-parametric tests Mann-Whitney (for two groups)
and Kruskal-Wallis (for multiple comparison) were used to compare
the median values of continuous variables. In case of a significant
result in Kruskal Wallis test multiple Mann-Whitney U Tests were
applied using Bonferroni correction. Furthermore, Spearman's rank
correlation (Rs) coefficient was used to identify the
correlation between continuous variables. Cox proportional hazard
model was used to analyse the prognostic value of RNA levels in
RCC. Test of normality was conducted using Shapiro-Wilk test, as
well as histograms, P-P plots and Q-Q plots. P<0.05 was
considered to indicate a statistically significant difference. All
reported P-values are two-sided. Data were analysed using SPSS 23.0
(IBM Corp.).
Results
Patients
Demographic data and clinicopathological
characteristics of the patients are summarised in Table I. The mean age of the patients was
63.05±12.29 years at the time of surgery and the mean follow-up
time was 51.78±25.02 months. Histology revealed 33 cases of ccRCC,
4 cases of pRCC and 4 cases of chRCC. At the end of follow-up, 13
patients had died, of which 7 were renal cancer-related. The median
OS time was 64 months (IQR, 1-82).
 | Table IPatient demographic data and
clinicopathologic characteristics. |
Table I
Patient demographic data and
clinicopathologic characteristics.
| Characteristic | Value |
|---|
| Number of
patients | 41 |
| Body mass index (mean
± SD) | 28.26±4.86 |
| Age, years (mean ±
SD) | 63.05±12.29
(34-82) |
| ECOG performance
status (n, %) |
|
0 | 30 (73.2) |
|
1 | 10 (24.4) |
|
2 | 1 (02.4) |
| Sex (n, %) |
|
Male | 25 (61.0) |
|
Female | 16 (39.0) |
| Follow-up, months
(mean ± SD) | 51.78±25.02 |
| Smoking (n, %) |
|
Yes | 14 (34.1) |
|
Former | 16 (39.0) |
|
No | 11 (26.8) |
| Tumour size, cm (mean
± SD) | 6.00±2.78 |
| Nephrectomy (n,
%) |
|
Partial | 5 (12.2) |
|
Radical | 36 (87.8) |
| Tumour histology (n,
%) |
|
ccRCC | 33 (80.5) |
|
pRCC | 4 (9.8) |
|
chRCC | 4 (9.8) |
| Tumour pathological
stage (n, %) |
|
pT1a | 12 (29.3) |
|
pT1b | 13 (31.7) |
|
pT2a | 3 (7.3) |
|
pT2b | 3 (7.3) |
|
pT3a | 10 (24.4) |
| Fuhrman nuclear
grade RCC (n, %) |
|
1 | 0 (0.0) |
|
2 | 12 (29.3) |
|
3 | 18 (43.9) |
|
4 | 11 (26.8) |
| Tumor necrosis (n,
%) |
|
Yes | 20 (48.8) |
|
No | 21 (51.2) |
ccRCC
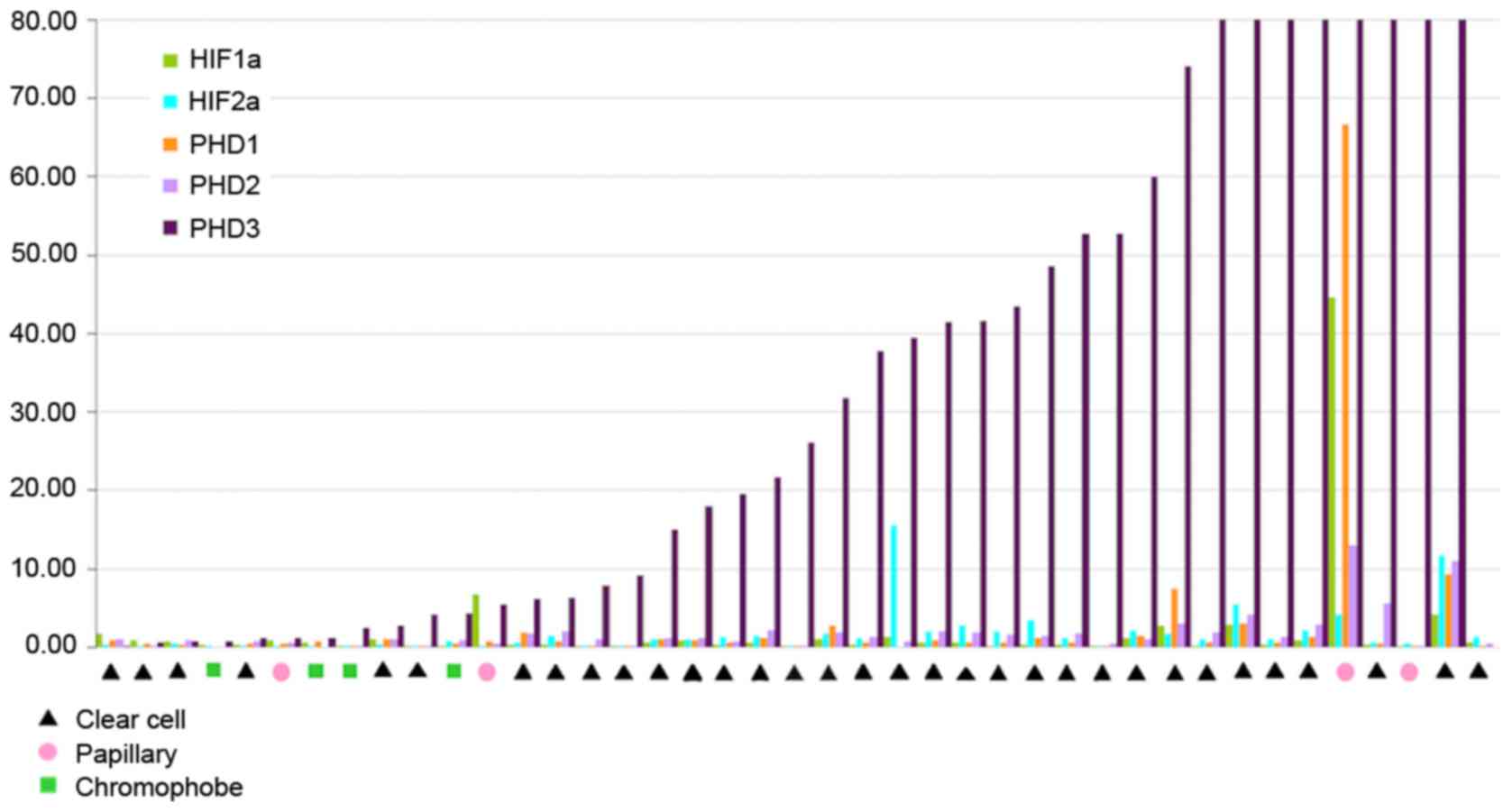
The results of RT-qPCR analysis for the relative
mRNA expression levels of the five genes in different tumours are
presented in Fig. 2. PHD3 mRNA
overexpression was recorded in 87.87% (29/33) of the ccRCC samples.
HIF-1α levels were decreased in 16/33 (48.48%) cases of ccRCC,
while HIF-2α did exhibit a specific pattern. In addition, PHD1 and
PHD2 transcript levels were similar in the majority of
patients.
HIF-1α was positively correlated with HIF-2α
(P=0.001), PHD1 (P<0.001) and PHD2 (P=0.035). HIF-2α levels were
significantly correlated with PHD1 (P<0.001), PHD2 (P<0.001)
and PHD3 (P<0.001), while PHD3 was significantly correlated with
PHD2 P<0.001).
chRCC and pRCC
PHD3 overexpression was observed in 2/4 of the chRCC
cases and in 3/4 of the pRCC cases (Fig. 2). Relative mRNA expression levels of
interrogated genes in different renal cell carcinomas are shown in
Table II.
 | Table IIRelative mRNA expression levels of
genes of interest. |
Table II
Relative mRNA expression levels of
genes of interest.
| Gene | Clear cell | Papillary | Chromophobe |
|---|
| HIF1a | 0.48
(0.28-0.95) | 3.79
(0.24-35.15) | 0.17
(0.07-0.26) |
| HIF2a | 1.17
(0.29-1.96) | 0.36
(0.16-3.26) | 0.42
(0.12-0.73) |
| PHD1 | 0.65
(0.43-1.19) | 0.63
(0.12-50.20) | 0.25
(0.05-0.46) |
| PHD2 | 1.34
(0.86-1.99) | 0.56
(0.26-9.89) | 0.48
(0.12-0.83) |
| PHD3 | 34.80
(6.69-58.19) | 59.41
(2.26-197.92) | 3.38
(2.48-4.30) |
Correlation of HIF-1a, HIF-2a, PHD1,
PHD2 and PHD3 expression levels with clinical parameters of ccRCC
samples
Relative mRNA expression levels of the analysed
genes were not associated with the majority of the clinical
parameters evaluated, including age, BMI, smoking history,
performance status, red blood cell count, haemoglobin, platelet
count, calcium, LDH and creatinine. The only positive correlation
identified was between PHD3 expression and pre-operative
haemoglobin level (P=0.032).
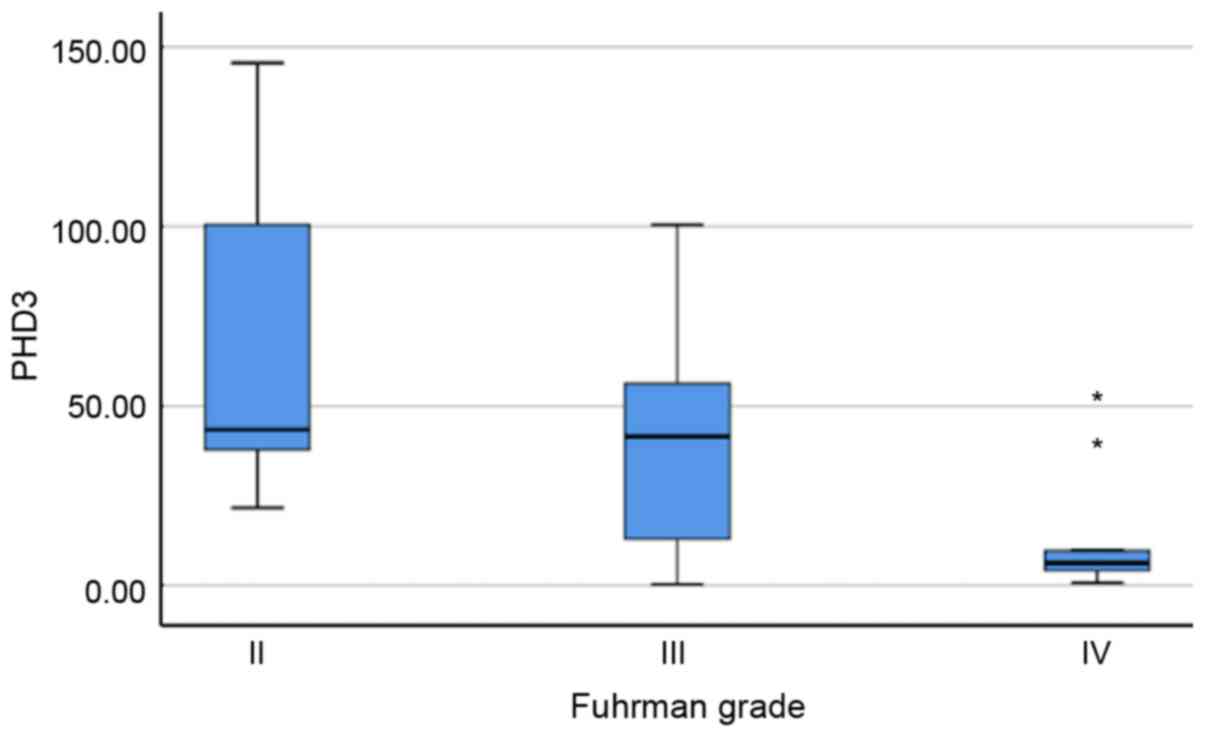
With regard to pathological parameters, PHD3
expression levels were inversely associated with Fuhrman grade
(P=0.015; Fig. 3). The median
relative expression level of PHD3 mRNA was 43.51 (31.93-123.09) in
grade II tumours, 41.53 (7.89-60.00) in grade III tumours and
significantly lower in grade IV tumours, which had a relative
expression level of 6.29 (3.43-24.59). No other association was
recorded between mRNA expression levels and tumour stage, presence
of necrotic or presence of sarcomatous elements.
All the aforementioned clinical and pathological
variables were evaluated for their prognostic value (Table III). Of these, stage, grade, serum
LDH, age and PHD2 mRNA expression demonstrated a statistically
significant correlation with OS in the univariate analysis.
 | Table IIIUnivariate Cox regression analysis
for overall survival. |
Table III
Univariate Cox regression analysis
for overall survival.
| Factor | HR | 95% CI | P-value |
|---|
| HIF1a | 0.39 | 0.10-1.44 | 0.158 |
| HIF2a | 0.98 | 0.81-1.20 | 0.863 |
| PHD1 | 0.56 | 0.22-1.48 | 0.244 |
| PHD2 | 0.46 | 0.22-0.99 | 0.049 |
| PHD3 | 0.99 | 0.97-1.01 | 0.449 |
| Stage | 2.45 | 1.32-4.54 | 0.004 |
| Grade | 4.41 | 1.73-11.25 | 0.002 |
| LDH | 1.01 | 1.00-1.01 | 0.042 |
| Calcium | 0.87 | 0.41-1.82 | 0.711 |
| PLT | 1.00 | 1.00-1.01 | 0.362 |
| Hb | 0.92 | 0.70-1.20 | 0.537 |
| RBC | 1.00 | 1.00-1.00 | 0.568 |
| ECOG performance
status | 2.13 | 0.88-5.15 | 0.093 |
| Age | 1.06 | 1.00-1.18 | 0.041 |
| Smoking
history | 2.12 | 0.96-4.66 | 0.062 |
Discussion
A key feature in the development of ccRCC, is the
inactivation of the tumour suppressor pVHL, which leads to
activation of the HIF pathway. Important regulators of HIF-α are
PHD1, PHD2 and PHD3, which mediate the degradation of HIF-1α and
HIF-2α. PHD3 has been shown to be the critical isoform that
demonstrates the most robust induction of expression under hypoxia,
while it is inactive in normoxic conditions (15,16).
Aside from its HIF-dependent functions, PHD3 has been suggested to
have the widest range of non-HIF targets and downstream effectors
(17). Recently, Högel et al
(18) demonstrated that PHD3
maintains cell growth and enhances cell cycle progression in renal
cancer by decreasing the stability of p27. Additionally, there is a
crucial involvement of PHD3 in the maintenance of key cellular
functions, including glycolysis and protein synthesis, in ccRCC.
PHD3 depletion significantly affects cellular processes associated
with glucose metabolism, post-transcriptional modification and
translation regulation in ccRCC cells (19).
In ccRCC, PHD3 is highly expressed. Sato et
al (20) reported that PHD3 is
frequently overexpressed in RCC tissue and demonstrated its
usefulness as a novel tumour antigen in immunotherapy for RCC
(20). In this previous study,
expression of PHD3 was quantified by RT-qPCR and immunostaining of
RCC cell lines and primary RCC tissues. Increased PHD3 mRNA
expression levels were observed in tumour tissues in 13/15 (87%)
cases, which is similar to the present result of 87.87%. The
previous study determined that PHD3 is selectively expressed in
cancerous tissue but not in non-cancerous tissue (20).
Tanaka et al (21) further confirmed the aforementioned
findings, demonstrating with RT-qPCR that PHD3 was overexpressed in
21/22 (95.4%) of the RCC tumour specimens (21). However, a notable finding of their
study was that PHD3 overexpression at the mRNA level was equally
observed in RCC tissues, with no associations with mutations or
epigenetic modifications of the VHL gene, which was also the case
in chromophobe and spindle cell carcinoma (21). An alternative mechanism that could
potentially explain this phenomenon is that tissue hypoxia may
induce PHD3 expression, as the catalytic activity of PHD3 is
oxygen-dependent, and hypoxia may induce accumulation of
unhydroxylated and undegraded HIF proteins (22).
Tanaka et al (23) also studied the mechanism and role of
PHD3 expression in RCC using RCC cell lines with and without VHL
gene mutation, as well as RCC tissues. High mRNA expression levels
of PHD3 were observed in both VHL-mutant cell lines and
VHL-wild-type cell lines. The aforementioned increased expression
levels were highly associated with activation of the PI3K/Akt
pathway in the VHL-wild-type RCC cell lines, independent of HIF
proteins (Fig. 1). On the other
hand, in the VHL-mutant RCC cell lines, PHD3 expression was more
strongly associated with HIF accumulation, likely due to inactive
VHL. The previous study also examined PHD3 expression by
immunohistochemistry in 116 patients with ccRCC who underwent
radical or partial nephrectomy between 2001 and 2009, and
correlation analysis was performed between the expression levels
and prognosis. PHD3-positive cells were observed in 82 of the 116
cases (70.7%), with the 5-year recurrence-free survival to be
statistically improved in patients with PHD3-positive tumours
compared with those with PHD3-negative tumours (P=0.003). However,
there was no significant difference in the cancer-specific survival
between the two groups (23).
The present mRNA expression data of five main
hypoxia genes in 33 cases are in agreement with the previous
studies. PHD3 mRNA expression levels in ccRCC were found to be
significantly high in comparison with other genes related to the
tumour hypoxia pathway.
In the ccRCC samples analysed in the current study,
PHD3 mRNA overexpression was inversely associated with nuclear
grade. To the best of our knowledge, this is the first study to
identify a correlation between PHD3 expression level and the
aggressiveness of tumour cells, suggesting a crucial involvement of
PHD3 in ccRCC. Kroeze et al (24) studied the same pathway by
immunohistochemistry of a tissue microarray containing tumours from
100 patients who underwent nephrectomy for ccRCC. Significant
correlations between Fuhrman grade and the expression levels of all
three PHD proteins were identified (PHD1, P=0.024; PHD2, P=0.0067;
PHD3, P=0.0012) (24). This
correlation was not confirmed for PHD1 and PHD2 in the present
study sample. However, both studies did not record any association
of PHD3 expression with survival outcomes. This may be due to the
small number of patients included in the current study. In the
study of Kroeze et al, nuclear factor-inhibiting HIF (FIH)
expression in the primary tumour exhibited a significant
independent prognostic value for patients with ccRCC in
multivariate analysis, suggesting that FIH may have an important
function as one of the final checks of HIF-α transcriptional
activity.
It is notable, that in ccRCC, the HIF pathway is
upregulated by inactivation of the pVHL tumour suppressor (25). Both HIF-1α and HIF-2α are important
regulators of angiogenesis and cell proliferation. In ccRCC, HIF-2α
has been reported to be involved in tumorigenesis and tumour
progression (24). Recent data from
Chen et al (26) and Wallace
et al (27) demonstrates
that HIF-2α can be specifically targeted by small molecule
antagonists, which may offer a novel approach for treating RCC. In
the present study, PHD3 mRNA levels were significantly associated
with HIF-2α levels and inversely associated with Fuhrman grade;
these findings may indicate that PHD3 potentially serves a negative
feedback role to decrease HIF-2α activity, representing a possible
therapeutic target in controlling HIF-2α. However, further
experiments are necessary to confirm the suggested correlation
between PHD3 and HIF-2α.
Finally, it would be negligent not to mention that
the present study has some limitations. The mutation status of the
VHL gene was not evaluated since extensive previous literature has
reported the absence of any correlation between VHL gene mutations
and clinicopathological parameters or survival of patients
(21,24). Additionally, the number of patients
included in the present study was small, but similar to other
studies in the field, which is expected given the low incidence of
renal carcinoma in the general population. However, the current
study presents, to the best of our knowledge, the largest cohort
for the evaluation of mRNA expression levels of five main
hypoxia-related genes by RT-qPCR and subsequent analyses.
In conclusion, the present study demonstrated that
PHD3 may play an important role in the valuable host molecular
adaptation that occurs during oxidative stress in ccRCC. Further
studies are required to validate the present results in view of
employing PHD3 as a prognostic and/or therapeutic target for
ccRCC.
Supplementary Material
Amplification plots of formalin-fixed
paraffin-embedded samples (tumour and normal) included in one run
for all markers. Samples with Ct values ≥36 were excluded from the
analysis.
Acknowledgements
The authors would like to thank Mrs. Aimilia
Daskalaki (Department of Pathology, Aristotle University of
Thessaloniki, School of Health Sciences, Faculty of Medicine,
Thessaloniki 54124, Greece) for providing support for reverse
transcription-quantitative PCR.
Funding
The present study was supported by the Hellenic
Urological Association.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SK, VK, VG and GD conceived and designed the study.
SK enrolled the patients, collected and interpreted patient data
and wrote the manuscript. VK supervised tissue processing, RNA
template preparation and RT-qPCR. VK and IV provided experimental
and clinical advice. VG contributed to the pathological review. SK,
IK, IV and SL analysed and interpreted the clinical and molecular
data. IV and SL helped to draft the manuscript. GD and VK
supervised the study and gave final approval of the version to be
published. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Gennimatas General Hospital, Aristotle University
of Thessaloniki, Greece (approval no. 18061/24-12-2013). Written
informed consent was obtained from all patients prior to
enrolment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sun M, Marconi L, Eisen T, Escudier B,
Giles RH, Haas NB, Harshman LC, Quinn DI, Larkin J, Pal SK, et al:
Adjuvant vascular endothelial growth factor-targeted therapy in
renal cell carcinoma: A systematic review and pooled analysis. Eur
Urol. 74:611–620. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kovacs G, Akhtar M, Beckwith BJ, Bugert P,
Cooper CS, Delahunt B, Eble JN, Fleming S, Ljungberg B, Medeiros
LJ, et al: The Heidelberg classification of renal cell tumours. J
Pathol. 183:131–133. 1997.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lane BR and Kattan MW: Prognostic models
and algorithms in renal cell carcinoma. Urol Clin North Am.
35:613–625, vii. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kimbro KS and Simons JW: Hypoxia-inducible
factor-1 in human breast and prostate cancer. Endocr Relat Cancer.
13:739–749. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Vaupel P, Kelleher DK and Höckel M: Oxygen
status of malignant tumors: Pathogenesis of hypoxia and
significance for tumor therapy. Semin Oncol. 28 (Suppl 8):29–35.
2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Maxwell PH, Wiesener MS, Chang GW,
Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER and
Ratcliffe PJ: The tumour suppressor protein VHL targets
hypoxia-inducible factors for oxygen-dependent proteolysis. Nature.
399:271–275. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Mandriota SJ, Turner KJ, Davies DR, Murray
PG, Morgan NV, Sowter HM, Wykoff CC, Maher ER, Harris AL, Ratcliffe
PJ and Maxwell PH: HIF activation identifies early lesions in VHL
kidneys: Evidence for site-specific tumor suppressor function in
the nephron. Cancer Cell. 1:459–468. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
De Luise M, Girolimetti G, Okere B,
Porcelli AM, Kurelac I and Gasparre G: Molecular and metabolic
features of oncocytomas: Seeking the blueprints of indolent
cancers. Biochim Biophys Acta Bioenerg. 1858:591–601.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gerard GF, D'Alessio JM, Kotewicz ML and
Noon MC: Influence on stability in Escherichia coli of the
carboxy-terminal structure of cloned Moloney murine leukemia virus
reverse transcriptase. DNA. 5:271–279. 1986.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kotewicz ML, D'Alessio JM, Driftmier KM,
Blodgett KP and Gerard GF: Cloning and overexpression of Moloney
murine leukemia virus reverse transcriptase in escherichia coli.
Gene. 35:249–258. 1985.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Marathe SV and McEwen JE: Vectors with the
gus reporter gene for identifying and quantitating promoter regions
in Saccharomyces cerevisiae. Gene. 154:105–107. 1995.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gourvas V, Sifakis S, Dalpa E, Soulitzis
N, Koukoura O and Spandidos DA: Reduced placental prolyl
hydroxylase 3 mRNA expression in pregnancies affected by fetal
growth restriction. BJOG. 117:1635–1642. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Aprelikova O, Chandramouli GV, Wood M,
Vasselli JR, Riss J, Maranchie JK, Linehan WM and Barrett JC:
Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors.
J Cell Biochem. 92:491–501. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rantanen K, Pursiheimo JP, Hogel H,
Miikkulainen P, Sundstrom J and Jaakkola PM: p62/SQSTM1 regulates
cellular oxygen sensing by attenuating PHD3 activity through
aggregate sequestration and enhanced degradation. J Cell Sci.
126:1144–1154. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jaakkola PM and Rantanen K: The
regulation, localization, and functions of oxygen-sensing prolyl
hydroxylase PHD3. Biol Chem. 394:449–457. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Högel H, Miikkulainen P, Bino L and
Jaakkola PM: Hypoxia inducible prolyl hydroxylase PHD3 maintains
carcinoma cell growth by decreasing the stability of p27. Mol
Cancer. 14(143)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Miikkulainen P, Hogel H, Rantanen K, Suomi
T, Kouvonen P, Elo LL and Jaakkola PM: HIF prolyl hydroxylase PHD3
regulates translational machinery and glucose metabolism in clear
cell renal cell carcinoma. Cancer Metab. 5(5)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sato E, Torigoe T, Hirohashi Y, Kitamura
H, Tanaka T, Honma I, Asanuma H, Harada K, Takasu H, Masumori N, et
al: Identification of an immunogenic CTL epitope of HIFPH3 for
immunotherapy of renal cell carcinoma. Clin Cancer Res.
14:6916–6923. 2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tanaka T, Kitamura H, Torigoe T, Hirohashi
Y, Sato E, Masumori N, Sato N and Tsukamoto T: Autoantibody against
hypoxia-inducible factor prolyl hydroxylase-3 is a potential
serological marker for renal cell carcinoma. J Cancer Res Clin
Oncol. 137:789–794. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Appelhoff RJ, Tian YM, Raval RR, Turley H,
Harris AL, Pugh CW, Ratcliffe PJ and Gleadle JM: Differential
function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the
regulation of hypoxia-inducible factor. J Biol Chem.
279:38458–38465. 2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tanaka T, Torigoe T, Hirohashi Y, Sato E,
Honma I, Kitamura H, Masumori N, Tsukamoto T and Sato N:
Hypoxia-inducible factor (HIF)-independent expression mechanism and
novel function of HIF prolyl hydroxylase-3 in renal cell carcinoma.
J Cancer Res Clin Oncol. 140:503–513. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kroeze SG, Vermaat JS, van Brussel A, van
Melick HH, Voest EE, Jonges TG, van Diest PJ, Hinrichs J, Bosch JL
and Jans JJ: Expression of nuclear FIH independently predicts
overall survival of clear cell renal cell carcinoma patients. Eur J
Cancer. 46:3375–3382. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Salama R, Masson N, Simpson P, Sciesielski
LK, Sun M, Tian YM, Ratcliffe PJ and Mole DR: Heterogeneous effects
of direct hypoxia pathway activation in kidney cancer. PLoS One.
10(e0134645)2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen W, Hill H, Christie A, Kim MS,
Holloman E, Pavia-Jimenez A, Homayoun F, Ma Y, Patel N, Yell P, et
al: Targeting renal cell carcinoma with a HIF-2 antagonist. Nature.
539:112–117. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wallace EM, Rizzi JP, Han G, When PM, Cao
Z, Du X, Cheng T, Czerwinski RM, Dixon DD, Goggin BS, et al: A
small-molecule antagonist of HIF2α is efficacious in preclinical
models of renal cell carcinoma. Cancer Res. 76:5491–5500.
2016.PubMed/NCBI View Article : Google Scholar
|