Introduction
Sentinel lymph node (SLN) biopsy is the standard
approach used for the locoregional staging of patients with
clinically T1-T2 invasive breast cancer (BC) with clinically
negative axillae (1).
Unfortunately, the conventional intraoperative histological
examination of frozen SLN sections has been associated with a
false-negative result rate of 10-30% for metastasis (2). To overcome this issue, an increasing
number of centers have adopted a novel molecular approach, namely
the one-step nucleic acid amplification (OSNA) assay (2-5).
OSNA is based on reverse transcription loop-mediated isothermal
amplification to quantify the content of tumor cells in the whole
SLN homogenate via evaluating the mRNA expression of cytokeratin
(CK)19 (2,3). The OSNA assay has several advantages,
as it assures the analysis of all SLNs and is a semi-quantitative,
reproducible, rapid and standardized method (2-5).
Another study supported the accuracy of the OSNA
assay for the staging of other types of cancer (6). A drawback of this approach is that
none of the tissue can be left for subsequent examination.
Consequently, it is impossible to carry out tissue structure
analysis or assessment of other biological markers (7-9).
Furthermore, several rare false-negative results may also occur in
cancers with decreased CK19 expression. In 2016, Martin-Sánchez
et al (10) suggested that
OSNA samples could be suitable for DNA molecular studies, including
the assessment of gene promoter methylation.
The aim of the present study was to verify whether
the OSNA-discarded samples could be used in gene expression
profiling studies of the SLN microenvironment in order to assess
host immunoinflammatory responses.
Materials and methods
Sample processing
Informed consent was obtained from all participants,
as recommended by the local Ethics Committee of the Coimbra
Hospital and University Centre (CHUC; Coimbra, Portugal) according
to the principles outlined in the Declaration of Helsinki (ethics
approval no. CHUC-045-20). The OSNA-remaining lysates from two
patients (samples 1 and 2) were randomly selected from samples
preserved at -80˚C at the Department of Pathology of CHUC. Both
patients suffered from stage I ductal invasive luminal A BC with a
clinically negative axilla. SLNs were identified through a
combination of techniques, using patent blue and radioisotopes or
superparamagnetic iron oxide, according to the established
department guidelines. After the extranodal tissue was removed, all
fresh SLNs were homogenized in 4 ml Lynorhag® solution
(Sysmex Corporation) using a Polytron® PT1300D
homogenizer (Kinematica AG). Briefly, 1 ml homogenate was
centrifuged at 10,000 x g for 1 min at room temperature and ~500 µl
of the intermediate phase were collected. A volume of 20 µl of the
intermediate phase was used for the OSNA assay utilizing the
Lynoamp™ BC kit on the RD-100i system (Sysmex
Corporation). The remaining volume was kept at -20˚C for subsequent
experiments.
Tsujimoto et al (3) determined the cut-off values for the
OSNA assay, suggesting that CK19 mRNA copies/µl <250 indicated
the absence of micrometastasis. Herein, based on the calculated
number of CK19 mRNA copies/µl, no metastasis was observed in either
SLN.
Furthermore, a total of 3 ml peripheral blood was
collected from a healthy volunteer in an EDTA tube, which served as
a positive control for the gene expression analysis. Subsequently,
peripheral blood mononuclear cells (PBMCs) were isolated by density
gradient centrifugation. Briefly, blood was slowly added to a
conical tube containing 3 ml Ficoll-Paque™ Plus solution (Cytiva)
using a polyethylene transfer pipet. The tube was then centrifuged
at 800 x g for 20 min at room temperature. Following
centrifugation, mononuclear cells at the interface were carefully
harvested and transferred into a 1.5-ml microtube. Subsequently,
half of the mononuclear cell suspension was centrifuged at 8,000 x
g for 5 min at room temperature, the supernatant was discarded, and
the pellet was resuspended in 300 µl NR buffer (NZYTech, Lda.)
supplemented with 1% β-mercaptoethanol (Sigma-Aldrich; Merck KGaA),
and preserved at -20˚C.
Total RNA was extracted from PBMCs using the NZY
Total RNA Isolation kit (NZYTech, Lda.), with a DNase
decontamination step, according to the manufacturer's
instructions.
The OSNA-remaining intermediate phase was subjected
to a DNA decontamination step, including the incubation of 87.5 µl
of the OSNA-remaining intermediate phase with 10 µl DNase and 2.5
µl digestion buffer for 15 min at room temperature. Subsequently, a
total of 100 µl of the aforementioned solution was subjected to an
RNA clean-up protocol using the NZY Total RNA Isolation kit.
Briefly, 350 µl NR buffer was added to the sample followed by
mixing. Then, the sample was supplemented with 250 µl 96% ethanol
followed by mixing using a pipette. The solution was then
transferred to a spin column placed in a 2-ml collection tube and
centrifuged at room temperature for 15 sec at ≥8,000 x g. The steps
were carried out according to the RNA extraction protocol. Finally,
30 µl RNAse-free water was added to the column to elute RNA. The
eluted RNA was quantified using a NanoDrop-1000 spectrophotometer
(Thermo Fisher Scientific, Inc.). RNA integrity and quality (IQ
score) were assessed using the Qubit™ RNA IQ Assay (Thermo Fisher
Scientific, Inc.) on a Qubit™ 4 Fluorometer (Invitrogen; Thermo
Fisher Scientific, Inc.).
Reverse transcription-quantitative PCR
analysis
The expression of three housekeeping control genes,
namely β2-microglobulin (B2M),
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and
ribosomal protein S18 (RPS18), and that of two genes
particularly expressed in lymphocytes, namely forkhead box P3
(FOXP3) and cluster of differentiation (CD4), were
analyzed. Total RNA from each sample (2 µg) was reverse-transcribed
into cDNA using the Omniscript RT kit (Qiagen GmbH) and random
hexamer primers in a 20-µl reaction volume according to the
manufacturer's instructions. The resultant cDNA was amplified using
the primers listed in Table I.
Amplification specificity was verified by Sanger sequencing. qPCR
was performed with a 20-µl reaction mixture containing 2 µl cDNA,
0.15 µM of each primer, and 1X iQM™ SYBR® Green Supermix
(Bio-Rad Laboratories, Inc.). Amplifications were carried out on
the CFX96 Touch Real-Time System (Bio-Rad Laboratories, Inc.) under
the following thermocycling conditions: One cycle at 95˚C for 5 min
followed by 40 cycles at 95˚C for 30 sec (denaturation), annealing
(the corresponding temperatures are listed in Table I) for 30 sec and 72˚C for 30 sec
(extension). To verify the absence of DNA contamination, RNA was
directly subjected to qPCR and no amplification curve was
detected.
 | Table IList of primer sequences used for
quantitative PCR analysis. |
Table I
List of primer sequences used for
quantitative PCR analysis.
| Gene (accession
no.) | Primer sequence
(5'→3') | Amplicon size
(bp) | Annealing T (˚C) |
|---|
| B2M
(NM_004048.4) | F:
GCATCATGGAGGTTTGAAGATG | 234 | 60 |
| | R:
TAAGTTGCCAGCCCTCCTAGAG | | |
| GAPDH
(NM_002046.7) | F:
AAGGTGAAGGTCGGAGTC | 229 | 56 |
| | R:
CCTGGAAGATGGTGATGG | | |
| RPS18
(NM_022551.3) | F:
GCAGACATTGACCTCACC | 207 | 56 |
| | R:
CTTCTTCAGTCGCTCCAG | | |
| CD4
(NM_001382706.1) | F:
CCATTTCTGTGGGCTCAGGT | 290 | 59 |
| | R:
TCAGCTTGGATGGACCTTTAGT | | |
| FOXP3
(NM_014009.4) | F:
CACATTTCATGCACCAGCTCT | 133 | 59 |
| | R:
TTGAGGGAGAAGACCCCAGT | | |
Statistical analysis
Three replicates were performed for each sample.
Data are presented as mean ± SD. Student's independent t-test was
used to compared mRNA concentrations and IQ scores. P<0.05 was
considered to indicate statistically significant differences. The
statistical package SPSS (version 19.0, IBM SPSS Statistics for
Windows; IBM Corp.) was used to perform the statistical
analysis.
Results
The RNA concentration, integrity and quality values,
as well as the RT-qPCR Cq values, are summarized in
Table II. Although the
concentrations of mRNA were statistically significantly different
among the three samples (P<0.01), values obtained for OSNA
sample 1 and the control sample (PBMCs) were very similar (1.2
difference). In addition, the high IQ values (7-8.8) indicated that
the samples mainly contained large or tertiary structured RNA
(>80%). Although the mRNA concentration of OSNA sample 2 was
more than 3 times higher compared with that of the other samples,
it exhibited a lower IQ score (P<0.05), indicating that this
sample consisted of a higher quantity of small and degraded RNA,
which was consistent with the Cq values obtained in the
RT-qPCR analysis. The RT-qPCR amplification plots are shown in
Fig. 1.
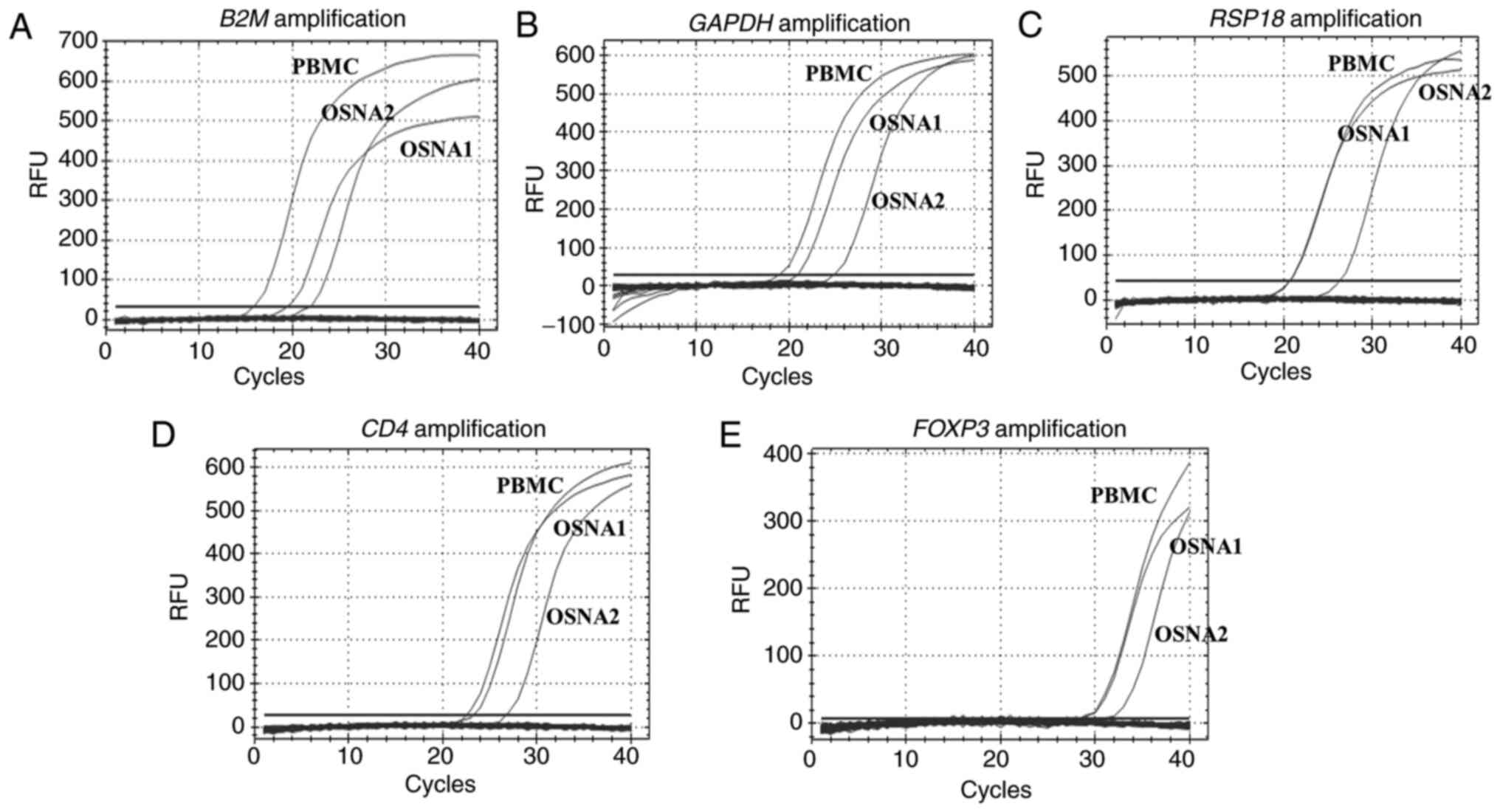 | Figure 1Quantitative PCR amplification curves
for (A) B2M, (B) GAPDH, (C) RPS18, (D)
CD4 and (E) FOXP3. OSNA, one-step nucleic acid
amplification; B2M, β2-microglobulin; GAPDH,
glyceraldehyde-3-phosphate dehydrogenase; RPS18, ribosomal
protein S18; CD4, cluster of differentiation 4;
FOXP3, forkhead box P3; PBMC, peripheral blood mononuclear
cell; RFU, relative fluorescence unit. |
 | Table IIRNA concentration, integrity and
quality and Cq values. |
Table II
RNA concentration, integrity and
quality and Cq values.
| Variables | OSNA 1 | OSNA 2 | PBMCs |
|---|
| RNA, mean ± SD | | | |
|
Concentration
(µg/µl) | 79.4±2.8 | 255±3.6 | 64.2±2.4 |
|
IQ
value | 8.8±0.21 | 7±0.25 | 8±0.31 |
| Cq from
qPCR | | | |
|
B2M | 19.35 | 21.83 | 15.75 |
|
GAPDH | 20.82 | 24.71 | 18.79 |
|
RPS18 | 20.72 | 26.34 | 20.70 |
|
CD4 | 21.83 | 26.12 | 21.78 |
|
FOXP3 | 29.47 | 32.20 | 29.30 |
Discussion
Lymph nodes are considered as the main escape route
for tumor cells from the primary site to other regions of the body.
Therefore, the evaluation of lymph nodes is crucial for the
prognosis of BC (1). Currently, the
OSNA assay is commonly used in clinical practice to detect macro-
and micrometastasis in early-stage BC with clinically negative
axillae (4,5). In addition, other potential
applications of the OSNA assay are under investigation (6). However, this diagnostic method, in its
current form, cannot be used in SLN microstructural studies, and
these studies could provide useful information regarding immune
responses and tumor aggressiveness (7-9).
Therefore, improving the prognostic value of OSNA may have a major
impact on the risk stratification of patients with cancer.
The present study suggested that OSNA-discarded
samples may be suitable for further gene expression analyses of the
SLN microenvironment. RNA quality and integrity play a key role in
qPCR experiments (11). The results
of the present study demonstrated that the IQ score for one of the
RNA samples was higher compared with the control sample (PBMCs). It
is often impossible to design RNA-specific primers for gene
expression profiling studies. Therefore, it would be useful to
include a DNA decontamination step, as was performed in the present
study. Furthermore, the RT-qPCR analysis results further verified
the feasibility of this method.
It has been suggested that a thorough selection of
reference genes for the normalization of gene expression is
crucial. Therefore, according to the current Minimum Information
for Publication of Quantitative Real-Time PCR Experiment
guidelines, the use of more than one reference gene is recommended
for all qPCR analyses (12).
Herein, three commonly used housekeeping genes, namely
RP18S, GAPDH and B2M, were assessed. To
perform quantification of gene expression, namely using the
2-∆∆Cq method (13),
experiments with OSNA-positive and OSNA-negative samples must be
performed to select the most suitable reference genes.
In addition to evaluating metastasis, SLNs are also
considered as natural targets for studying tumor-immune system
interactions. Therefore, in the present study, two genes, namely
CD4 and FOXP3, were analyzed. CD4 is a glycoprotein
expressed on the surface of immune cells, such as T helper cells,
monocytes, macrophages and dendritic cells. In addition, CD4 is
highly expressed in PBMCs and lymph nodes (13). FOXP3 is a regulatory T-cell
lineage-specific transcription factor, consequently exhibiting
reduced expression in PBMCs (14,15).
Martin-Sánchez et al (10) demonstrated that the OSNA-remaining
homogenate could be used in DNA-based studies, particularly in
methylation analysis. The authors revealed an association between
the hypermethylation of the Ras association domain family member 1
gene and macrometastasis, micrometastasis and the number of
isolated tumor cells in BC SLNs, suggesting that the prognostic
value of the OSNA assay could be improved.
Although the study of the SLN microenvironment using
an OSNA assay is not feasible during the surgical procedure, it may
provide important information regarding tumor-immune system
interactions that could previously only be partially assessed by
standard pathological evaluation (7-9).
This technical report was not designed to search for
new markers and no comparisons were established between
OSNA-positive and -negative patients, or regarding pathological or
clinical characteristics. Furthermore, no comparisons were
performed between gene expression levels in the three samples. The
control that was used (PBMCs from a healthy donor) served as a
positive control to access the feasibility of determining mRNA
expression from OSNA samples. Lymph nodes comprise a complex cell
population, different from PBMCs, and may not exhibit the same gene
expression profile.
This new approach may help provide a combined test
identifying both metastasis and new prognostic markers associated
with immunoinflammatory response. We are currently performing
further studies to support this hypothesis.
The results of the present preliminary study
demonstrated that residual OSNA lysates could be used for further
gene expression analysis, suggesting that this material could be
employed as a bank of biological molecules for identifying novel
biomarkers associated with the interplay between cancer and immune
responses. However, future prospective studies should be performed
to further evaluate the association between the gene expression
profile and prognosis of patients with BC.
Acknowledgements
Not applicable.
Funding
The present study was funded by GenomePT-National Laboratory for
Genome Sequencing and Analysis (grant no.
POCI-01-0145-FEDER-022184) and national funds through the FCT –
Foundation for Science and Technology, within the scope of the
project UIDB/04539/2020.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HCS, MFD and IG conceived and designed the study; CR
and HCS performed the RT-qPCR analysis; AG performed the OSNA
assay; HCS and CR analysed the data and confirm the authenticity of
all the raw data; CR and IG, with the participation of all other
authors, wrote the original draft of the manuscript; CR, MFD and
HCS reviewed and edited the manuscript. All the authors have read
and approved that final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from the participants
or their legal representatives (CHUC-045-20), as recommended by the
local Ethics Committee, following the tenets of the Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cardoso F, Kyriakides S, Ohno S,
Penault-Llorca F, Poortmans P, Rubio IT, Zackrisson S and Senkus E:
ESMO Guidelines Committee. Electronic address: simpleclinicalguidelines@esmo.org:.
Early breast cancer: ESMO Clinical Practice Guidelines for
diagnosis, treatment and follow-up+. Ann Oncol.
30:1194–1220. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jimbo K, Kinoshita T, Suzuki J, Asaga S,
Hojo T, Yoshida M and Tsuda H: Sentinel and nonsentinel lymph node
assessment using a combination of one-step nucleic acid
amplification and conventional histological examination. Breast.
22:1194–1199. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tsujimoto M, Nakabayashi K, Yoshidome K,
Kaneko T, Iwase T, Akiyama F, Kato Y, Tsuda H, Ueda S, Sato K, et
al: One-step nucleic acid amplification for intraoperative
detection of lymph node metastasis in breast cancer patients. Clin
Cancer Res. 13:4807–4816. 2007.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Banerjee SM, Michalopoulos NV, Williams
NR, Davidson T, El Sheikh S, McDermott N, Tran-Dang MA, Davison S
and Keshtgar MR: Detailed evaluation of one step nucleic acid
(OSNA) molecular assay for intra-operative diagnosis of sentinel
lymph node metastasis and prediction of non-sentinel nodal
involvement: Experience from a London teaching hospital. Breast.
23:378–384. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hunter-Smith AE and Rayter Z: One-step
nucleic acid amplification: The possible value in assessing
sentinel lymph node metastasis during mastectomy. Breast Cancer
(Dove Med Press). 10:13–21. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Brito MJ, Honavar M, Cipriano MA, Lopes J,
Coelho H, Silva AR, Silva M, Guimarães S, Frutuoso A, Gomes A, et
al: Molecular staging of patients with colon cancer. The
C-Closer-II Study: A multicentre study in Portugal. Acta Med Port.
31:661–669. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gibert-Ramos A, López C, Bosch R, Fontoura
L, Bueno G, García-Rojo M, Berenguer M and Lejeune M: Immune
response profile of primary tumour, sentinel and non-sentinel
axillary lymph nodes related to metastasis in breast cancer: An
immunohistochemical point of view. Histochem Cell Biol.
152:177–193. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Grigoriadis A, Gazinska P, Pai T, Irhsad
S, Wu Y, Millis R, Naidoo K, Owen J, Gillett CE, Tutt A, et al:
Histological scoring of immune and stromal features in breast and
axillary lymph nodes is prognostic for distant metastasis in lymph
node-positive breast cancers. J Pathol Clin Res. 4:39–54.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Shiota T, Miyasato Y, Ohnishi K,
Yamamoto-Ibusuki M, Yamamoto Y, Iwase H, Takeya M and Komohara Y:
The Clinical Significance of CD169-Positive lymph node macrophage
in patients with breast cancer. PLoS One.
11(e0166680)2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Martín-Sánchez E, Pernaut-Leza E, Mendaza
S, Cordoba A, Vicente-Garcia F, Monreal-Santesteban I, Vizcaino JP,
De Cerio MJ, Perez-Janices N, Blanco-Luquin I, et al: Gene promoter
hypermethylation is found in sentinel lymph nodes of breast cancer
patients, in samples identified as positive by one-step nucleic
acid amplification of cytokeratin 19 mRNA. Virchows Arch.
469:51–59. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gallego Romero I, Pai AA, Tung J and Gilad
Y: RNA-seq: Impact of RNA degradation on transcript quantification.
BMC Biol. 12(42)2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fagerberg L, Hallström BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lu L, Barbi J and Pan F: The regulation of
immune tolerance by FOXP3. Nat Rev Immunol. 17:703–717.
2017.PubMed/NCBI View Article : Google Scholar
|















