Introduction
The presence of epidermal growth factor receptor
(EGFR) mutations determines the efficacy of EGFR tyrosine kinase
inhibitor (EGFR-TKI) therapy in patients with non-small cell lung
cancer (NSCLC). EGFR-TKIs are used as the first-line therapy for
advanced NSCLC cases with EGFR mutations (1-4).
Therefore, detecting these mutations is crucial for patients with
NSCLC. Previous clinical studies have demonstrated favorable
outcomes of patients with common mutations receiving EGFR-TKI
therapy, such as those with selected exon 19 deletions and a L858R
point mutation in exon 21 (1-4).
In EGFR exons 18-21, the frequency of common mutations is 90% among
all EGFR mutations. The remaining 10% includes uncommon mutations,
which are a heterogeneous group of molecular alterations (4-6).
The variant mutations consist of >400 types, which were
registered in Catalogue of Somatic Mutation in Cancer database
version 84 in 2018(6). The
response to EGFR-TKIs in cases with uncommon mutations has been
debated, but remains largely unknown at present.
PCR methods are used clinically for the detection of
EGFR mutations. Two widely used PCR methods have been approved as
in vitro diagnostic methods, namely the Scorpion
Amplification Refractory Mutation System (Scorpion-ARMS; Qiagen
therascreen® EGFR; Qiagen, Inc.) and the
cobas® EGFR Mutation Test v2 (cobas v2; Roche
Diagnostics). These methods are beneficial for clinical use due to
their short detection time and cost-effectiveness. However, these
PCR methods only selectively detect certain EGFR mutations, such as
exon 19 deletions and a L858R point mutation in exon 21, and not
others (7,8). Therefore, the aim of the present
study was to identify the frequency of the mutations not detected
by these PCR methods. To achieve this aim, the current study
evaluated EGFR mutations using PCR methods and direct
sequencing.
Patients and methods
Patients
The present study included 70 Japanese patients with
NSCLC, from whom informed consent was obtained for the use of their
samples in this research. These patients underwent surgical lung
resection at the Tokyo Medical University Ibaraki Medical Center
(Ami, Japan) between September 2016 and March 2019 (Table I). A total of 61.4% of the patients
were male and 38.6% were female. The patient age ranged between 42
and 84 years. PCR methods and direct sequencing were performed to
analyze EGFR status in the surgical samples. EGFR-TKIs were
administered to the patients who developed NSCLC recurrence after
surgery. The protocol of the present study received ethics approval
from the Institutional Review Board of Tokyo Medical University
(Tokyo, Japan; approval no. 17-56).
 | Table IBackground information of the 70
patients with non-small cell lung cancer. |
Table I
Background information of the 70
patients with non-small cell lung cancer.
| Variables | Total, n (%) |
|---|
| Age, years [median
(range)] | 71 (42-84) |
| Sex, male/female | 43 (61.4)/27
(38.6) |
| Histology,
ADC/SCC/LCC/PC | 61 (87.1)/6 (8.6)/1
(1.4)/2 (2.9) |
| Pathological stage,
0/I/II/III/IV | 5 (7.1)/41 (58.6)/14
(20.0)/5 (7.2)/5 (7.1) |
| Smoking status,
never/former + current | 26 (37.1)/44
(62.9) |
PCR method as the first EGFR mutation
testing method
All 70 surgical samples were formalin-fixed and
paraffin-embedded (FFPE). This PCR method was performed with cobas
v2, which is an allele-specific real-time PCR system used to
identify 42 types of EGFR nucleotide mutations in exons 18-21 using
FFPE NSCLC tissues. According to the package insert, cobas v2
requires ≥3.13 ng DNA, which includes 5% mutated DNA, to detect the
mutation. DNA isolation, amplification/detection and result
reporting can be performed in <8 h, with up to 30 specimens
processed simultaneously (7). DNA
was subjected to cobas v2 at SRL, Inc.
DNA extraction and direct sequencing
analysis as the second EGFR testing method
Direct sequencing was performed to further analyze
the presence of EGFR mutations in the FFPE surgical samples
determined as wild-type EGFR by cobas v2. Tumor cells were
harvested via macrodissection, and DNA was extracted from tumor
cells using a DNeasy Tissue kit (Qiagen, Inc.). PCR was performed
using a primer set flanking exons 18-21 as the TKI domain. PCR
products of different sizes were treated with ExoSAP-IT™ PCR
Product Cleanup (Thermo Fisher Scientific, Inc.) according to the
manufacture's protocol. The PCR primers were as fllows: Exon 18:
Forward, 5'-TGAGGTGACCCTTGTCTCTG-3' and reverse,
5'-CCTGTGCCAGGGACCTTAC-3' (180 nucleotides); exon 19: Forward,
5'-ATGTGGCACCATCTCACAAT-3' and reverse, 5'-CCACACAGCAAAGCAGAAAC-3'
(178 nucleotides); exon 20: Forward, 5'-ATGCGAAGCCACACTGAC-3' and
reverse, 5'-CCTTCCCTGATTACCTTTGC-3' (238 nucleotides); and exon 21:
Forward, 5'-GCAGGGTCTTCTCYGTTTCA-3' and reverse,
5'-TGACCTAAAGCCACCTCCTT-3' (199 nucleotides). These kits were used
according to the manufacturer's instructions. Sequencing analysis
was performed using an Applied Biosystems™ 3130xl Genetic Analyzer
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol.
PCR method as the third EGFR mutation
testing method
Scorpion-ARMS, which is a real time-PCR assay that
combines the ARMS and Scorpions fluorescent primer/probe system,
was performed to further validate the samples shown as ‘wild-type’
on cobas v2 but ‘EGFR mutations’ on direct sequencing.
Scorpion-ARMS can detect 29 somatic mutations in exons 18-21 of
EGFR. The limit of detection (LOD) is ~1% (8). DNA was subjected to Scorpion-ARMS at
SRL, Inc.
EGFR-TKI treatment for NSCLC
recurrence after surgery
EGFR-TKIs were administered to patients who
developed recurrence after surgery. The determination of the
efficacy of EGFR-TKI therapy was based on the Response Evaluation
Criteria in Solid Tumors (RECIST) (9) using CT, and Positron Emission
Tomography (PET) Response Criteria In Solid Tumors (PERCIST)
(10) using fluorodeoxyglucose
(FDG) uptake on PET/CT. The cases with partial or complete response
were confirmed through changes of tumor measurements, according to
RECIST. The cases with complete metabolic response were confirmed
using the 100% standardized uptake value (SUV) normalized to lean
body mass (LBM) decline, according to PERCIST. In PERCIST, the
LBM-corrected SUV (SUL) is proposed as a more appropriate
quantitative method. Fat contributes to body weight (BW), and the
accumulation of FDG and the SUV are affected by BW. The SUL was
calculated according to the Janmahasatian formula (11). Stable disease was defined as
fulfilling the criteria for ≥8 weeks following the start of
EGFR-TKI therapy. Patients receiving treatment for >4 weeks were
considered as assessable, and patients who had received EGFR-TKI
therapy for <4 weeks were considered as non-assessable.
Results
EGFR mutation status
The first PCR method, cobas v2, detected 29
mutations among all 70 patients with NSCLC. The remaining 41
patients were identified as NSCLC with EGFR wild-type by cobas v2,
among whom direct sequencing as the second EGFR mutation testing
method detected mutations in 3 patients (Table II). These 3 cases were validated
using Scorpion-ARMS as the third EGFR mutation testing method.
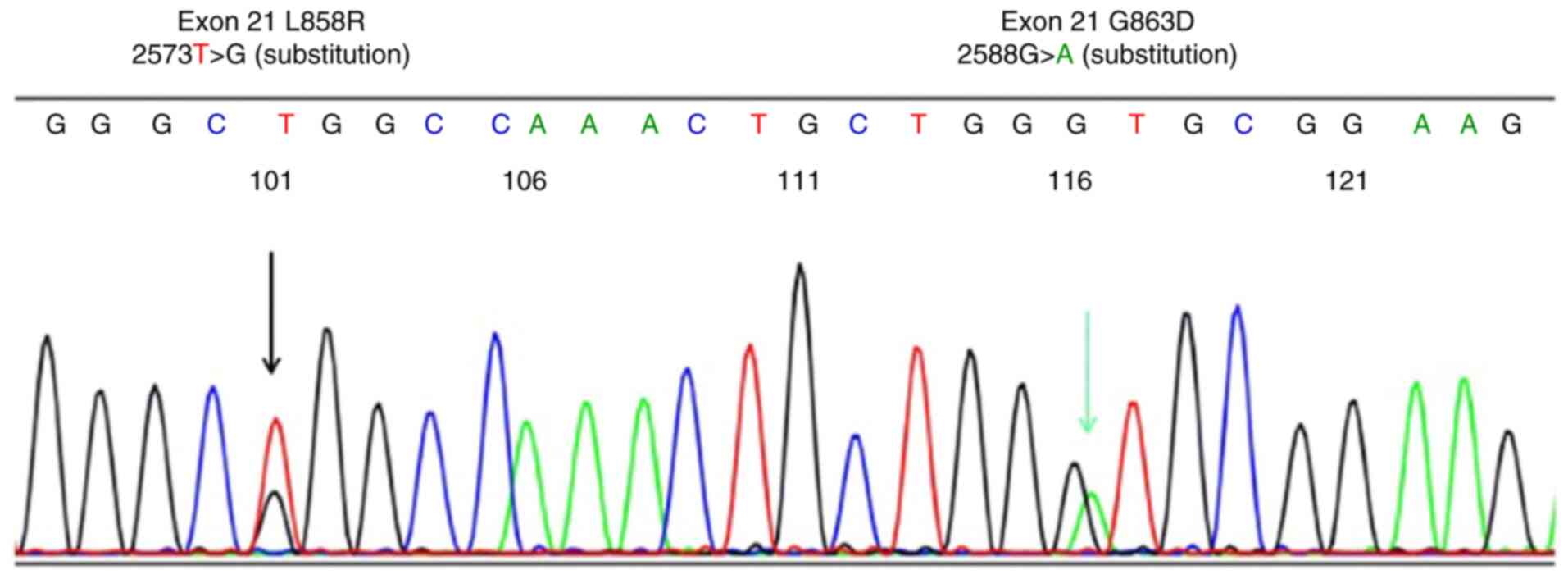
L858R + G863D compound mutations in direct sequencing (Fig. 1) were identified as a L858R single
mutation, and the others were identified as wild-type by
Scorpion-ARMS. Thus, two mutations were undetectable by both PCR
methods (Table II).
 | Table IIInformation on 32 EGFR mutations. |
Table II
Information on 32 EGFR mutations.
| Exon/amino acid | No. | cobas v2 | Direct
sequencing | Scorpion ARMS |
|---|
| Exon 18 | | | | |
|
G719X | 1 | 1 | | |
| Exon 19 deletion | 12 | 12 | | |
| Exon 20 | | | | |
|
H773_V774
ins TH | 1 | Wild-type | 1 | Wild-type |
| Exon 21 | | | | |
|
L833V +
H835L | 1 | Wild-type | 1 | Wild-type |
|
L858R | 14 | 14 | | |
|
L858R +
L861Q | 1 | 1 | | |
|
L858R +
G863D | 1 | Wild-type | 1 | L858Ra |
|
L861Q | 1 | 1 | | |
| Total EGFR
mutations | 32 | 29 | 3 | 1 |
EGFR-TKI treatment for NSCLC
recurrence after surgery
Among the 32 patients with EGFR mutations, EGFR-TKIs
were administered to 3 patients with postoperative recurrence. In
total, one patient with an exon 19 deletion detected by cobas v2
partially responded to osimertinib (third-generation EGFR-TKI),
according to RECIST. Another patient with exon 21 L858R + L861Q
detected by cobas v2 was treated with gefitinib (first-generation
EGFR-TKI) but was not deemed as assessable due to the development
of an adverse event (interstitial lung disease). The other patient
with exon 21 L858R + G863D detected by direct sequencing responded
to afatinib (second-generation EGFR-TKI); this was the case shown
as ‘wild-type’ on cobas v2 but ‘EGFR mutation’ on direct
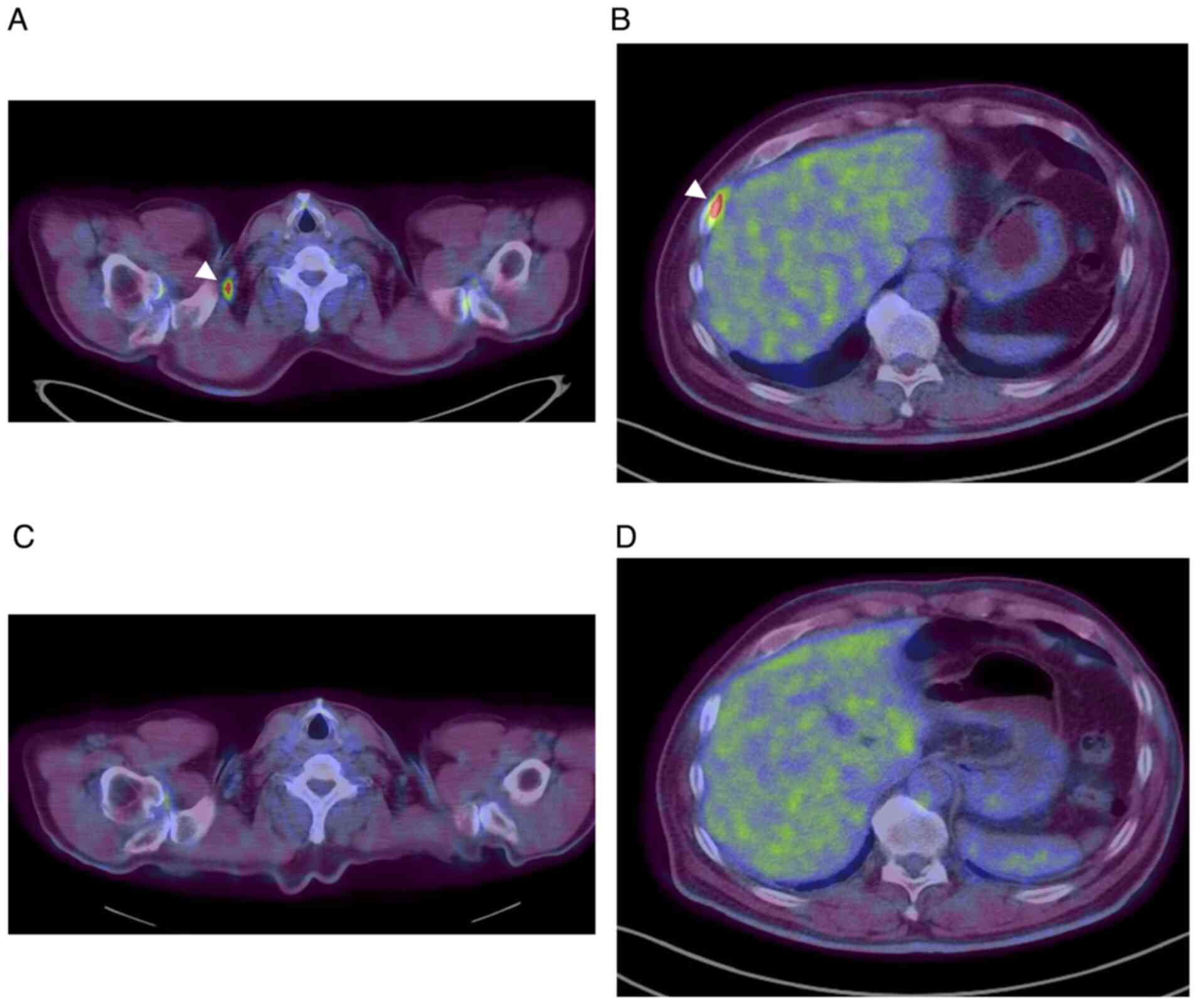
sequencing. At recurrence, SUL was >5.0 in the right
supraclavicular lymph node and the right pleural dissemination, as
detected via PET/CT (Fig. 2A and
B). Moreover, the level of
carcinoembryonic antigen (CEA), a tumor marker of lung
adenocarcinoma, was increased to 28.3 ng/ml (normal reference value
<5.0 ng/ml). After EGFR-TKI therapy, a complete metabolic
response was observed, as evident by the FDG uptake on PET/CT,
according to PERCIST (Fig. 2C and
D), and CEA levels decreased to
4.3 ng/ml, which was within the normal range.
Discussion
Previous studies have shown that PCR methods are
unable to detect 10% of all EGFR mutations (4-6).
In the present study, the rate of undetectable mutations was 9.4%
(3/32) when using cobas v2, and this result was similar to that
reported by previous studies (4-6).
Among the undetectable mutations with cobas v2, the L858R + G863D
compound mutation was detected by direct sequencing, and was
identified as a single L858R mutation by Scorpion-ARMS; this was a
patient with invasive adenocarcinoma, a cancer type that was solid
enough to be analyzed by PCR methods. It was found that L858R was
selected in cobas v2 and Scorpion-ARMS methods, while G863D was not
selected in either method. It was suggested that the detection of
L858R depended on the difference in the LOD between cobas v2 and
Scorpion-ARMS. Moreover, the LOD of L858R in cobas v2 was 4.0-5.3
when using the cobas® EGFR Mutation Test v2 for in
vitro diagnostic use, as per the description provided by Roche
Diagnostics, whereas that in Scorpion-ARMS was 1.26, as shown in
the therascreen® EGFR RGQ PCR Kit Handbook from Qiagen,
Inc.
The reason why only Scorpion-ARMS could detect L858R
in this case was likely because the LOD of L858R in Scorpion-ARMS
was lower than that in cobas v2.
Mutations that were detected by direct sequencing
but not by Scorpion-ARMS in the present study, including H773_V774
ins TH in exon 20, L833V, H835L and G863D in exon 21, were not
selected by Scorpion-ARMS, which is why this method was unable to
detect these mutations. The cases with H773_V774 ins TH in exon 20
and L833V + H835L compound mutation in exon 21 did not develop
recurrence. Thus, the efficacy of EGFR-TKIs against these mutations
was not assessable.
Recently, next generation sequencing (NGS) has been
used for the comprehensive detection of gene mutations. The types
of selected genes and mutations depend on each NGS package.
Moreover, NGS is not a type of whole genome or exon sequencing
(12). FoundationOne CDx is one of
the most widely used NGS methods, and its LOD for EGFR mutations is
2.4-5.1%, as per the ‘Technical Information’ description provided
by Foundation Medicine, Inc.
In a previous study, the frequency of compound
mutations, including L858R, was found to be higher than other types
of mutations, and compound mutations have properties that may be
associated with poor clinical outcome compared with simple
mutations (13). The present study
identified two compound mutations, including L858R, and one that
was detected by direct sequencing was identified as a single L858R
mutation by Scorpion-ARMS (Table
II). It was suggested that some cases with a single L858R
mutation identified by selective PCR methods may harbor a compound
mutation containing L858R. Furthermore, a previous study revealed
that NSCLC cases with tumors harboring exon 19 deletions had a
longer survival compared with those harboring L858R during EGFR-TKI
therapy (4). In this previous
study, the response in L858R cases may include those with a L858R
compound mutation.
A previous post hoc analysis combining phase III
studies suggested that second-generation EGFR-TKIs were effective
in patients with uncommon mutations, particularly those involving
point mutations or duplications in exons 18-21(5). Cases with uncommon mutations in the
present study, including undetectable mutations by cobas v2, such
as the L858R + G863D compound mutation, showed a response to
second-generation EGFR-TKI therapy. In the present study, among 32
patients with EGFR mutations, 3 patients received EGFR-TKI
treatment for recurrence after surgery. Of those 3 patients, 1
patient partially responded to osimertinib; 1 patient was treated
with gefitinib, but was not assessable due to the adverse events;
and 1 patient responded to afatinib. The statistical significance
of findings such as overall response in the present study was
difficult to analyze due to the limited sample size.
The results of the present study suggested that EGFR
testing using a selective method may result in patients with
uncommon mutations, including those with compound mutations,
missing the opportunity to receive EGFR-TKI therapy. As there are
numerous variants of EGFR mutations, it is difficult to evaluate
the response rate to EGFR-TKI therapy, with regards to each
uncommon mutation. However, there is the possibility that cases
with these mutations will respond to treatment. However, direct
sequencing, including that of whole exons 18-21, is not appropriate
for clinical use in all patients with NSCLC, due to the long
detection time and high cost. Thus, a suitable screening test
should be compatible with diverse EGFR mutations, including
compound mutations, and have a lower LOD for EGFR-TKI therapy.
In conclusion, the present study demonstrated that
certain mutations that were not selected by PCR methods were
detectable by direct sequencing, including cases of mutations that
responded to EGFR-TKI therapy. A suitable screening test should be
compatible with diverse EGFR mutations, in order to avoid losing
the opportunity of patients with uncommon mutations to receive
EGFR-TKI therapy.
Acknowledgements
Not applicable.
Funding
Funding: The authors' department received research grants from
Boehringer-Ingelheim, Chugai Pharmaceutical, Astra Zeneca, Pfizer,
Taiho Pharmaceutical, MSD, Eli Lilly, Ono Pharmaceutical, Teijin
and Nihon Mediphysics.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
EN and NI conceived and designed the study; TMa, HN
and TMi performed analysis of DNA data; SO, IT and KF performed the
interpretation of clinical data; YM performed the pathological
diagnosis; TMa, EN and TO performed the interpretation of DNA data.
EN and TMa confirm the authenticity of the raw data. All authors
have participated in the writing of the manuscript. All authors
have read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study obtained ethics approval from the
Institutional Review Board of Tokyo Medical University (approval
no. 17-56), and patients provided written consent to
participate.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests and they had no association with the promotion of PCR
methods or EGFR-TKI therapy. The author's department received
research grants from Boehringer-Ingelheim, Chugai Pharmaceutical,
Astra Zeneca, Pfeizer, Taiho Pharmaceutical, MSD, Eli Lilly, Ono
Pharmaceutical, Teijin and Nihon Mediphysics.
References
|
1
|
Yoshioka H, Shimokawa M, Seto T, Morita S,
Yatabe Y, Okamoto I, Tsurutani J, Satouchi M, Hirashima T, Atagi S,
et al: Final overall survival results of WJTOG3405, a randomized
phase III trial comparing gefitinib versus cisplatin with docetaxel
as the first-line treatment for patients with stage IIIB/IV or
postoperative recurrent EGFR mutation-positive non-small-cell lung
cancer. Ann Oncl. 30:1978–1984. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa
K, Niho S, Tsuji F, Linke R, Rosell R, Corral J, et al: Dacomitinib
versus gefitinib as first-line treatment for patients with
EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): A
randomized, open-label, phase 3 trial. Lancet Oncol. 18:1454–1466.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Soria JC, Ohe Y, Vansteenkiste J,
Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura
F, Nogami N, Kurata T, et al: Osimertinib in untreated EGFR-mutated
advanced non-small cell lung cancer. N Engl J Med. 378:113–125.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yang JC, Wu YL, Schuler M, Sebastian M,
Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, et al:
Afatinib versus cisplatin-based chemotherapy for EGFR
mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6):
Analysis of overall survival data from two randomized, phase 3
trials. Lancet Oncol. 16:141–151. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yang JC, Sequist LV, Geater SL, Tsai CM,
Mok TS, Schuler M, Yamamoto N, Yu CJ, Ou SI, Zhou C, et al:
Clinical activity of afatinib in patients with advanced
non-small-cell lung cancer harbouring uncommon EGFR mutations: A
combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung
6. Lancet Oncl. 16:830–838. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Nakajima E, Sugita M, Furukawa K,
Takahashi H, Uchida O, Kawaguchi Y, Ohira T, Matsubayashi J, Ikeda
N, Hirsch FR and Franklin WA: Frequency and significance of
epidermal growth factor receptor mutations detected by PCR methods
on patients with non-small cell lung cancer. Oncol Lett.
17:5125–5131. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kimura H, Ohira T, Uchida O, Matsubayashi
J, Shimizu S, Nagao T, Ikeda N and Nishio K: Analytical performance
of the cobas EGFR mutation assay for Japanese non-small-cell lung
cancer. Lung Cancer. 83:329–333. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Nicola T, Stephen M, Antonio S, James B
and Tom B: Mode of action and application of Scorpion primers to
mutation detection. Nucleic Acids Res. 28:3752–3761.
2000.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumors:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Richard L, Heather J, Yvette K and Martin
A: From RECIST to PERCIST: Evolving considerations for PET response
criteria in solid tumors. J Nucl Med. 50 (Suppl 1):122S–150S.
2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Tahari AK, Chien D, Azadi JR and Wahl RL:
Optimum lean body formulation for correction of standardized uptake
value in PET imaging. J Nucl Med. 55:1481–1484. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Umbarger MA, Kennedy CJ, Saunders P,
Breton B, Chennagiri N, Emhoff J, Greger V, Hallam S, Maganzini D,
Micale C, et al: Next-generation carrier screening. Genet Med.
16:132–140. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kim EY, Cho EN, Park HS, Hong JY, Lim S,
Youn JP, Hwang SY and Chang YS: Compound EGFR mutation is
frequently detected with co-mutations of actionable genes and
associated with poor clinical outcome in lung adenocarcinoma.
Cancer Biol Ther. 17:237–245. 2016.PubMed/NCBI View Article : Google Scholar
|