Introduction
Malignant melanoma (MM) is a rare refractory tumor
that originates from melanocyte stem cells and is associated with
aggressive behavior and poor prognosis. MM is divided into two
categories according to the primary lesions, mucosal malignant
melanoma (mMM) and cutaneous malignant melanoma (cMM) (1,2),
which differ in epidemiology and etiology. While cMM is more common
among Caucasians, mMM is relatively more common among Asians.
Ultraviolet light exposure is a major risk factor for cMM but not
for mMM, and mMM develops in later life stages than cMM.
Investigation of mMM has been difficult due to the rarity of this
disease and little is known about its pathogenesis and genetics.
Understanding the physiology of melanocyte stem cells may be
central to elucidating the carcinogenesis of MM.
The transmembrane protein collagen type XVII (COL17)
is mainly expressed in epidermal basal keratinocytes and is a
structural component of the hemidesmosome, an adhesion complex at
the epidermal-dermal junction. COL17 is involved in skin diseases,
with germline mutations in COL17 resulting in junctional
epidermolysis bullosa (JEB) and autoimmunity towards COL17 leading
to bullous pemphigoid (3). Altered
COL17 expression has been reported in various skin cancer types,
including melanoma (4). Recent
experiments in rodent models have revealed that a truncation
mutation in the Col17a1 gene promoted melanoma progression
(5).
In the present study, COL17 α1 (COL17A1) was
selected as a candidate gene and it was examined whether its
variants were related to skin cancers and other cancer types using
catalogued autopsy cases in the Japanese Geriatric Single
Nucleotide Polymorphism (JG-SNP) database.
Materials and methods
Study population
The present study included subjects registered in
the JG-SNP (6), which comprised
consecutive autopsy cases collected at Tokyo Metropolitan Geriatric
Hospital (Tokyo, Japan) between 1995 and 2012. Autopsy procedures
were performed on ~29% of the patients who died in the hospital.
The JG-SNP included a total of 2,343 subjects (1,298 males and
1,045 females). The mean age at the time of death was 80.6±8.8
years. The presence or absence of any disease was determined by a
thorough examination upon autopsy. Details of the JG-SNP database
are described elsewhere (6). The
distributions of diseases in the study group did not substantially
differ from those reported in a survey by the Ministry of Health,
Labor and Welfare of Japan (7).
For panel sequencing, two cases of mMM were selected.
Genotyping and statistical
analysis
Genomic DNA was extracted from the renal cortex
using a standard phenol-chloroform procedure and kept at -80˚C
until use. All samples were genotyped using the Infinium Human
Exome Bead Chip Version 1.1 (Illumina, Inc.) by iScan, in
accordance with the supplier's protocol. Genotype calling was
performed for all samples, as a single project using the Genotyping
Module (version 1.9) of the GenomeStudio data analysis software
package. Initial genotype clustering was performed using the
default Illumina cluster file (HumanExome 12v1-1_A.egt) and
manifest file (HumanExome-12v1-1_A.bpm), applying the GenTrain2
clustering algorithm. Specimens with a per-sample call rate of
>98% were considered eligible. Associations between the variants
and cancer status of the patients were displayed on a contingency
table and Fisher's exact tests were performed by using SPSS
Statistics 25.0 (IBM Corporation). To minimize bias, the
pathological assessment, genotyping and statistical analysis had
been performed at different institutions in a double-blinded
manner.
Panel sequencing
A total of two cases of mMM were selected for panel
sequencing analysis. AmpliSeq 7.0.2 (Thermo Fisher Scientific,
Inc.) was used to design an AmpliSeq Custom panel that covers the
exons, exon-intron boundaries and 5'UTR and 3'UTR of the genes
encoding AKT1, APC, ATM, AXIN2, BMPR1A, BRCA1, BRCA2, BARD1, BRIP1,
BUB1, BUB1B, BUB3, CDH1, CDKN1B, CHEK2, contactin 6 (CNTN6), ENG,
EPCAM, EPHX1, FANCC, FANCE, FAN1, GALNT12, LRP6, MBD4, MCM9, MYH11,
MLH1, MLH3, MSH2, MSH3, MSH6, MUTYH, NFKBIZ, NTHL1, PIK3CA, PMS1,
PMS2, POLD1, POLE, POLQ, PTEN, RAD52, RBL1, REV3 like, DNA directed
polymerase ζ catalytic subunit (REV3L), RNF43, RPS20, SCG5-GREM1,
SDHB, SDHD, SMAD4, SMAD9, SMARCA4, STK11, TDRD3, TGFBR2, TP53,
UIMC1, XAF1 and XRCC4, as well as the upstream regions of the genes
encoding GREM1, APC and MSH2(8).
An Ion Chef instrument was used for emulsion PCR, bead enrichment
and chip loading onto the Ion 530 chip (Thermo Fisher Scientific,
Inc) using the Ion 520 and 530 kits (Thermo Fisher Scientific,
Inc.). The loaded chips were subjected to sequencing on an Ion
GeneStudio S5 Plus sequencer (Thermo Fisher Scientific, Inc.). The
obtained data were analyzed using Ion Reporter server 5.10 (Thermo
Fisher Scientific, Inc.). The called variants were reviewed by
visual inspection on the Integrative Genomics Viewer (version 4.2,
15 September 2009; Broad Institute).
Only variants including missense, nonsense, indel
and splice site variants were retained. Variants with a minor
allele frequency (MAF) of <1% according to the JMorp database,
which catalogues variations in 8.6K Japanese individuals (9), were selected. The mutation databases
of InterVar (https://wintervar.wglab.org/) and the Human Gene
Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) professional
version as of April 2021 were used to annotate variants as
pathogenic, likely pathogenic, of unknown significance, likely
benign or benign. Polyphen-2 was used to predict the functional
consequence of the amino acid substitution (10).
Results
Exome chip analysis
The JG-SNP database included a total of 12 patients
with skin cancer, comprising two with mMM (81 years, male; 83
years, male), three with extramammary Paget's disease (EMPD) (93
years, female; 91 years, male; 76 years, male), three with squamous
cell carcinoma (SCC) (85 years, male; 87 years, female; 92 years,
female), two with basal cell carcinoma (BCC) (77 years, female; 92
years, female), one with apocrine carcinoma (83 years, male) and
one with unknown histology origin (90 years, male). The database
included no cases of cMM.
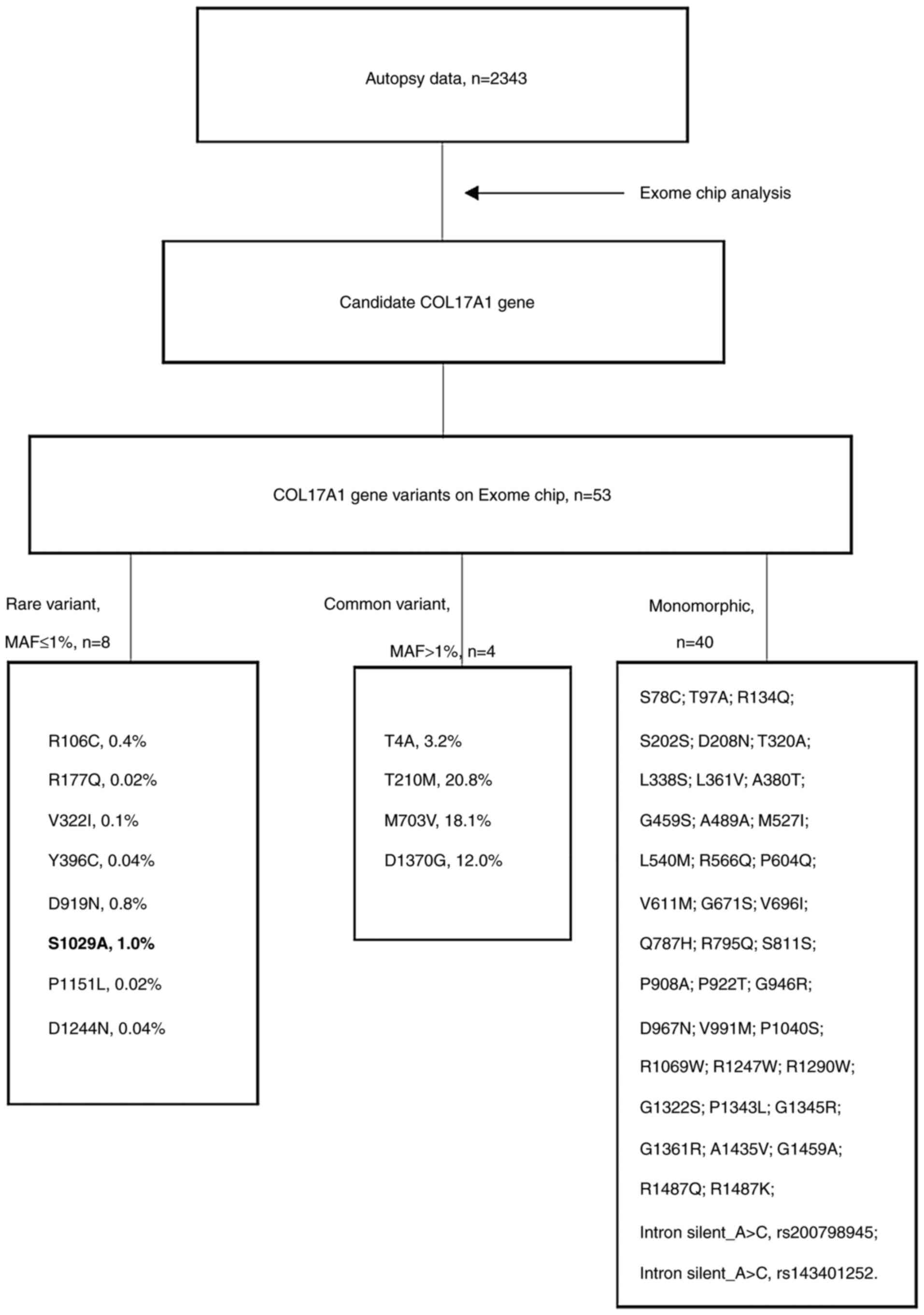
Fig. 1 presents a
flow chart of the analysis to determine germ line variants in the
candidate COL17A1 gene. Altogether, 53 COL17A1 missense single
nucleotide variants (SNVs) were analyzed on the Exome Chip, of
which 52 were successfully genotyped. Analysis of 2,343 subjects
revealed that 40 SNVs were monomorphic and 12 were bimorphic. These
SNVs were analyzed with regard to association with skin cancer and
other cancer types. Only one nonsynonymous variant, p.Ser1029Ala
(rs118166857), exhibited a nominal sign of association with skin
cancer [Fisher's exact P=0.002; odds ratio (OR)=16.93, 95% CI:
4.44-64.64; Table I]. The other 11
variants did not exhibit any positive sign of association (data not
shown).
 | Table ICharacteristics of p.Ser1029Ala
carriers. |
Table I
Characteristics of p.Ser1029Ala
carriers.
| Characteristic | p.Ser1029Ala (+)
(n=48) | p.Ser1029Ala (-)
(n=2,295) | P-value | OR (95% CI) |
|---|
| Sex, F/M | 22/26 | 1023/1272 | 0.884 | 1.05 (0.59-1.87) |
| Age, years | 82.0±9.2 | 80.6±8.8 | 0.217a | NC |
| Total cancer,
+/- | 34/14 | 1412/883 | 0.230 | 1.52 (0.81-2.85) |
| Skin cancer | 3 | 9 | 0.002 | 16.93
(4.44-64.64) |
| Mucosal melanoma | 2 | 0 | 0.001 | NC |
| Paget's disease | 1 | 2 | 0.060 | 24.39
(2.17-273.70) |
| Other skin
cancer | 0 | 7 | 0.997 | 1 (0.99-1.00) |
| Blood cancer | 7 | 205 | 0.197 | 1.74 (0.77-3.93) |
| Breast cancer | 6 | 76 | 0.006 | 4.17
(1.72-10.11) |
| Colorectal
cancer | 6 | 389 | 0.559 | 0.7 (0.3-1.66) |
| Myelodysplastic
syndrome | 3 | 39 | 0.053 | 3.86
(1.15-12.94) |
| Urinary tract
cancer | 3 | 43 | 0.066 | 3.49
(1.04-11.67) |
| Prostatic
cancer | 3 | 210 | 0.798 | 0.66
(0.20-2.15) |
| Gastric cancer | 3 | 259 | 0.358 | 0.52
(0.16-1.70) |
| Malignant
lymphoma | 3 | 122 | 0.740 | 1.19
(0.36-3.88) |
| Thyroid cancer | 2 | 52 | 0.304 | 1.88
(0.44-7.93) |
| Lung cancer | 2 | 272 | 0.114 | 0.32
(0.08-1.34) |
| Biliary tract
cancer | 2 | 62 | 0.380 | 1.57
(0.37-6.60) |
| Esophageal
cancer | 1 | 31 | 0.487 | 1.55
(0.21-11.62) |
| Mesothelioma | 1 | 4 | 0.098 | 12.19
(1.34-111.10) |
| Myeloma | 1 | 39 | 0.566 | 1.23
(0.17-9.15) |
| Small intestine
cancer | 1 | 11 | 0.220 | 4.42
(0.56-34.92) |
| Kidney cancer | 1 | 33 | 0.508 | 1.46
(1.20-10.89) |
It was indicated that the p.Ser1029Ala variant was
carried by two of the two patients with mMM and one of the three
patients with EMPD, but not by any of the patients with other skin
cancers, including SCC and BCC. Among the two cases of mMM, case
one was an 81-year-old male who had mMM that originated in the anal
canal, with massive metastasis to the lymph nodes and brain. The
tumor cells exhibited melanin granules and immunohistochemistry
revealed positive staining for S100A and HMB-45 (anti-melanoma).
Case two was an 83-year-old male with a large lung tumor with
necrosis in the center, as well as brain metastasis, both of which
were histologically confirmed as MM. Close inspection for the
primary lesion at the skin, esophagus, oral cavity, pharynx,
larynx, trachea, anus and external genital organs did not reveal
any relevant lesions. Thus, this case was most likely rare lung MM
(11), although it was not
possible to exclude the possibility of MM with an undetected or
diminished primary nest.
The database included three cases of EMPD. The one
patient with EMPD who carried the p.Ser1029Ala variant was a
76-year-old male with lesions at the external genital organs. This
case exhibited infiltration in the epidermis without distant
metastasis.
The p.Ser1029Ala variant appeared in 47 heterozygote
carriers and one homozygote and the MAF was 1.0%. These 48 carriers
were surveyed for other cancers (Table
I) and it was indicated that most cancers were not associated
with this variant. However, a nominal positive sign of association
with breast cancer was obtained (P=0.006, OR=4.17, 95% CI:
1.72-10.11).
Targeting sequencing of two mMM
cases
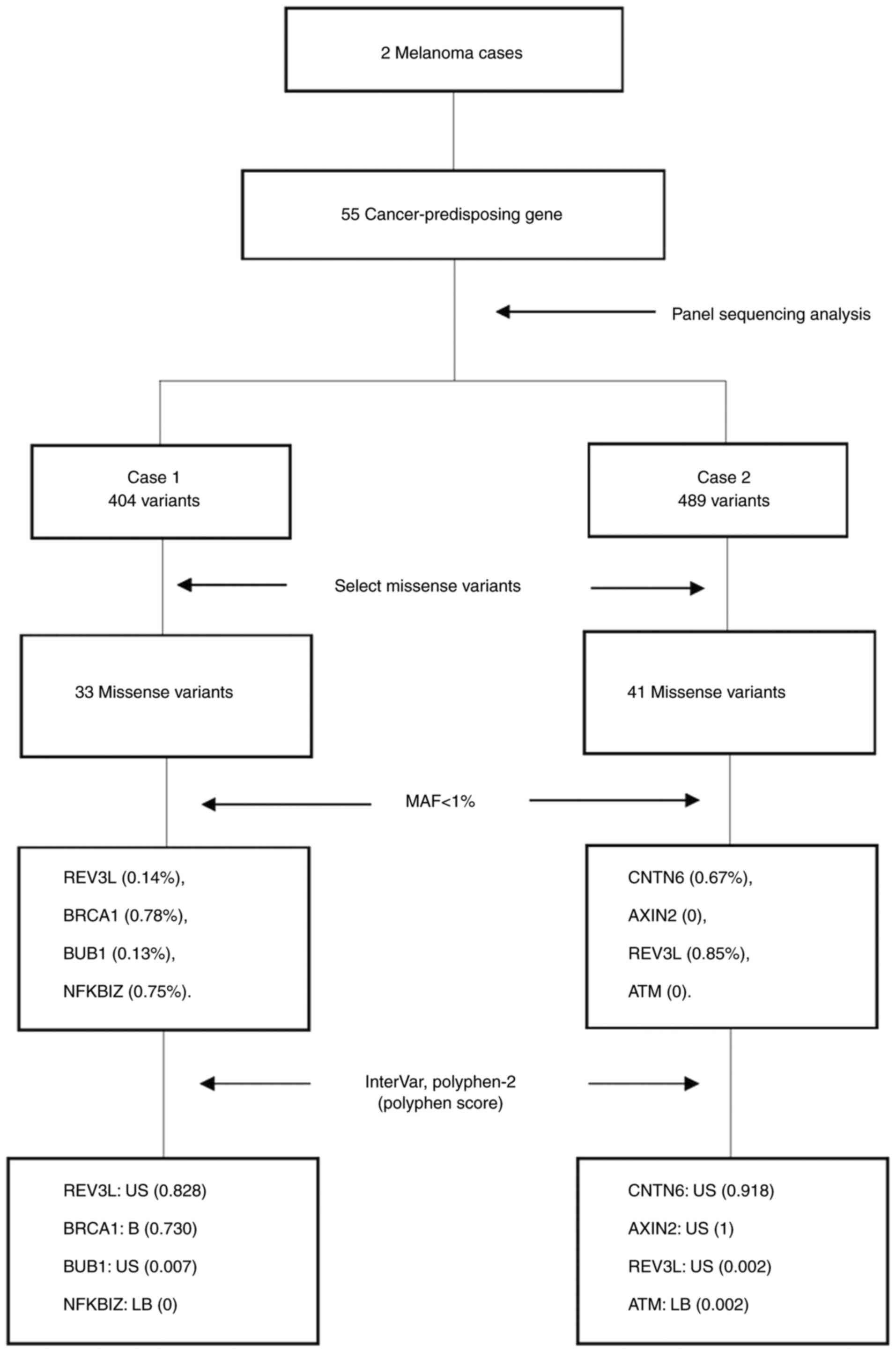
To discover further potential germline variants,
panel sequencing of 55 cancer-predisposing genes (including
BRCA1/2, TP53 and mismatch repair genes) was performed in the two
patients with mMM. Among the 55 genes, 404 variants were identified
in case 1 and 489 variants in case 2. These comprised both common
and rare variants and included single-nucleotide and
multi-nucleotide variants, as well as insertion/deletions. From the
pathogenic variants, the synonymous variant types and unknown types
were removed. Finally, 33 missense variants were identified in case
1 and 41 missense variants in case 2, including both common and
rare variants (Fig. 2).
Table II presents
the rare variants, which had MAF values of <1% according to the
database of ToMMo 8.3KJPN (https://jmorp.megabank.tohoku.ac.jp/202102/). There
were 4 rare variants in case 1 (REV3L p.His1319Tyr, MAF=0.14%;
BRCA1 p.Tyr856His, MAF=0.78%; BUB1 p.Cys698Tyr, MAF=0.13%; NFKBIZ
p.Thr307Ser, 0.75%) and 4 rare variants in case 2 (CNTN6
p.Arg471Thr, MAF=0.67%; AXIN2 p.His513Tyr MAF=0%; REV3L
p.Arg1970His, MAF=0.85%; ATM p.His683Gln, MAF=0%). The AXIN2
p.His513Tyr and ATM p.His683Gln variants were novel variants
without accession numbers in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/).
 | Table IIRare variants in patients with
mucosal melanoma. |
Table II
Rare variants in patients with
mucosal melanoma.
| A, Patient 1 |
|---|
| Gene | SNV | VE | dbSNP | InterVar |
Predictiona | MAFb (%) |
|---|
| REV3L | p.His1319Tyr | Missense | rs763112147 | US | Possibly
damaging | 0.14 |
| BRCA1 | p.Tyr856His | Missense | rs80356892 | B | Possibly
damaging | 0.78 |
| BUB1 | p.Cys698Tyr | Missense | rs764154158 | US | Benign | 0.13 |
| NFKBIZ | p.Thr307Ser | Missense | rs3821727 | LB | Benign | 0.75 |
| B, Patient 2 |
| Gene | SNV | VE | dbSNP | InterVar |
Predictiona | MAFb (%) |
| CNTN6 | p.Arg471Thr | Missense | rs763596062 | US | Possibly
damaging | 0.67 |
| AXIN2 | p.His513Tyr | Missense | NV | US | Probably
damaging | 0 |
| REV3L | p.Arg1970His | Missense | rs3218606 | US | Benign | 0.85 |
| ATM | p.His683Gln | Missense | NV | LB | Benign | 0 |
InterVar and Polyphen-2 were used to annotate the
pathogenicity of the variants. Variants of unknown significance
included REV3L p.His1319Tyr and BUB1 p.Cys698Tyr in case 1, and
CNTN6 p.Arg471Thr, AXIN2 p.His513Tyr and REV3L p.Arg1970His in case
2. The functional significance of variants was predicted using
Polyphen-2 and three possibly damaging predictions and one probably
damaging prediction were obtained. In case 1, the Polyphen score
was 0.828 for REV3L p.His1319Tyr and 0.730 for BRCA1 p.Tyr856His,
although the InterVar results indicated benign features. The
Polyphen score was 0.007 for BUB1 p.Cys698Tyr and 0 for NFKBIZ
p.Thr307Ser. In case 2, the Polyphen score was 0.918 for CNTN6
p.Arg471Thr, 1 for the novel variant AXIN2 p.His513Tyr, 0.002 for
REV3L p.Arg1970His and 0.002 for ATM p.His683Gln.
Discussion
A nationwide registry of MM in Japan included 5,566
patients with MM during the three years of 2011-2013(12). Among these patients, one in seven
had mMM. In Japan, MM is a rare cancer type and mMM accounts for a
higher proportion of MM cases compared to that in Caucasians
(13). In mMM cases, lesions are
most common in the head and neck (51.6%), followed by the
gastrointestinal tract (28.3%). Primary mMM in the lung is rare,
accounting for only 1.7% of mMM (12). Thus, the patients analyzed in the
present study were rare refractory cancer cases. The two melanoma
patients in the JG-SNP database had the mucosal rather than the
cutaneous type. The expression of col17 is ~30-50% higher in
mucosal keratinocytes than in skin keratinocytes, suggesting that
col17 may also have an important role in mucosal keratinocytes
(14).
In the present study, a rare missense variant,
COL17A1 p.Ser1029Ala (MAF=1.0%), was detected in two of the two
patients with mMM and one of the three patients with EMPD. This
variant was not observed in SCC and BCC patients. This may be
explained by the notion that the pathogenesis of non-melanoma skin
cancers predominantly involves other adhesion molecules, such as
cadherins, integrins and selectins (15). It was also observed that the
COL17A1 p.Ser1029Ala variant was nominally associated with breast
cancer. Considering the recent findings that Col17A1 functions as
an essential molecule for the stem cell niche and cell competition
(16,17), this variant may be worth studying
for its potential cancer-predisposing role.
COL17A1 is one of the triple helical collagen genes,
which encodes a type 2 oriented transmembrane protein with
collagenous domains in the extracellular domain. COL17A1 is a
component of the hemidesmosome, a multi-protein adhesion complex
that maintains stable attachment at the dermal-epidermal junction.
COL17A1 is well known as a causative gene for JEB, which leads to
fragile-desmosome and skin-blistering phenotypes (18). Over 40 rare variants of COL17A1
have been reported [Human Genome Mutation Database professional
(http://www.hgmd.cf.ac.uk/ac/index.php), as of April
2021]. Most are nonsense variants, while certain variants are
missense variants, which are generally associated with milder
phenotypes, such as more localized lesions and late disease onset.
This list does not include the p.Ser1029Ala mutation identified in
the present study. This variant is c.3085 T>G on the COL17A1
cDNA. It is located on exon 45 and the residue resides in the
non-collagen (NC) 10 region in the ectodomain. Functional
prediction using polyphen-2 and InterVar suggested that the
consequence of amino acid substitution at this position is benign,
indicating that it is not a null functional mutation.
COL17A1 expression is upregulated in association
with the invasion of melanoma (4)
and its role in early carcinogenesis was recently uncovered. In a
mouse model, Col17a1 dysfunction, caused by the introduction of
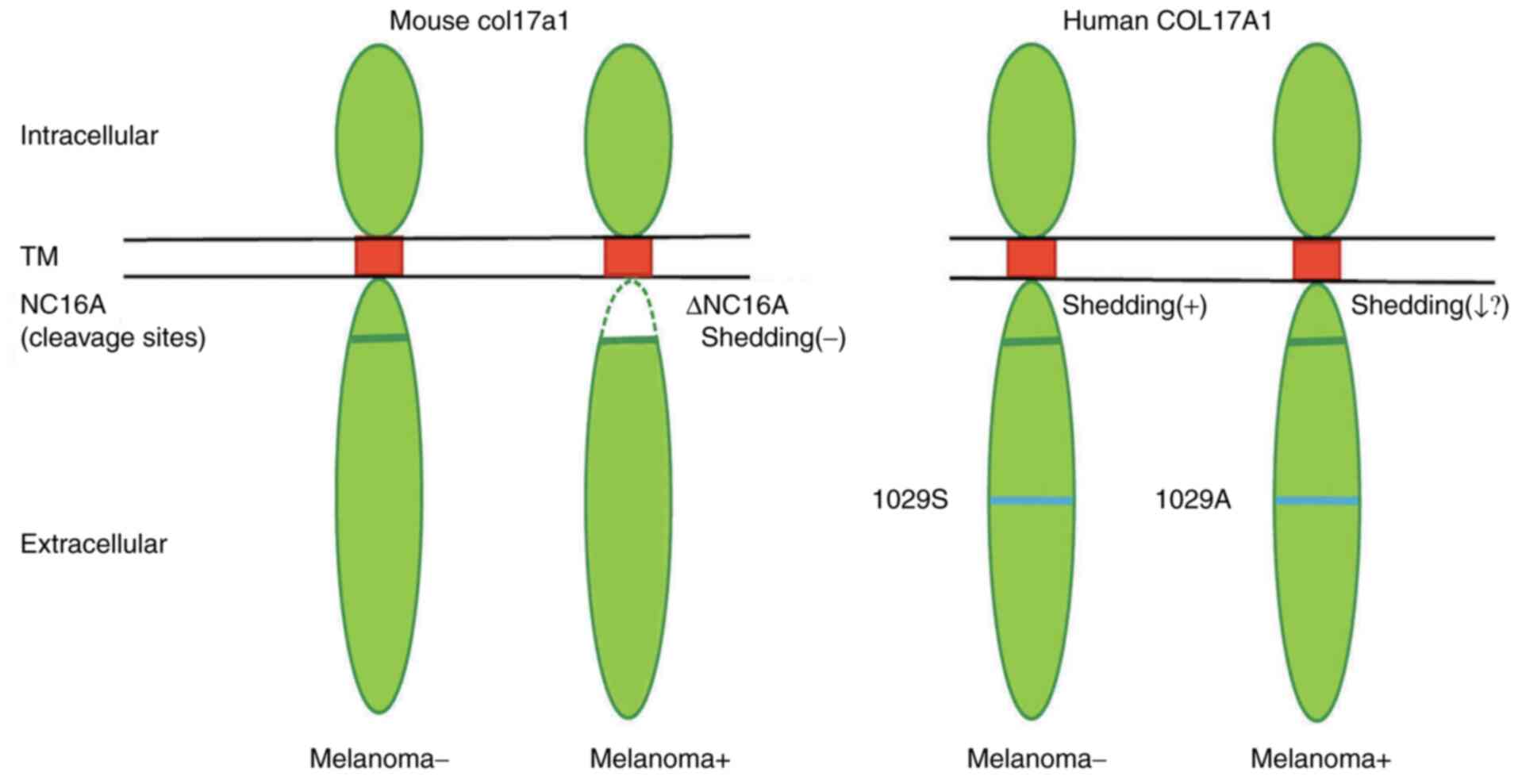
NC16 deletion, supported melanoma progression through modulation of
the skin tumor microenvironment by basal keratinocytes (5). Col17a1 is not expressed in
melanocytes but is expressed in adjacent keratinocytes and provides
a niche for melanocyte stem cells (16). Col17a1 is dynamically shed by
proteases at the NC16A domain, which has an important role in cell
motility, adhesion and cell differentiation at the epidermal site.
It is tempting to speculate that the variant p.Ser1029Ala in the
ectodomain may be less efficiently shed compared to the wild-type,
thereby assisting melanoma progression (Fig. 3).
Recent results obtained with Col17a1-null mice
revealed that this protein functions as an essential molecule for
the proliferation of melanocyte stem cells and has essential roles
in cell development, aging and carcinogenesis at epidermal sites
(19). These results appear to be
in line with the present observation that p.Ser1029Ala of COL17A1
may have a role in melanoma progression (5). COL17A1 has recently been demonstrated
to have an important role in cell competition (17), a newly described mechanism that
allows tumor cells to outcompete and eliminate less fit adjacent
normal cells (20). It is tempting
to speculate that p.Ser1029Ala in the ectodomain may change the
microenvironment to favor normal cells being easily
outcompeted.
A potential link between COL17A1 and breast cancer
has been suggested by findings that COL17A1 expression is
suppressed in breast cancer cells (21) and that replenishing COL17A1
expression has an anti-proliferative effect (22). In breast cancer cells, COL17A1
participates in regulating the transition from monolayered to
multilayered epithelial cells in precancerous lesions. This may be
related to its role in tumorigenesis of the monolayered epithelia
of the mammary gland (23).
Although the two patients with mMM in the present
study had no familial history, other potential cancer-predisposing
genes and variants were searched by examining the sequences of 55
cancer-related genes. No pathogenic germline mutations in BRCA1/2
or P53 were identified, but only variants of unknown significance
were found in genes possibly related to melanoma.
REV3L, also known as DNA polymerase ζ, is the
largest catalytic subunit of DNA polymerase and is important in
translation synthesis, which is usually the first line of defense
against ultraviolet radiation or chemical adducts in DNA (24). Somatic mutation of REV3L is
frequently identified in cancer, including melanoma (25). In melanoma cells, REV3L constitutes
an important part of the response to cisplatin, resulting in
improved survival and growth (26). Of note, REV3L also binds and is
down-regulated by microRNA (miR)-340(27), which is a modulator of RAS-RAF-MAPK
signaling in melanoma (28). It
may be worthwhile investigating the mutual regulation between REV3L
and miR-340 in melanoma.
AXIN2 acts as a negative regulator in the Wnt
signaling pathway. It forms a complex with GSK-3β and β-catenin and
promotes GSK-3β-dependent phosphorylation of β-catenin. The novel
AXIN2 p.His513Tyr variant identified in the present study appears
to be localized at the GSK-3β interaction site (29,30).
Wnt/β-catenin dysregulation is the key to melanocyte deterioration.
The melanoma cell line PR-Mel carries an AXIN2 deletion mutation
(31). Specifically, AXIN2 is a
negative regulator of Wnt/β-catenin signaling, which acts to
stimulate hair growth in the hair follicle stem cell (32). Loss of function of
Drosophila Axn may cause cell competition by inducing cell
death in the surrounding wild-type cells (19). Thus, an AXIN2 variant may have a
fundamental role in melanoma.
CNTN6 is a ligand of NOTCH receptor 1 (NOTCH1). The
NOTCH1 signaling pathway is a determinant of stemness and
plasticity in melanoma stem/initiating cells, through regulation of
melanoma aggressiveness (33).
NOTCH1 is also involved in cell competition (19). BUB1 is related to melanoma, in that
BUB1 expression distinguishes melanoma, benign nevi and lymph
nodes, thus contributing to the detection of melanoma
micrometastasis in sentinel lymph nodes. High BUB1 expression is
found in metastatic melanoma. BUB1 is downstream of sirtuin 1,
which is upregulated in melanoma and its inhibition exerts an
anti-proliferative effect in melanoma cells (34). The rare variants found in these
genes may relate to melanoma predisposition by affecting cell
competition.
The present study has certain limitations.
Consecutive autopsy cases from geriatric hospitals were used as
cancer and control subjects. Obviously, there is a survival bias
due to the high age of the subjects. Indeed, this sample does not
represent subjects who died prematurely. Due to being an autopsy
study, the present study was also limited in terms of access to
clinical data related to cancer progression and tumor stages.
Relevant pathological samples to perform an immunohistochemical
analysis are not available, so that it is not possible to determine
whether the p.Ser1029Ala variant affected the shedding of COL17A1.
The small sample size for melanoma may be a hindrance to
determining a significant association.
In conclusion, the present study reported on a
COL17A1 p.Ser1029Ala variant identified in rare malignancies of
epithelial origin in an autopsy database. The present study was
retrospective and the statistical analyses were based on a small
number of patients; thus, the present observations only serve for
hypothesis generation. There remains a requirement for further
studies including larger numbers of patients and molecular
pathogenesis.
Acknowledgements
Not applicable.
Funding
Funding: This study was funded by NANKEN KYOTEN TMDU.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MM designed the present study. TA performed the
pathological analysis. MT acquired and analysed clinical data. HE
and YO generated the targeting sequencing data. DT performed
genotyping and statistical analyses. MM, TA and MT confirm the
authenticity of all the raw data. DT and MM wrote the manuscript.
All authors read and approved the manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committees of
Tokyo Medical and Dental University (Tokyo, Japan; approval no.
02016-011-03) and the Tokyo Metropolitan Geriatric Hospital (Tokyo,
Japan; approval no. 230405). Written informed consent was obtained
from a family member of each study participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mikkelsen LH, Larsen AC, von Buchwald C,
Drzewiecki KT, Prause JU and Heegaard S: Mucosal malignant
melanoma-a clinical, oncological, pathological and genetic survey.
APMIS. 124:475–486. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yde SS, Sjoegren P, Heje M and Stolle LB:
Mucosal melanoma: A literature review. Curr Oncol Rep.
20(28)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Nishie W: Collagen XVII processing and
blistering skin diseases. Acta Derm Venereol.
12(adv00054)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Krenacs T, Kiszner G, Stelkovics E, Balla
P, Teleki I, Nemeth I, Varga E, Korom I, Barbai T, Plotar V, et al:
Collagen XVII is expressed in malignant but not in benign
melanocytic tumors and it can mediate antibody induced melanoma
apoptosis. Histochem Cell Biol. 138:653–667. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hwang BJ, Zhang Y, Brozowski JM, Liu Z,
Burette S, Lough K, Smith CC, Shan Y, Chen J, Li N, et al: The
dysfunction of BP180/collagen XVII in keratinocytes promotes
melanoma progression. Oncogene. 38:7491–7503. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sawabe M, Arai T, Kasahara I, Esaki Y,
Nakahara K, Hosoi T, Orimo H, Takubo K, Murayama S and Tanaka N:
Tokyo Metropolitan Geriatric Medical Center; Japan Science and
Technology Agency. Developments of geriatric autopsy database and
Internet-based database of Japanese single nucleotide polymorphisms
for geriatric research (JG-SNP). Mech Ageing Dev. 125:547–552.
2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yamada M, Sato N, Ikeda S, Arai T, Sawabe
M, Mori S, Yamada Y, Muramatsu M and Tanaka M: Association of the
chromodomain helicase DNA-binding protein 4 (CHD4) missense
variation p.D140E with cancer: Potential interaction with smoking.
Genes Chromosomes Cancer. 54:122–128. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Eguchi H and Okazaki Y: Next-generation
sequencing for genetic diagnosis of hereditary colorectal cancer
and polyposis syndrome. In: Recent Advances in the Treatment of
Colorectal Cancer. Ishida H (eds). Springer, pp115-125, 2019.
|
|
9
|
Tadaka S, Hishinuma E, Komaki S, Motoike
IN, Kawashima J, Saigusa D, Inoue J, Takayama J, Okamura Y, Aoki Y,
et al: jMorp updates in 2020: Large enhancement of multi-omics data
resources on the general Japanese population. Nucleic Acids Res.
49:D536–D544. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Adzhubei I, Jordan DM and Sunyaev SR:
Predicting functional effect of human missense mutations using
PolyPhen-2. Curr Protoc Hum Genet Chapter 7: Unit7.20, 2013.
|
|
11
|
Peng J, Han F, Yang T, Sun J, Guan W and
Guo X: Primary malignant melanoma of the lung: A case report and
literature review. Medicine (Baltimore). 96(e8772)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tomizuka T, Namikawa K and Higashi T:
Characteristics of melanoma in Japan: A nationwide registry
analysis 2011-2013. Melanoma Res. 27:492–497. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Carvajal RD, Spencer SA and Lydiatt W:
Mucosal melanoma: A clinically and biologically unique disease
entity. J Natl Compr Canc Netw. 10:345–356. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kamaguchi M and Iwata H: The diagnosis and
blistering mechanisms of mucous membrane pemphigoid. Front Immunol.
24(34)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pogorzelska-Dyrbus J and Szepietowski JC:
Adhesion molecules in non-melanoma skin cancers: A comprehensive
review. In Vivo. 35:1327–1336. 2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Tanimura S, Tadokoro Y, Inomata K, Binh
NT, Nishie W, Yamazaki S, Nakauchi H, Tanaka Y, McMillan JR,
Sawamura D, et al: Hair follicle stem cells provide a functional
niche for melanocyte stem cells. Cell Stem Cell. 8:177–187.
2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu N, Matsumura H, Kato T, Ichinose S,
Takada A, Namiki T, Asakawa K, Morinaga H, Mohri Y, De Arcangelis
A, et al: Stem cell competition orchestrates skin homeostasis and
ageing. Nature. 568:344–350. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Natsuga K, Watanabe M, Nishie W and
Shimizu H: Life before and beyond blistering: The role of collagen
XVII in epidermal physiology. Exp Dermatol. 28:1135–1141.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Vishwakarma M and Piddini E: Outcompeting
cancer. Nat Rev Cancer. 20:187–198. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ferrari AJ, Drapkin R and Gogna R: Cell
fitness: More than push-ups. Int J Mol Sci. 22(518)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yodsurang V, Tanikawa C, Miyamoto T, Lo
PHY, Hirata M and Matsuda K: Identification of a novel p53 target,
COL17A1, that inhibits breast cancer cell migration and invasion.
Oncotarget. 8:55790–55803. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lothong M, Sakares W, Rojsitthisak P,
Tanikawa C, Matsuda K and Yodsurang V: Collagen XVII inhibits
breast cancer cell proliferation and growth through deactivation of
the AKT/mTOR signaling pathway. PLoS One.
16(e0255179)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kozawa K, Sekai M, Ohba K, Ito S, Sako H,
Maruyama T, Kakeno M, Shirai T, Kuromiya K, Kamasaki T, et al: The
CD44/COL17A1 pathway promotes the formation of multilayered,
transformed epithelia. Curr Biol. 26:3086–3097.e7. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Martin SK and Wood RD: DNA polymerase ζ in
DNA replication and repair. Nucleic Acids Res. 19:8348–8361.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chae YK, Anker JF, Carneiro BA, Chandra S,
Kaplan J, Kalyan A, Santa-Maria CA, Platanias LC and Giles FJ:
Genomic landscape of DNA repair genes in cancer. Oncotarget.
26:23312–23321. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Song L, McNeil EM, Ritchie AM, Astell KR,
Gourley C and Melton DW: Melanoma cells replicate through
chemotherapy by reducing levels of key homologous recombination
protein RAD51 and increasing expression of translesion synthesis
DNA polymerase ζ. BMC Cancer. 18(864)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Arivazhagan R, Lee J, Bayarsaikhan D, Kwak
P, Son M, Byun K, Salekdeh GH and Lee B: MicroRNA-340 inhibits the
proliferation and promotes the apoptosis of colon cancer cells by
modulating REV3L. Oncotarget. 26:5155–5168. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Strong AM, Setaluri V and Spiegelman VS:
MicroRNA-340 as a modulator of RAS-RAF-MAPK signaling in melanoma.
Arch Biochem Biophys. 1:118–124. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Julius MA, Schelbert B, Hsu W, Fitzpatrick
E, Jho E, Fagotto F, Costantini F and Kitajewski J: Domains of axin
and disheveled required for interaction and function in wnt
signaling. Biochem Biophys Res Commun. 276:1162–1169.
2000.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ikeda S, Kishida S, Yamamoto H, Murai H,
Koyama S and Kikuchi A: Axin, a n9gative regulator of the Wnt
signaling pathway, forms a complex with GSK-3beta and beta-catenin
and promotes GSK-3beta-dependent phosphorylation of beta-catenin.
EMBO J. 17:1371–1384. 1998.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Castiglia D, Bernardini S, Alvino E,
Pagani E, Luca ND, Falcinelli S, Pacchiarotti A, Bonmassar E,
Zambruno G and D'Atri S: Concomitant activation of Wnt pathway and
loss of mismatch repair function in human melanoma. Genes
Chromosomes Cancer. 47:614–624. 2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Smith AA, Li J, Liu B, Hunter D, Pyles M,
Gillette M, Dhamdhere GR, Abo A, Oro A and Helms JA: Activating
hair follicle stem cells via R-spondin2 to stimulate hair growth. J
Invest Dermatol. 136:1549–1558. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Du Y, Shao H, Moller M, Prokupets R, Tse
YT and Liu ZJ: Intracellular notch1 signaling in cancer-associated
fibroblasts dictates the plasticity and stemness of melanoma
stem/initiating cells. Stem Cells. 37:865–875. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen J, Wu F, Shi Y, Yang D, Xu M, Lai Y
and Liu Y: Identification of key candidate genes involved in
melanoma metastasis. Mol Med Rep. 20:903–914. 2019.PubMed/NCBI View Article : Google Scholar
|