Introduction
Worldwide, colorectal cancer (CRC) is considered one
of the most common malignancies and a significant threat to health
(1,2). In 2020, CRC was ranked the fourth
most common type of cancer and the third most common cause of
cancer deaths worldwide with an incidence rate of 24.8 and a
mortality rate of 12 cases per 100,000, according to the World
Health Organization (3,4). In Asia, the incidence rate is
increasing, making CRC a notable health burden (5). In the Lebanese population, CRC ranked
fourth among men for the most common types of cancer with an
expected incidence rate of 17.5 cases per 100,000 and ranked second
among women with an expected incidence rate of 10.6 cases per
100,000 in 2020, making it one of the highest reported types of
cancer in the country (6).
CRC is initially characterized by polyps (Ps), which
are classified according to their growth pattern as adenomatous or
serrated. Ps progress to the adenoma (Ad) stage within 10-20 years,
followed by invasive adenocarcinoma (ADC). Since there is a slow
progression from the precancerous P to invasive ADC stage, early
detection can decrease the mortality rate and increase the chances
of recovery and survival (7,8). A
study demonstrated that early stage diagnosis in patients with CRC
led to a decrease in the CRC mortality rate in Australia (9). Furthermore, in the United Kingdom,
diagnosis at early stages has been shown to increase the survival
rate compared with diagnosis at later stages (9).
A number of screening tools have been utilized for
early detection of CRC; these can be invasive and non-invasive.
Usually, CRC is identified via endoscopic imaging and
histopathological examination of removed specimens (biopsies)
(10). Colonoscopy is an invasive
visual screening tool used to visualize and detect Ps. Other
screening tools include fecal occult blood and immunochemical
tests, which detect blood in the stool (8,11,12).
However, these screening tools have limitations in terms of
sensitivity, specificity, cost and degree of invasiveness.
Therefore, the development of novel diagnostic biomarkers is
crucial for early diagnosis of CRC (13,14).
CRC is considered to be a multifactorial disease
that is characterized by a sequence of genetic and epigenetic
alterations in the different stages. These alterations are
classified into three classes: Chromosomal and microsatellite
instability and CpG island methylation phenotype (13,15).
In addition, epigenetic regulators, including long non-coding RNAs
and microRNAs (miRNAs or miRs), have been implicated in different
types of cancer, including CRC (15). miRNAs, short non-coding RNAs
composed of 18-24 nucleotides, are known as gene regulatory
molecules for their ability to suppress translational activity by
using other miRNAs as cleavage targets, thereby altering the
function of protein-coding genes (13,16).
miRNAs can be characterized as oncogenic or tumor suppressor
miRNAs. Oncogenic miRNAs are upregulated in cancer and mediate
downregulation of tumor suppressor genes. By contrast, tumor
suppressor miRNAs are downregulated in cancer, which is accompanied
by upregulation of oncogenes (17,18).
These oncogenes are highly expressed and contribute to cancer
development by increasing cellular proliferation and tumor
progression (19).
Furthermore, there is an increasing evidence that
certain miRNAs are dysregulated in cancer (17,18).
Studies have revealed that numerous miRNAs (including miR-21,
miR-17-92 cluster, miR-143 and miR-145) are deregulated in patients
with CRC and serve an important role in its initiation,
development, and progression (20-22).
One of the most commonly upregulated miRNAs in CRC is miR-21, which
plays an essential role in tumor progression (23).
The role of miR-31, miR-146b, miR-145 and miR-186
have been studied in different types of cancer. Several studies
have reported the oncogenic role of miR-31 and miR-146b in
different populations and types of cancer, including CRC (24-27).
Accordingly, miR-31 has been shown to be upregulated in CRC and is
associated with tumor progression, as well as survival rate.
However, miR-145 and miR-186 are downregulated in colorectal
cancer, which is accompanied by activation of oncogenic mRNAs, such
as twist family bHLH transcription factor 1, Fascin Actin-Bundling
Protein 1 and Zinc Finger E-Box Binding Homeobox 1 (28-30).
The aim of the present retrospective study was to
investigate the expression levels of miRNAs (miR-146b, miR-31,
miR-186 and miR-145) in 222 formalin-fixed paraffin-embedded (FFPE)
colorectal tissue samples taken from Lebanese patients with
different precancerous (P) and cancer stages (Ad and ADC) and to
identify miRNAs that may be used for diagnosis, particularly at P
stage.
Materials and methods
Colon tissue specimens
The present study was approved by the Medical
Committee at Bahman Hospital (Beirut, Lebanon; approval no. 27).
Retrospective FFPE specimens from 222 patients with CRC at
different stages [P (n=68), Ad (n=78), ADC (n=76)], as well as
specimens of normal healthy controls (N; n=81) were obtained from
the Anatomy and Pathology Department, Bahman Hospital, Haret Hreik,
Lebanon from patients that underwent colonoscopy and resection
between 2011 and 2019, excluding patients with hereditary
disease.
Total RNA extraction
Total RNA, including small RNA fraction, was
extracted by processing four 20 µm thick ribbons from each FFPE
tissue sample using a RecoverAll Total Nucleic Acid Isolation kit
(Ambion; Thermo Fisher Scientific, Inc.; cat. no. AM1975) according
to the manufacturer's instructions. Briefly, deparaffinization of
FFPE tissue was performed using xylene at 50˚C followed by washing
with ethanol twice to remove xylene. Proteins were digested by
incubating the samples with protease enzyme for 15 min at 50˚C and
15 min at 80˚C. Total RNA was isolated in glass-fiber filter
cartridges using an isolation additive mixture and washing with
high ethanol-wash buffers. DNA digestion was then performed using
DNase and RNA was purified by washing and elution using reagents
provided in the kit. RNA concentration and quality were detected
using a NanoDrop ND1000 spectrophotometer (Thermo Fisher
Scientific, Inc.) by measuring the optical density at 260 and 280
nm, then samples were stored at -80˚C. Only samples of high quality
and integrity (A260/A280 ratio of 1.8-2.1) were used for subsequent
experiments.
miRNA reverse transcription (RT) and
expression level analysis
A total of 10 ng total RNA was reverse transcribed
using TaqMan® MicroRNA RT kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
recommendations. The hsa-miR-146b (cat. no. 001097), hsa-miR-186
(cat. no. 002285), hsa-miR-145 (cat. no. 002278), hsa-miR-31 (cat.
no. 002279), small nucleolar RNA RNU48 (cat. no. 001006) primers
and probes were used as part of the TaqMan Small RNA Assays™ kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). RT was
performed in a multiplex reaction setup using two miRNA primers
with an endogenous control (for example, miR-31 and miR-145 primers
were used together with the endogenous control RNU48).
miRNA expression was assessed by RT-quantitative
(q)PCR using Bio-Rad CFX96 Real-Time System, C1000 Thermal Cycler
(Bio-Rad Laboratories, Inc.). Reactions were performed in duplicate
for each miRNA probe using 5.0 TaqMan® Universal Master
Mix without Amperase Uracil N-glycosylase (Applied Biosystems;
Thermo Fisher Scientific, Inc.; 4324018), 0.5 20X microRNA probe,
1.0 diethyl pyrocarbonate-treated water and 3.5 µl prepared cDNA.
The following probes were used: hsa-miR-31 (cat. no. 002279),
hsa-miR-145 (cat. no. 002278), hsa-miR-186 (cat. no. 002285),
hsa-miR-146b (cat. no. 001097), RNU48 (cat. no. 001006; all Applied
Biosystems). Each plate contained the samples, no template control,
no RT control and healthy tissue samples. The thermocycling
conditions were 95˚C for 10 min, followed by 40 cycles of 95˚C for
15 sec and annealing at 60˚C for 60 sec. Only samples with average
miRNA Cq<35 or endogenous control Cq of 22-29 were included. The
relative expression of target miRNAs at the different stages (P, Ad
and ADC) was then normalized to RNU48 and compared with N using the
2-ΔΔCq method (31).
Normalization of tumor tissues was based on N present on the
RT-qPCR plate to ensure inter-run calibration.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism (GraphPad Software, Inc; version 8) and SPSS version 25
software (IBM, Corp.). Patient demographics are presented as mean ±
standard deviation (SD) for continuous variables and as numbers or
percentages for categorical variables. Due to the lack of
normality, Kruskal-Wallis test was performed to compare expression
levels each miRNA at P, Ad, and ADC stages with healthy controls,
followed by Dunn's test to perform pairwise comparison. The
diagnostic value of miRNAs at P stage was detected by receiver
operating characteristic (ROC) curve analysis using the ΔCq values
of the samples and the predicted probability was calculated using
binary logistic regression. The ROC curve was plotted to indicate
the area under the curve (AUC) and P-value, as well as to calculate
the positive predictive and negative predictive values and
diagnostic accuracy between P and N to determine the diagnostic
value of miR-31, miR-145, miR-186 and miR-146b. Youden's index was
calculated in addition to the cut-off value, sensitivity and
specificity of miRNAs. P<0.05 was considered to indicate a
statistically significant difference. Independent set of 20 samples
was also used to confirm the diagnostic accuracy of miR-31 and
miR-145.
Results
Clinicopathological characteristics of
patients
The features of 303 biopsies are presented in
Table I, including 222 patients in
the precancerous and cancerous stages with a mean age of
60.15±15.13, in addition to 81 N samples with a mean age of
57.1±18.66 years. The male to female ratio was 1.09 for patients
and 0.7 for N samples. P and Ad samples were all non-malignant
(hyperplastic Ps and low-grade Ads). In addition, 93.4% of ADC
samples were grade 2 (moderately differentiated). A total of 0.9%
had Crohn's disease while 0.45% had a family history of CRC (data
not shown).
 | Table IClinicopathological characteristics
of patients. |
Table I
Clinicopathological characteristics
of patients.
| Characteristic | Healthy
control | Polyp | Adenoma | Adenocarcinoma | Precancerous and
cancerous stages |
|---|
| Sample, n (%) | 81.00 (36.40) | 68.00 (30.70) | 78.00 (35.10) | 76.00 (34.38) | 222.00 (73.26) |
| Age at diagnosis,
years (mean ± SD) | 57.10±18.66 | 58.06±15.34 | 58.46±15.22 | 63.47±14.55 | 60.15±15.13 |
| Sex | | | | | |
|
Male
(%) | 35.00 (43.20) | 43.00 (63.20) | 37.00 (47.40) | 36.00 (47.30) | 116.00 (52.20) |
|
Female
(%) | 46.00 (56.70) | 25.00 (36.70) | 41.00 (52.50) | 40.00 (52.60) | 106.00 (47.70) |
| Colon margin
(%) | 81(100) | | | | |
|
Colon
(%) | | 48.00 (70.50) | 73.00 (93.50) | 61.00 (80.20) | 182.00 (81.90) |
|
Right sided
colon (%) | | 13.00 (27.00) | 11.00 (15.00) | 14.00 (22.90) | 38.00 (20.80) |
|
Left sided
colon (%) | | 30.00 (62.50) | 57.00 (78.00) | 45.00 (73.70) | 132.00 (72.50) |
|
Transverse
colon (%) | | 5.00 (10.40) | 5.00 (6.80) | 2.00 (3.20) | 12.00 (6.50) |
|
Rectum
(%) | | 20.00 (29.40) | 5.00 (6.40) | 15.00 (19.70) | 40.00 (18.00) |
| Tumor grade stage
(%) | | | | G1, 3.00
(3.90); | |
| | | | | G2, 71.00
(93.40); | |
| | | | | G3/4, 2.00
(2.60) | |
Overexpression of miR-31 at different
stages of CRC
The mean expression of miR-31 across all stages was
significantly upregulated (P<0.0001). miR-31 was significantly
increased in the P, Ad and ADC stages compared with N (P<0.0001,
P=0.0007 and P<0.0001, respectively; Fig. 1A; Table II).
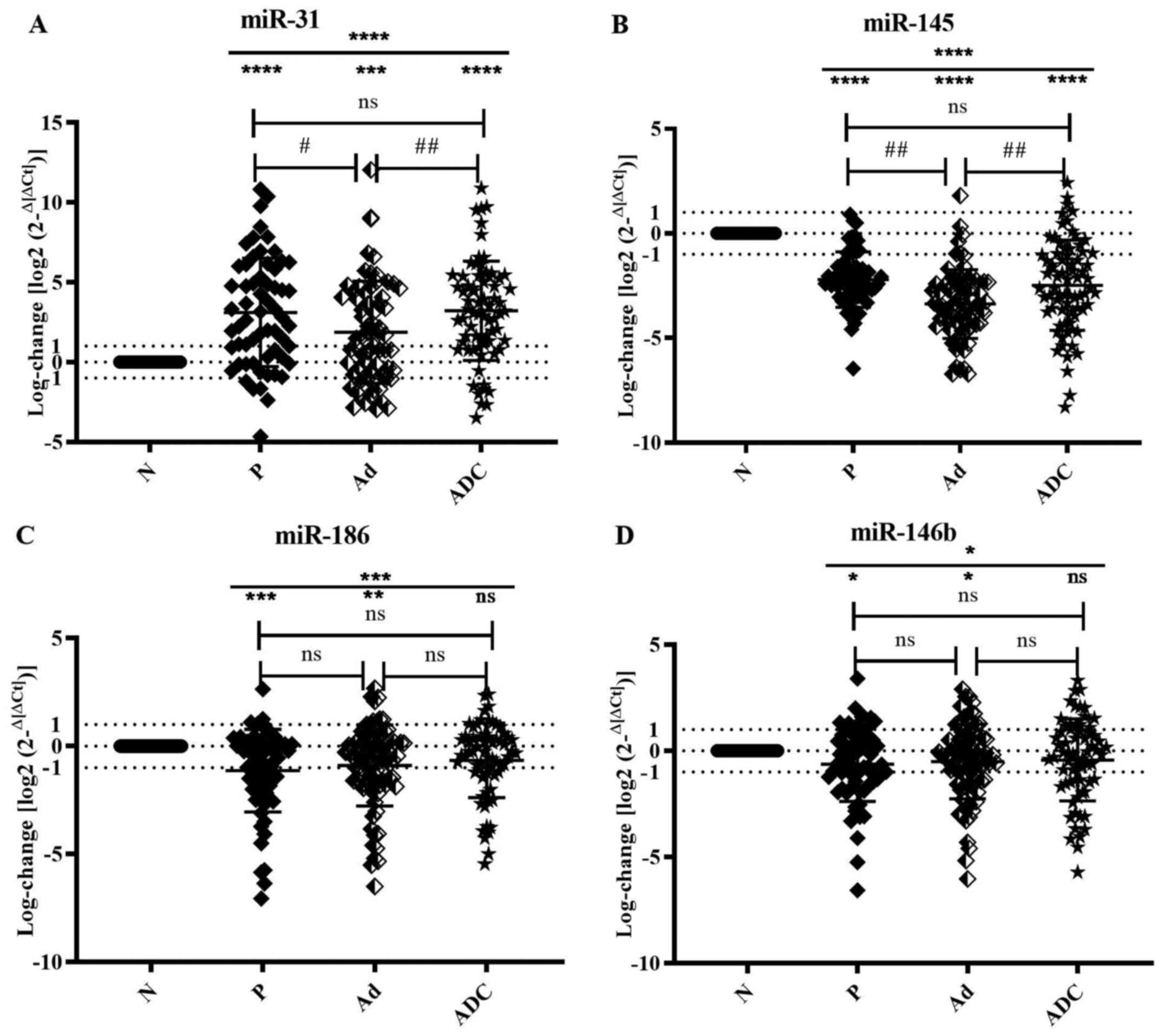 | Figure 1Dysregulation of miR-31, 145, 186 and
146b expression at different stages. Scatter plots show expression
profiles of (A) miR-31, (B) miR-145, (C) miR-186 and (D) miR-146b,
represented by log-changes at all stages relative to the average
ΔCq value of healthy controls (measured by reverse
transcription-quantitative PCR with RNU48 as an endogenous
control). Data are presented as the mean ± SEM. Data were analyzed
by Kruskal-Wallis (*P<0.05, **P<0.01,
***P<0.001 and ****P<0.001) and Dunn's
test (#P<0.05 and ##P<0.01). miR,
microRNA; N, normal; P, polyp; Ad, adenoma; ADC, adenocarcinoma;
ns, not significant. |
 | Table IIExpression levels of miRs-31, -145,
-186 and -146b compared with healthy controls. |
Table II
Expression levels of miRs-31, -145,
-186 and -146b compared with healthy controls.
| miRNA | Stage | Number | Mean ± SD | 95% CI | P-value |
|---|
| miR-31 | P | 61 | 3.1150±3.3940 | 2.2460-3.9850 | <0.0001 |
| | Ad | 67 | 1.8750±3.2030 | 1.0930-2.6560 | 0.0007 |
| | ADC | 70 | 3.2200±3.1050 | 2.4800-3.9610 | <0.0001 |
| miR-145 | P | 60 | -2.2090±1.3250 | -2.5510-1.8670 | <0.0001 |
| | Ad | 72 | -3.3740±1.6470 | -3.7610-2.9870 | <0.0001 |
| | ADC | 74 | -2.4780±2.1590 | -2.9780-1.9770 | <0.0001 |
| miR-186 | P | 64 | -1.1270±1.9140 | -1.6050-0.6491 | 0.0006 |
| | Ad | 78 | -0.8968±1.8780 | -1.3200-0.4733 | 0.0053 |
| | ADC | 65 | -0.6588±1.7240 | -1.0860-0.2316 | 0.1470 |
| miR-146b | P | 64 | -0.6320±1.7520 | -1.0700-0.1943 | 0.0260 |
| | Ad | 77 | -0.5011±1.7560 | -0.8997-0.1025 | 0.0263 |
| | ADC | 66 | -0.4283±1.9330 |
-0.9035-0.04695 | 0.2614 |
In addition, miR-31 was significantly downregulated
in Ad compared with P stage (P=0.0244) but significantly
upregulated in ADC compared with Ad (P=0.0023). However, there was
no significant change in its expression at ADC compared with P
stage (P=0.4816) Therefore, miR-31 expression increased
significantly at all stages with respect to healthy controls and at
P and ADC stages compared with Ad (Fig. 1A).
Downregulation of miR-145 in different
stages of CRC
Notably, miR-145 was significantly downregulated
across P, Ad and ADC stages when compared with N (P<0.0001;
Fig. 1B; Table II).
miR-145 expression was significantly downregulated
at Ad compared with P (P=0.0014) and at ADC compared with Ad stage
(P=0.0044). However, no significant change in the expression of
this miRNA between P and ADC stage (P=0.6184) was noted (Fig. 1B).
Dysregulated expression of miR-186 in
different stages of CRC
miR-186 was significantly deregulated (P=0.0009)
according to the Kruskal Wallis test. Significant downregulation of
miR-186 was observed at P and Ad (P=0.0006 and P=0.0053,
respectively) compared with N. ADC stage showed a slight but not
significant downregulation of miR-186 (P=0.147; Fig. 1C; Table II).
Evaluation of the expression profile of miR-186
between the different stages showed no significant change between
Ad and P (P=0.4439), ADC and Ad stages (P=0.319) and ADC and P
stages (P=0.0923; Fig. 1C).
Expression profile of miR-146b in
different stages of CRC
Compared with N, miR-146b demonstrated a significant
decrease expression (P=0.0241). Dunn's test revealed a significant
downregulation at P (P=0.026) and Ad stages (P=0.0263; Fig. 1D, Table II). No significant change was
observed between expression levels of miR-146b in Ad and P
(P=0.8971), ADC and P (P=0.3774) and ADC and Ad stages (P=0.4279;
Fig. 1D). Therefore, miR-146b
expression did not significantly change in the different
precancerous and cancerous stages of the disease.
ROC for miR-31, miR-145, miR-186 and
miR-146b
The positive and negative predictive value and
diagnostic accuracy of miR-31, miR-145, miR-186, and miR-146b were
assessed (Table III). miR-31 and
miR-145 were found to significantly differentiate between P and N
(AUC=0.7771; 95% CI, 0.6972-0.8570; P<0.0001 and AUC=0.8269; 95%
CI, 0.7566-0.8972; P<0.0001, respectively; Fig. 2). At the optimal cut-off values of
2.533 for miR-31 and 2.857 for miR-145, the sensitivity and
specificity were 76.67 and 69.74 vs. 80 and 72%, respectively.
miR-31 and miR-145 exhibited a diagnostic accuracy of 71.3 and
78.5% respectively. Moreover, the diagnostic accuracy of miR-31 and
miR-145 was 77.4 and 76.7% respectively, when applied on an
independent set of samples (Table
SI). On the other hand, miR-186 and miR-146b showed poor
discrimination between P and N (AUC=0.6875; 95% CI, 0.5987-0.7763;
P=0.0002 and AUC=0.6313; 95% CI, 0.5372-0.7253; P=0.0086,
respectively). Therefore, miR-31 and miR-145 can be used as early
diagnostic markers to differentiate between P and N tissue
(Fig. 2).
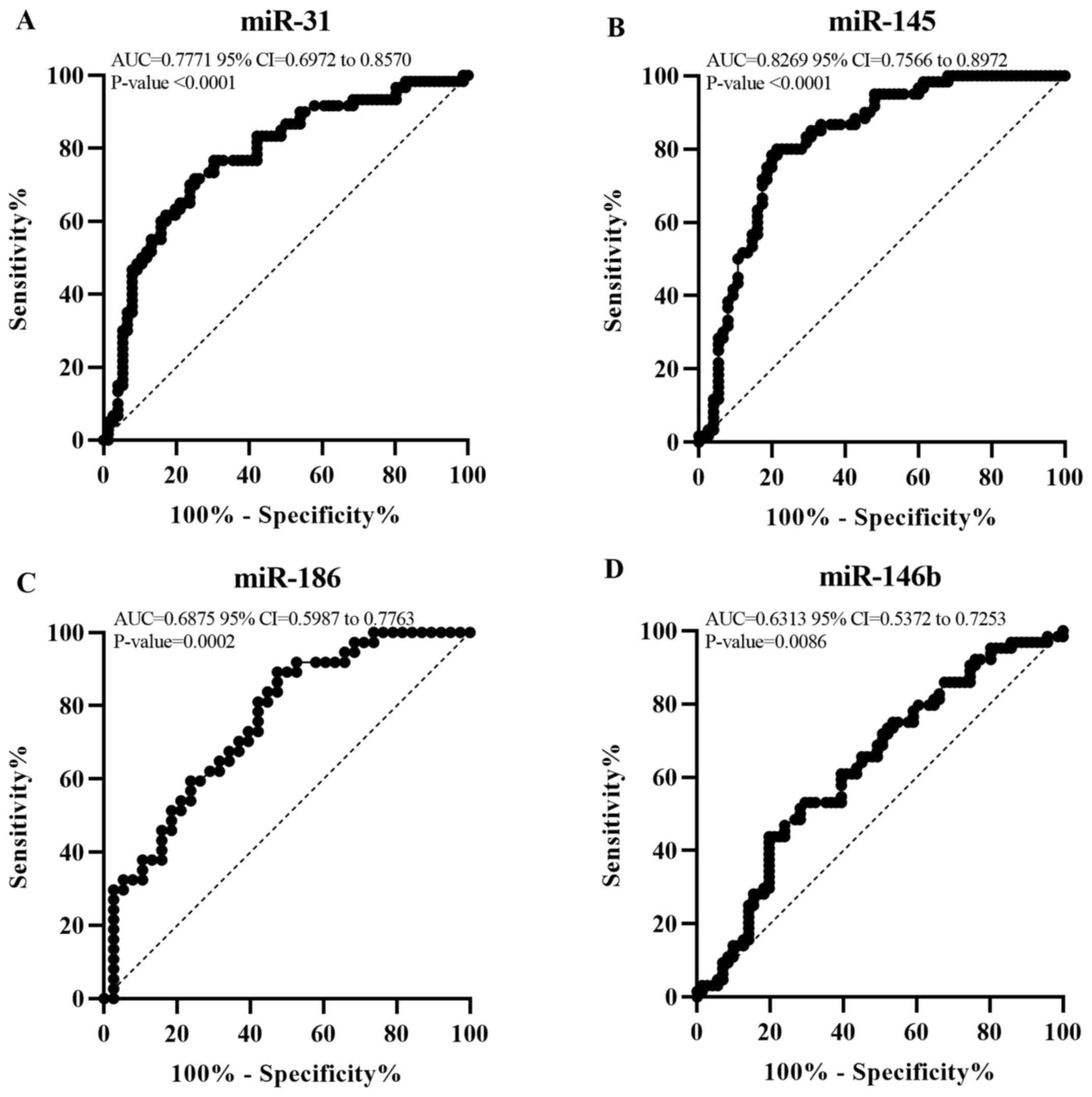 | Figure 2miR-31 and miR-145 show valuable
diagnostic potential at P stage. Receiver operator characteristic
curve for (A) miR-31, (B) miR-145, (C) miR-186 and (D) miR-146b
expression. This graph represents the AUC for miR-31, miR-145,
miR-186 and miR-146b to reveal their diagnostic value at P stage.
AUC<0.5, no discrimination; 0.5≤AUC<0.7, poor;
0.7≤AUC<0.8, acceptable; 0.8≤AUC<0.9, excellent; AUC≥0.9,
outstanding discrimination. AUC, area under the curve; P, polyp;
miR, microRNA. |
 | Table IIIDiagnostic parameters to evaluate the
diagnostic potential of miRs at polyp stage. |
Table III
Diagnostic parameters to evaluate the
diagnostic potential of miRs at polyp stage.
| miR | AUC | SEM | P-value | 95% CI | Sensitivity, % | Specificity, % | Cut-off value | Youden's index | PPV | NPV | DA, % |
|---|
| miR-31 | 0.7771 | 0.04078 | <0.0001 | 0.6972-0.8570 | 76.67 | 69.74 | 2.533 | 0.4641 | 55.0 | 84.2 | 71.3 |
| miR-145 | 0.8269 | 0.03586 | <0.0001 | 0.7566-0.8972 | 80.00 | 72.00 | 2.857 | 0.5200 | 78.3 | 78.7 | 78.5 |
| miR-186 | 0.6875 | 0.04529 | 0.0002 | 0.5987-0.7763 | 67.19 | 60.56 | 1.704 | 0.2775 | 48.4 | 78.9 | 64.4 |
| miR-146b | 0.6313 | 0.04799 | 0.0086 | 0.5372-0.7253 | 60.94 | 60.56 | 1.545 | 0.2150 | 43.8 | 76.1 | 60.7 |
Discussion
CRC is a major worldwide health burden with a high
mortality rate (32). Despite the
availability of several screening techniques for CRC diagnosis,
such as colonoscopy and blood- and stool-based biomarkers, these
tests are not ideal and require improvement for effective early
diagnosis to increase survival rate (13,14).
Considering the contribution of miRNA in the
initiation and progression of cancer, researchers have investigated
their role as an early detection biomarker in several types of
cancer, including CRC (20,33,34).
miRNA expression varies within populations primarily
due to specific genetic variation and single nucleotide
polymorphism in mature miRNA (35,36).
The present study assessed the expression profile of miRNAs
(miR-31, miR-145, miR-186 and miR-146b) in FFPE tissue of Lebanese
patients at different stages (P, Ad and ADC) to identify miRNAs
that are aberrantly expressed in CRC samples and highlight miRNAs
that may have early diagnostic value. When the different CRC stages
were compared with healthy controls, miR-31 was upregulated at all
stages (P<0.0001). By contrast, miR-145, miR-186 and miR-146b
were significantly downregulated at all stages (P<0.0001, 0.0009
and 0.0241, respectively). Of these four miRNAs, the present study
identified miR-31 and miR-145 as potentially useful diagnostic
factors with AUC=0.7771 for miR-31 and 0.8269 for miR-145, and
diagnostic accuracy of 71.3 and 78.5%, respectively.
In CRC, dysregulation of miR-31 and miR-145 have
been implicated in cell proliferation, invasion and migration in
vitro and in tumorigenesis and metastasis in CRC tissue
(37,38). miR-31 is reported to be upregulated
in CRC tissue suppressing Special AT-rich sequence-binding protein
2 gene and regulating v-Raf murine sarcoma viral oncogene homolog B
activation, serving a role in the signaling pathway downstream of
epidermal growth factor receptor (39,40).
Furthermore, miR-145 is downregulated in CRC tissue and induces
tumorigenesis by acting on its target, Kirsten rat sarcoma viral
oncogene homolog (41). Consistent
with the present results, the upregulation of miR-31 and
downregulation of miR-145 have been reported in CRC tissue in
different populations such as the Chinese, Japanese and European
populations (38,42,43).
Cui (44) reported
that miR-31 upregulation has a high diagnostic value between normal
and tumor stages. In addition, Peng et al (45) stated that miR-145 is a potential
biomarker for the early diagnosis of CRC. Here, miR-31 and miR-145
significantly differentiated between P and N stages (P<0.0001;
diagnostic accuracy, 71.3 and 78.5%, respectively). These results
indicate that miR-31 and miR-145 detection may be valuable for the
early-stage diagnosis of CRC.
miR-186 exhibits contradicting effects in certain
types of cancer, including CRC, indicating its role as an
onco-miRNA and tumor suppressor miRNA based on its targets
(46). Islam et al
(47) reported that miR-186 is
significantly upregulated in colon tissue and cell lines in a study
performed in Australia. Conversely, Li et al (30) reported a significant downregulation
of miR-186 in colon tissues in the Chinese population and colon
cancer cell lines. Here, miR-186 expression was significantly
downregulated at P stage, accompanied by non-significant
downregulation at Ad and ADC stages compared with N. Additionally
miR-186 was significantly upregulated at ADC compared with P and Ad
stages. This suggests a role for miR-186 as a potential biomarker
at the early stages of multistage CRC carcinogenesis. When the
diagnostic role of miR-186 was investigated using ROC analysis, it
showed a poor diagnostic role between P and N stages with an AUC of
0.6875 and diagnostic accuracy of 64.4%. Hence, miR-186 was shown
to be downregulated at P stage in Lebanese patients but exhibited
poor diagnostic value.
miR-146b has been reported to be deregulated in
different types of cancer. For example, miR-146b has been shown to
be upregulated in papillary thyroid carcinoma and lung and gastric
cancer (48-50).
However, several studies have reported that miR-146b serves a tumor
suppressor function in solid tumors, such as osteosarcoma,
pancreatic and breast cancer and glioma (51-54).
A study in 2012 reported that miR-146b expression is upregulated
from non-neoplastic to dysplastic stage in CRC but downregulated
from the dysplasia stage to the cancerous stage in Crohn's disease
(27). Zhu et al (26) reported that elevated expression of
miR-146b is associated with high prognosis of CRC at the tumor
nodule metastasis stage. In the present study, there was no
significant change in miR-146b expression at different stages of
CRC compared with N. In addition, the ROC curve analysis showed
that miR-146b cannot be used as a diagnostic biomarker between P
stage and N. These results are not consistent with previous studies
suggesting that miR-146b expression is variable between the
Lebanese population and others (26,27).
Thus, miR-146b cannot be considered a biomarker to assess the
progression of CRC in the present samples.
Collectively, there is a growing appreciation for
the role of miRNA expression in CRC. Among the studied miRNAs, only
miR-31 was upregulated in the different stages whereas miR-145,
miR-186, and miR-146b were downregulated. In addition, miR-31 and
miR-145 can be considered as novel predictive tools for diagnosis
of CRC at the early stages of tumorigenesis. Novel miRNA candidates
can be added to established screening tools according to the
expression levels and hence decrease the burden of this disease and
improve survival rate.
Supplementary Material
Diagnostic parameters to evaluate the
diagnostic potential of miR-31 and miR-145 on an independent set of
samples at the polyp stage.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Lebanese University
(grant no. 46693).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SAS and RAM conceived and designed the study. LA
provided FFPE tissue and interpreted the clinicopathological
characteristics of patients. SAS, RAM, RAK analyzed and interpreted
the data. ZAS and BH performed statistical analysis. RN conceived
the study and revised the manuscript. SAS and RAM wrote the
manuscript and confirmed the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Medical Committee at
Bahman Hospital (Beirut, Lebanon; approval no. 27).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang J, Raju GS, Chang DW, Lin S-H, Chen
Z and Wu X: Global and targeted circulating microRNA profiling of
colorectal adenoma and colorectal cancer. Cancer. 124:785–796.
2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rawla P, Sunkara T and Barsouk A:
Epidemiology of colorectal cancer: Incidence, mortality, survival,
and risk factors. Prz Gastroenterol. 14:89–103. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
International Agency for Research on
Cancer: Estimated crude incidence and mortality rates in 2020,
worldwide, both sexes, all ages. IARC, Lyon, 2020.
|
|
5
|
Sung JJY, Ng SC, Chan FKL, Chiu HM, Kim
HS, Matsuda T, Ng SS, Lau JY, Zheng S, Adler S, et al: Asia Pacific
Working Group: An updated Asia Pacific Consensus Recommendations on
colorectal cancer screening. Gut. 64:121–132. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Shamseddine A: Cancer trends in Lebanon
and projections to 2020. Human & Health, 2015. https://www.syndicateofhospitals.org.lb/Content/uploads/SyndicateMagazinePdfs/8217_8-11.pdf.
|
|
7
|
Nagy ZB, Wichmann B, Kalmár A, Galamb O,
Barták BK, Spisák S, Tulassay Z and Molnár B: Colorectal adenoma
and carcinoma specific miRNA profiles in biopsy and their
expression in plasma specimens. Clin Epigenetics.
9(22)2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
American Cancer Society: Colorectal Cancer
Facts and Figures 2020-2022. American Cancer Society, Atlanta, GA,
2020.
|
|
9
|
Cole SR, Tucker GR, Osborne JM, Byrne SE,
Bampton PA, Fraser RJ and Young GP: Shift to earlier stage at
diagnosis as a consequence of the National Bowel Cancer Screening
Program. Med J Aust. 198:327–330. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ni Y, Xie G and Jia W: Metabonomics of
human colorectal cancer: New approaches for early diagnosis and
biomarker discovery. J Proteome Res. 13:3857–3870. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Knudsen AB, Zauber AG, Rutter CM, Naber
SK, Doria-Rose VP, Pabiniak C, Johanson C, Fischer SE,
Lansdorp-Vogelaar I and Kuntz KM: Estimation of benefits, burden,
and harms of colorectal cancer screening strategies: Modeling Study
for the US Preventive Services Task Force. JAMA. 315:2595–2609.
2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gellad ZF, Stechuchak KM, Fisher DA, Olsen
MK, McDuffie JR, Østbye T and Yancy WS: Am J. Gastroenterol.
106:1125–1134. 2011.
|
|
13
|
Brînzan C, Aşchie M, Cozaru G, Dumitru E
and Mitroi A: The diagnostic value of miR-92a, -143, and -145
expression levels in patients with colorectal adenocarcinoma from
Romania. Medicine (Baltimore). 99(e21895)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Al-Sheikh YA, Ghneim HK, Softa KI,
Al-Jobran AA, Al-Obeed O, Mohamed MA, Abdulla M and Aboul-Soud MA:
Expression profiling of selected microRNA signatures in plasma and
tissues of Saudi colorectal cancer patients by qPCR. Oncol Lett.
11:1406–1412. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jung G, Hernández-Illán E, Moreira L,
Balaguer F and Goel A: Epigenetics of colorectal cancer: Biomarker
and therapeutic potential. Nat Rev Gastroenterol Hepatol.
17:111–130. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Xiao Z, Chen S, Feng S, Li Y, Zou J, Ling
H, Zeng Y and Zeng X: Function and mechanisms of microRNA-20a in
colorectal cancer. Exp Ther Med. 19:1605–1616. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Forterre A, Komuro H, Aminova S and Harada
M: A comprehensive review of cancer MicroRNA therapeutic delivery
strategies. Cancers (Basel). 12(1852)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gasparello J, Papi C, Allegretti M,
Giordani E, Carboni F, Zazza S, Pescarmona E, Romania P, Giacomini
P, Scapoli C, et al: A distinctive microRNA (miRNA) signature in
the blood of colorectal cancer (CRC) patients at surgery. Cancers
(Basel). 12(12)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Vishnoi K, Viswakarma N, Rana A and Rana
B: Transcription factors in cancer development and therapy. Cancers
(Basel). 12(E2296)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Schetter AJ, Okayama H and Harris CC: The
role of microRNAs in colorectal cancer. Cancer J. 18:244–252.
2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Meng W-J, Yang L, Ma Q, Zhang H, Adell G,
Arbman G, Wang ZQ, Li Y, Zhou ZG and Sun XF: MicroRNA expression
profile reveals miR-17-92 and miR-143-145 cluster in synchronous
colorectal cancer. Medicine (Baltimore). 94(e1297)2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Schee K, Boye K, Abrahamsen TW, Fodstad Ø
and Flatmark K: Clinical relevance of microRNA miR-21, miR-31,
miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC
Cancer. 12(505)2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Saberinia A, Alinezhad A, Jafari F,
Soltany S and Akhavan Sigari R: Oncogenic miRNAs and target
therapies in colorectal cancer. Clin Chim Acta. 508:77–91.
2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Eslamizadeh S, Heidari M, Agah S,
Faghihloo E, Ghazi H, Mirzaei A and Akbari A: The role of MicroRNA
signature as diagnostic biomarkers in different clinical stages of
colorectal cancer. Cell J. 20:220–230. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Slattery ML, Pellatt AJ, Lee FY, Herrick
JS, Samowitz WS, Stevens JR, Wolff RK and Mullany LE: Infrequently
expressed miRNAs influence survival after diagnosis with colorectal
cancer. Oncotarget. 8:83845–83859. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhu Y, Wu G, Yan W, Zhan H and Sun P:
miR-146b-5p regulates cell growth, invasion, and metabolism by
targeting PDHB in colorectal cancer. Am J Cancer Res. 7:1136–1150.
2017.PubMed/NCBI
|
|
27
|
Kanaan Z, Rai SN, Eichenberger MR, Barnes
C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE,
et al: Differential microRNA expression tracks neoplastic
progression in inflammatory bowel disease-associated colorectal
cancer. Hum Mutat. 33:551–560. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Shen X, Jiang H, Chen Z, Lu B, Zhu Y, Mao
J, Chai K and Chen W: MicroRNA-145 inhibits cell migration and
invasion in colorectal cancer by targeting TWIST. OncoTargets Ther.
12:10799–10809. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Feng Y, Zhu J, Ou C, Deng Z, Chen M, Huang
W and Li L: MicroRNA-145 inhibits tumour growth and metastasis in
colorectal cancer by targeting fascin-1. Br J Cancer.
110:2300–2309. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H
and Cai J: MiR-186-5p upregulation inhibits proliferation,
metastasis and epithelial-to-mesenchymal transition of colorectal
cancer cell by targeting ZEB1. Arch Biochem Biophys. 640:53–60.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-ΔΔC(T)) Method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
To KK, Tong CW, Wu M and Cho WC: MicroRNAs
in the prognosis and therapy of colorectal cancer: From bench to
bedside. World J Gastroenterol. 24:2949–2973. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: Opportunities and challenges.
BioMed Res Int. 2015(125094)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hrašovec S and Glavač D: MicroRNAs as
novel biomarkers in colorectal cancer. Front Genet.
3(180)2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Jarry J, Schadendorf D, Greenwood C, Spatz
A and van Kempen LC: The validity of circulating microRNAs in
oncology: Five years of challenges and contradictions. Mol Oncol.
8:819–829. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Rawlings-Goss RA, Campbell MC and Tishkoff
SA: Global population-specific variation in miRNA associated with
cancer risk and clinical biomarkers. BMC Med Genomics.
7(53)2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yu T, Ma P, Wu D, Shu Y and Gao W:
Functions and mechanisms of microRNA-31 in human cancers. Biomed
Pharmacother. 108:1162–1169. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang D, Liu Q, Ren Y, Zhang Y, Wang X and
Liu B: Association analysis of miRNA-related genetic polymorphisms
in miR-143/145 and KRAS with colorectal cancer susceptibility and
survival. Biosci Rep. 41(41)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Mi B, Li Q, Li T, Liu G and Sai J: High
miR-31-5p expression promotes colon adenocarcinoma progression by
targeting TNS1. Aging (Albany NY). 12:7480–7490. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Igarashi H, Kurihara H, Mitsuhashi K, Ito
M, Okuda H, Kanno S, Naito T, Yoshii S, Takahashi H, Kusumi T, et
al: Association of MicroRNA-31-5p with clinical efficacy of
anti-EGFR therapy in patients with metastatic colorectal cancer.
Ann Surg Oncol. 22:2640–2648. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
42
|
Toiyama Y, Takahashi M, Hur K, Nagasaka T,
Tanaka K, Inoue Y, Kusunoki M, Boland CR and Goel A: Serum miR-21
as a diagnostic and prognostic biomarker in colorectal cancer. J
Natl Cancer Inst. 105:849–859. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Cui Q: Significance of miR-27a and miR-31
in early diagnosis and prognosis of colorectal cancer. Oncol Lett.
18:3092–3096. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Peng J, Xie Z, Cheng L, Zhang Y, Chen J,
Yu H, Li Z and Kang H: Paired design study by real-time PCR:
miR-378* and miR-145 are potent early diagnostic biomarkers of
human colorectal cancer. BMC Cancer. 15(158)2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Xiang Y, Tian Q, Guan L and Niu S-S: The
dual role of miR-186 in cancers: Oncomir battling with tumor
suppressor miRNA. Front Oncol. 10(233)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Islam F, Gopalan V, Vider J, Wahab R,
Ebrahimi F, Lu CT, Kasem K and Lam AKY: MicroRNA-186-5p
overexpression modulates colon cancer growth by repressing the
expression of the FAM134B tumour inhibitor. Exp Cell Res.
357:260–270. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Geraldo MV, Fuziwara CS, Friguglieti CU,
Costa RB, Kulcsar MA, Yamashita AS and Kimura ET: MicroRNAs
miR-146-5p and let-7f as prognostic tools for aggressive papillary
thyroid carcinoma: A case report. Arq Bras Endocrinol Metabol.
56:552–557. 2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Patnaik SK, Kannisto E, Mallick R and
Yendamuri S: Overexpression of the lung cancer-prognostic miR-146b
microRNAs has a minimal and negative effect on the malignant
phenotype of A549 lung cancer cells. PLoS One.
6(e22379)2011.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yoon SO, Kim EK, Lee M, Jung WY, Lee H,
Kang Y, Jang YJ, Hong SW, Choi SH and Yang WI: NOVA1 inhibition by
miR-146b-5p in the remnant tissue microenvironment defines occult
residual disease after gastric cancer removal. Oncotarget.
7:2475–2495. 2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Al-Khalaf HH and Aboussekhra A:
MicroRNA-141 and microRNA-146b-5p inhibit the prometastatic
mesenchymal characteristics through the RNA-binding protein AUF1
targeting the transcription factor ZEB1 and the protein kinase AKT.
J Biol Chem. 289:31433–31447. 2014.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Lin F, Wang X, Jie Z, Hong X, Li X, Wang M
and Yu Y: Inhibitory effects of miR-146b-5p on cell migration and
invasion of pancreatic cancer by targeting MMP16. J Huazhong Univ
Sci Technolog Med Sci. 31:509–514. 2011.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Hurst DR, Edmonds MD, Scott GK, Benz CC,
Vaidya KS and Welch DR: Breast cancer metastasis suppressor 1
up-regulates miR-146, which suppresses breast cancer metastasis.
Cancer Res. 69:1279–1283. 2009.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Katakowski M, Zheng X, Jiang F, Rogers T,
Szalad A and Chopp M: MiR-146b-5p suppresses EGFR expression and
reduces in vitro migration and invasion of glioma. Cancer Invest.
28:1024–1030. 2010.PubMed/NCBI View Article : Google Scholar
|
















