Introduction
Cervical cancer is considered one of the leading
causes of cancer-associated mortality among women globally
(1,2). In particular, >85% of new cases
and 90% of cervical cancer-related deaths occur in developing
countries (3-5).
Despite efforts made to improve cervical cancer therapy, the 5-year
survival rate remains <50% (6,7). In
China, the incidence rate of cervical cancer is estimated to be
~15.4/100,000, with a relatively high mortality rate (8-10).
Due to the huge population and high rate of human papillomavirus
(HPV) infection, precise diagnosis and treatment are required for
cervical cancer in China.
HPV, a highly prevalent sexually transmitted virus,
is a circular dsDNA virus containing six early genes (E1, E2, E4,
E5, E6 and E7) and two late genes named L1 and L2 (11-16).
Currently, ~200 HPV genotypes have been identified based on the
nucleotide diversity of the L1 gene, and 15 of these are regarded
as ‘high risk’, contributing to the development of cervical cancer,
including HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -53, -56,
-58, -59, -66 and -68 (17-21).
It has been reported that HPV16 is the ‘highest-risk’ HPV genotype
with HPV-18 being second, according to their oncogenic potential
(22-27).
More than 1 year of persistent HPV infection may be
an important risk factor for the progression of cervical cancer and
its precursors (28,29). HPVs can establish their progeny,
spread their viral genes, infect basal cells and further promote
epithelial-mesenchymal transition (30,31).
However, HPV infection itself is not sufficient for the initiation
and establishment of malignant cell transformation (32,33).
Accordingly, HPV tests have a poor positive predictive value, since
HPV infection will progress to cervical cancer in only a few women
(34,35). Clinical data have indicated that
HPV infection is self-limiting and regresses in several cases,
suggesting that other biomolecular mechanisms are involved in the
progression of cervical cancer. A number of studies have
demonstrated that DNA methylation is involved in the carcinogenic
process of cervical cancer (36-40).
The hypermethylation of CpG islands in the promoter regions of
specific genes, including tumour suppressor genes, leads to the
silencing of the gene and inhibits the downstream pathways. By
contrast, disruption of epigenetic processes can lead to the
activation of oncogenes, and the accumulation of epigenetic changes
is an essential step in the development of cervical cancer
(41,42).
An increasing number of studies have indicated that
DNA methylation is an early event in tumorigenesis and plays a
major role in tumour initiation and the progression of cervical
cancer (43). Therefore, it is
crucial to identify reliable prognostic and predictive DNA
methylation-related biomarkers that may help in the early diagnosis
and treatment strategies for cervical cancer. In the present study,
to elucidate the effect of the combination of HPV genotypes and DNA
methylation, these methylation biomarkers in HPV-16- and
HPV-18-positive women with cervical carcinoma were analysed using a
human Illumina Human Methylation 850 K BeadChip. The purpose of the
present study was to identify the hypermethylation of CpG islands
of genetic variability from samples tested positive HPV-16 and
HPV-18 and to elucidate possible methylation biomarkers of
HPV-positive women with a risk of developing cervical cancer.
Materials and methods
Human tissue specimens
A total of six paraffin-embedded specimens,
including three cases of HPV 18 (HPV-18 group)- and 3 cases of
HPV-16 (HPV-16 group)-positive cervical carcinoma tissues, were
collected and diagnosed on a pathological basis according to the
FIGO (2009) clinical staging criteria (44). Normal cervical tissues were
obtained from three women with hysteromyoma who underwent total
hysterectomy (normal group) from July, 2014 through December, 2017
at the Department of Obstetrics and Gynecology of the First
People's Hospital of Lanzhou and Gansu Provincial Hospital. All
experiments performed in the present study were approved by the
Ethics Committee of The First People's Hospital of Lanzhou and
Gansu Provincial Hospital (approval no. (2016-02). Written informed
consent was obtained from all the patients, and the time of signing
the agreement was the time of sample collection.
HPV genotyping and grouping
DNA extraction was performed using a QIAGEN QIAamp
DNA Mini kit (cat. no. 51304; Qiagen GmbH) from formalin-fixed and
paraffin-embedded tissue sections according to the manufacturer's
instructions. The DNA samples were identified and quantified using
a NanoDrop™ 8000 spectrophotometer (Thermo Fisher Scientific, Inc.)
and agarose gel electrophoresis. Genotyping with positive samples
was performed by using the HPV Genotyping Detection kit, the Assay
Kit for Genotyping Human Papillomavirus (PCR-reverse dot blot)
(cat. no. CP.008.022; Guangzhou, LBP Medicine Science &
Technology Co., Ltd.) for subtypes HPV-16 and HPV-18. Samples were
tested following the manufacturer's instructions (45). The HPV-positive control and
-negative controls were set in each experiment. In total, 12
samples were genotyped with the HPV Genotyping Detection kit, and
three HPV-16-positive specimens and three HPV-18-positive specimens
were randomly selected (Table
SI).
DNA methylation chip
An Illumina Human Methylation 850K BeadChip
(Illumina, Inc.) was used to detect the whole genome methylation
status of HPV-16- and HPV-18-positive tissues. Genomic DNA of the
normal group (three cases normal cervical tissues), HPV-16 group
(HPV-16-positive specimens with cervical cancer), and HPV-18 group
(three cases of HPV-18-positive specimens with cervical cancer) was
extracted using the QIAamp DNA Mini kit (cat. no. 51304; Qiagen
GmbH) and bisulfite-converted using the EZ DNA Methylation kit
(cat. no. D5001; Zymo Research Corp.). The converted DNA was
hybridized to an Infinium Human Methylation 850 K BeadChip. The
subsequent bioinformatics analysis was performed by Genergy Co. The
Illumina 850 K methylation chip analysis data have been uploaded in
the GEO public database repository (accession no. GSE169622,
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169622).
Pyrosequencing
Through pyrosequencing, nine candidates of the
HPV-16 group screened by the 850K methylation chip were verified.
An EZ 96-DNA methylation kit (Zymo Research Corp.) was used for
bisulfite conversion in accordance with the manufacturer's standard
procedures, with fully methylated and unmethylated samples as test
controls. PyroMark Assay Design 2.0 was used for the synthesis of
bisulfite-PCR primers, which were synthesized by The Beijing
Genomics Institute (BGI). A list of bisulfite PCR primers is
presented in Table SII. The
bisulfite PCR amplification conditions were as follows:
Pre-denaturation at 95˚C for 3 min; 40 cycles at 94˚C for 30 sec,
52˚C for 30 sec, and 72˚C for 1 min; and a final elongation at 72˚C
for 7 min. The HotStarTaq DNA polymerase (Qiagen Ltd.) was used for
disulfide PCR amplification. Compared to the PyroMark Q96 ID
(Qiagen Ltd.), Pyro Q CpG software (version 2.0.6, Qiagen GmbH)
automatically analysed the methylation status of each site.
Bioinformatics analysis
The 850K chip data analysis w implemented in R
language. The whole analysis model is based on the highly
integrated R analysis package ChAMP (Version: 2.8.9), which
inherits the methods of Minfi, Limma, Sva, and IMA analysis
packages. The graph was acquired using self-written R script, the
basic function in GGplot2 implementation. Microarray data were
normalized using BMIQ (Beta MIXture Quantile dilation). SVD
(Singular Value Decomposition) was applied to evaluate the major
components of variables in the data set, and then a Bayesian
model-based Combat method was used to eliminate the batch effect.
Quality control is achieved through a set of functions provided by
ChAMP, such as CpG.GUI, champ.QC, and QC.GUI, and then MVP
(Methylation variable position), DMR (Differently methylated
regions), Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) analysis are also implemented with ChAMP. Among
them, MVP uses the method of Limma R package. Limma firstly
establishes multiple linear regression model for all data as a
whole, and then applies the regression model to each probe line and
calculates the modified T-statistic and P-value in combination with
the Bayesian model. The sites with P≤0.05 after the FDR correction
were considered as differential methylation sites.
Cervical cancer data sets
The DNA methylation data from The Cancer Genome
Atlas (TCGA)-Cervical Squamous Cell Carcinoma and Endocervical
Adenocarcinoma (CESC) were downloaded from TCGA website (https://portal.gdc.cancer.gov/). β-values were
extracted to evaluate the DNA methylation level of each probe. The
candidate significantly differentially methylated CpG sites of the
HPV-18 group were screened using the 850 k methylation chip were
verified in (TCGA-CESC).
Statistical analysis
Statistical analysis was performed using SPSS
software (Release 13.0, SPSS Inc.) Statistical analysis data
comparisons between two groups were analysed using the Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Methylation analysis and general
characteristics
The 850K methylation sites in the HPV 16- and HPV
18-positive cervical carcinoma and normal cervical tissues were
analysed using the Illumina 850K methylation chip (GSE169622). The
data from cervical carcinoma and normal cervical tissue samples
were normalized (negative and positive controls were provided by
Genergy Co.) and processed. To analyse DNA methylation differences
between the cervical cancer group and the normal group, Δ β and
P-values were used to construct a volcanic map of CpG sites, in
order to reflect the magnitude and statistical significance of
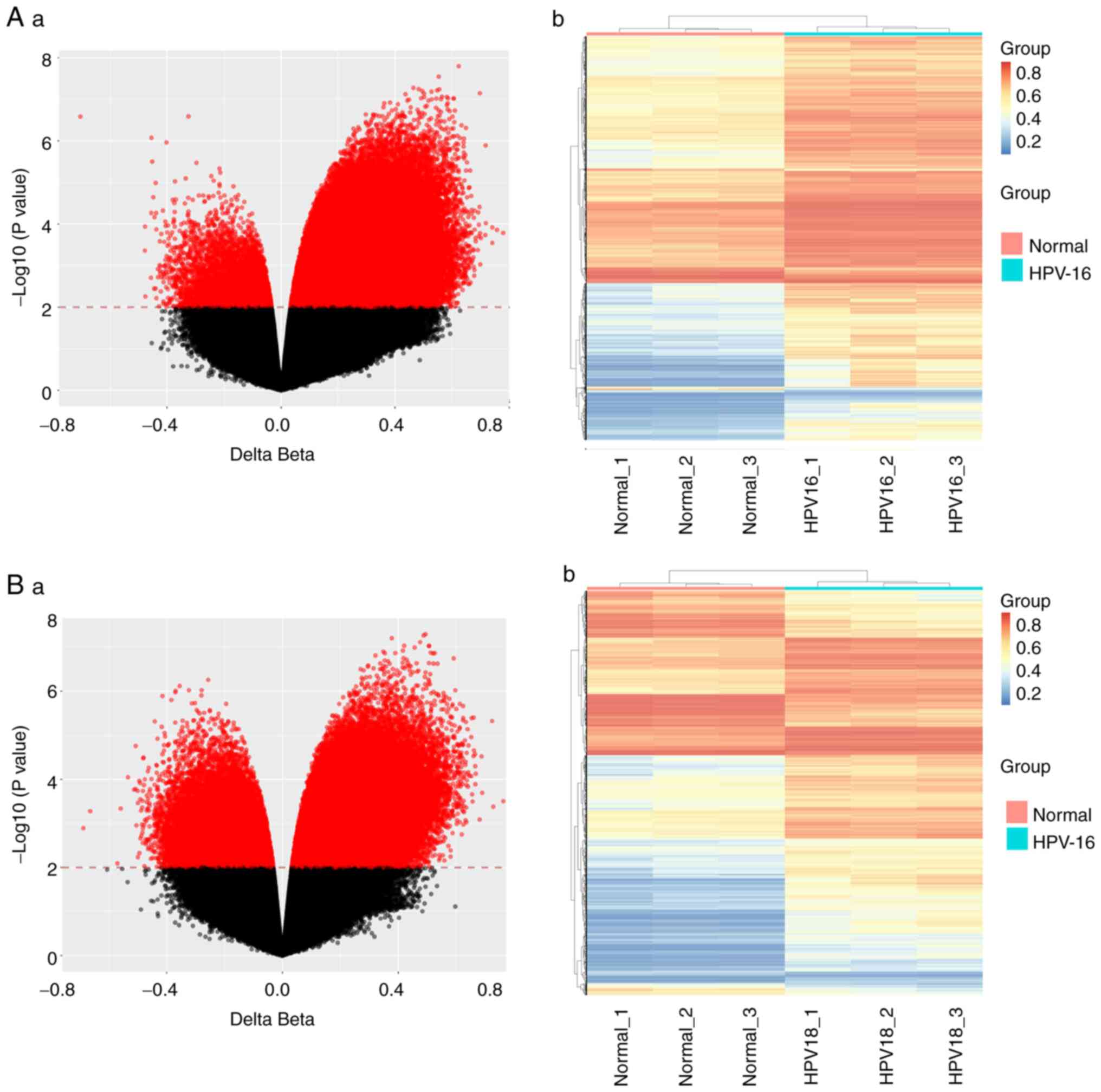
differences (P<0.01). As depicted in Fig. 1, the gene methylation profiles of
HPV-16 (HPV-16 group) and HPV-18 (HPV-18 group) cervical carcinoma
samples differed significantly from those of the control samples
(normal group). In total, it was observed that 106,378 sites
demonstrated differential expression in HPV 16-positive cervical
carcinoma tissues compared to normal cervical tissues, of which
101,152 were hypermethylated and 5,226 were hypomethylated
(Fig. 1A-a). In addition, 70,744
sites showed differential expression in HPV 18-positive cervical
carcinoma tissues compared to normal cervical tissues, of which
53,168 were hypermethylated and 17,576 were hypomethylated
(Fig. 1B-a). In addition, a
cluster analysis map was generated to show the methylation status
of different groups. As demonstrated in Fig. 1, there were significant differences
in methylation patterns and states between cervical cancer and
normal samples in the HPV-16 group and HPV-18 group. Thus, there
may be biomarkers suitable for cervical cancer screening among
these differential methylation sites.
GO functional analysis
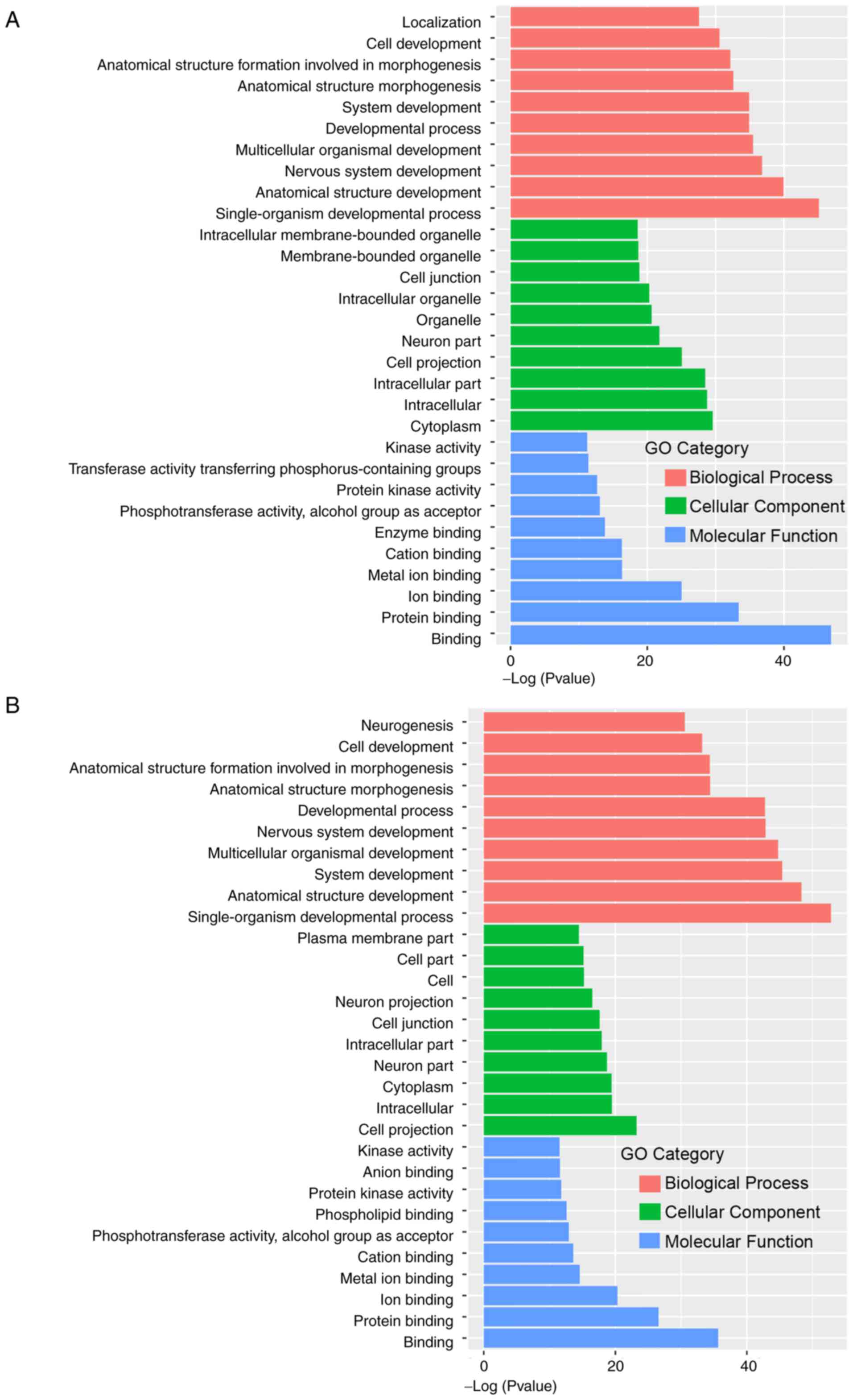
The GO functional annotation analysis of the HPV-16
differentially expressed methylation sites revealed that these
enriched genes were mainly involved in Biological Process, Cellular
Component and Molecular Function (Fig.
2A). The results of cellular component analyses revealed that
molecules distributed in the cell periphery, plasma membrane, cell
junction and cell membrane components were significantly enriched.
Important functions, such as cell migration, transport and
synthesis of substances all occur place at these sites. At the
molecular level, functional annotation analysis revealed that the
highly enriched genes were related to calcium binding, protein
binding, cytoskeletal protein binding, metal ion transmembrane
transport activity and phosphotransferase activity (Fig. 2A). The GO functional annotation
analysis of the HPV-18-differentially expressed methylation sites
demonstrated that these genes were mainly enriched in Biological
Processes, Cellular Component and Molecular Function (Fig. 2B). The biological processes of the
two groups of methylation differential genes were mainly
concentrated in the biological development process and anatomical
structure development.
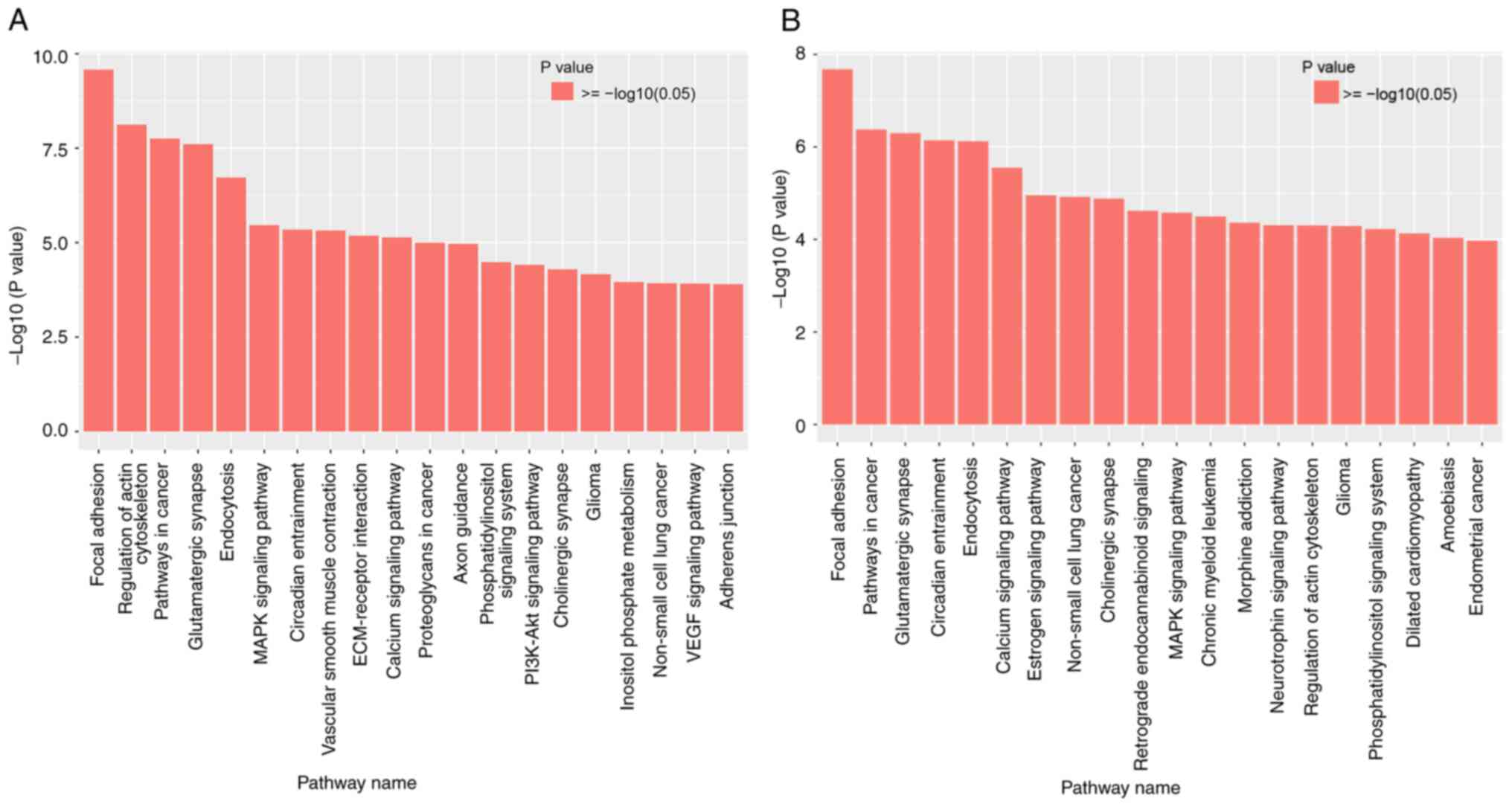
KEGG signalling pathway analysis
KEGG pathway functional analysis annotates and
classifies the functions of pathways in the KEGG database according
to whole genes and differential genes. The signalling pathways were
further investigated using the KEGG database. According to the
criteria of P<0.01 and FDR <0.05, the top 20 related
signalling pathways of different methylation sites were selected
(Fig. 3). The most prominent major
signalling pathways for the HPV-16-positive samples were focal
adhesion, pathways in cancer, glutamatergic synapse and the
regulation of actin cytoskeleton, suggesting that these pathways
are major regulatory factors of cancer behaviours (Fig. 3A). Additionally, focal adhesion,
pathways in cancer, glutamatergic synapse and circadian entrainment
signalling pathways were significantly upregulated in the
HPV-18-positive samples (Fig. 3B).
It was observed that focal adhesion and pathways in cancer are
among the top pathways in the comparison of the two groups. This
suggests that it may be possible to monitor the degree of cervical
lesions by detecting gene differential methylation sites in these
pathways.
Identification of different
methylation sites in the HPV-16 and HPV-18 groups
Subsequently, the 3,000 methylation variable
positions in genes with the most significant difference were
further analysed by KEGG according to the selected significantly
different signalling pathways, including focal adhesion and
pathways in cancer from HPV-16- and HPV-18-positive samples. The
results indicated that a total of 10 genes, including CHRM2, GNG4,
LAMA4, CHAD, ITGA8, COL11A1, FGF10, IGF1, TEK and COL11A2, were
screened from the focal adhesion and PI3K-AKT signalling pathways
from the HPV-16 group. Moreover, the Gene network analysis revealed
that the most significantly different CpG sites, cg24575234,
cg20818778, cg14289461, cg06818777, cg08361126, cg06381931,
cg08976810, cg20881548, cg08264401 and cg25459558, can be found in
the CHRM2, GNG4, LAMA4, CHAD, ITGA8, COL11A1, FGF10, IGF1, TEK and
COL11A2 genes, indicating their possible diagnostic roles in
cervical cancer development (Table
I). Some of these genes have already exhibited critical roles
in several types of cancer, such as thymic, gastric, ovarian,
breast, pancreatic and colorectal cancer. For examples, it has been
shown that DNA methylation of GNG4 is a common epigenetic
alteration in thymic carcinoma (46). LAMA4 and COL11A1 are associated
with tumour invasion and metastasis (47,48).
 | Table IThe top 10 methylation difference
sites of HPV-16 group screened using the 850 k methylation
chip. |
Table I
The top 10 methylation difference
sites of HPV-16 group screened using the 850 k methylation
chip.
| Sequence | Probe ID | Normal group
average methylation level | HPV-16 group
average methylation level | Difference between
two groups | P-value | FDR | Gene |
|---|
| 1 | cg24575234 | 0.0700±0.0234 | 0.6615±0.1325 | 0.592 | 0.000318 |
0.00888b | CHRM2 |
| 2 | cg20818778 | 0.1310±0.0074 | 0.6913±0.1583 | 0.560 | 0.000888 |
0.01466a | GNG4 |
| 3 | cg14289461 | 0.1687±0.0852 | 0.7237±0.1104 | 0.555 | 0.000511 |
0.011113a | LAMA4 |
| 4 | cg06818777 | 0.0654±0.0250 | 0.6105±0.1142 | 0.545 | 0.000241 |
0.007846b | CHAD |
| 5 | cg08361126 | 0.0901±0.0376 | 0.6207±0.1280 | 0.531 | 0.000514 |
0.011149a | ITGA8 |
| 6 | cg06381931 | 0.1690±0.0502 | 0.6946±0.0608 | 0.526 | 4.34E-05 |
0.004157b | COL11A1 |
| 7 | cg08976810 | 0.0369±0.0098 | 0.5484±0.1995 | 0.511 | 0.003815 |
0.033664a | FGF10 |
| 8 | cg20881548 | 0.2406±0.0908 | 0.7451±0.0314 | 0.505 | 0.000138 |
0.006232b | IGF1 |
| 9 | cg08264401 | 0.2143±0.0175 | 0.7006±0.0315 | 0.486 | 1.74E-06 |
0.002313b | TEK |
| 10 | cg25459558 | 0.0932±0.0189 | 0.5783±0.1569 | 0.485 | 0.00171 |
0.02099a | COL11A2 |
For the HPV-18 group, the 10 most significantly
different CpG sites, cg03520644, cg25792518, cg06958829,
cg00172849, cg19707040, cg02501779, cg19679123, cg14427009,
cg25993718 and cg27423357 was selected. The gene network analysis
indicated that they were located in COL11A1, CHAD, CHAD, COL11A1,
CTNNA2, CBLN4, SMAD3, PCDH17, CBLN4 and FLT1 (Table II). In addition, the 10 selected
methylation significantly different sites were verified in
TCGA-CESC (Table III). All 10
sites exhibited statistically significant differences (P<0.05),
and seven sites demonstrated highly significant differences
(P<0.01, Table III).
 | Table IIThe top 10 methylation difference
sites of HPV-18 group screened using the 850 k methylation
chip. |
Table II
The top 10 methylation difference
sites of HPV-18 group screened using the 850 k methylation
chip.
| Sequence | Probe ID | G5 normal group
average methylation level | G3 cervical cancer
HPV-18-positive group average methylation level | Difference between
two groups | P-value | FDR | Gene |
|---|
| 1 | cg03520644 | 0.1225±0.0342 | 0.3966±0.0241 | 0.2741 | 5.98E-05 |
0.009034a | COL11A1 |
| 2 | cg25792518 | 0.3841±0.0564 | 0.7650±0.0315 | 0.3809 | 8.44E-05 |
0.009827a | CHAD |
| 3 | cg06958829 | 0.1182±0.0121 | 0.6611±0.0797 | 0.5430 | 4.17E-05 |
0.008166a | CHAD |
| 4 | cg00172849 | 0.1219±0.0440 | 0.3997±0.0141 | 0.2778 | 8.70E-05 |
0.009885a | COL11A1 |
| 5 | cg19707040 | 0.1011±0.0136 | 0.4713±0.0504 | 0.3702 | 3.63E-05 |
0.007918a | CTNNA2 |
| 6 | cg02501779 | 0.1815±0.0145 | 0.7214±0.0734 | 0.5399 | 3.00E-05 |
0.007761a | CBLN4 |
| 7 | cg19679123 | 0.4993±0.0466 | 0.8142±0.0246 | 0.3149 | 8.42E-05 |
0.009826a | SMAD3 |
| 8 | cg14427009 | 0.2460±0.0267 | 0.7325±0.0381 | 0.4865 | 5.43E-06 |
0.006396a | PCDH17 |
| 9 | cg25993718 | 0.0971±0.0343 | 0.5181±0.0515 | 0.4211 | 4.20E-05 |
0.00819a | CBLN4 |
| 10 | cg27423357 | 0.3920±0.0364 | 0.7782±0.0226 | 0.3862 | 1.20E-05 |
0.006597a | FLT1 |
 | Table IIIThe top 10 methylation difference
sites of HPV-18 group screened using the 850 k methylation chip
were verified in TCGA. |
Table III
The top 10 methylation difference
sites of HPV-18 group screened using the 850 k methylation chip
were verified in TCGA.
| Sequence | Probe ID | Normal group | Cervical cancer
group HPV-18-positive | t-test | P-value | Gene |
|---|
| 1 | cg03520644 | 0.616±0.0312 | 0.7381±0.0467 | -20.847 |
<0.001c | COL11A1 |
| 2 | cg25792518 | 0.2926±0.1046 | 0.8885±0.0626 | -8.464 | 0.001b | CHAD |
| 3 | cg06958829 | 0.1529±0.0501 | 0.7040±0.0587 | -12.366 |
<0.001c | CHAD |
| 4 | cg00172849 | 0.1886±0.0464 | 0.6807±0.0938 | -8.146 | 0.001b | COL11A1 |
| 5 | cg19707040 | 0.0350±0.084 | 0.5199±0.0944 | -8.523 | 0.001b | CTNNA2 |
| 6 | cg02501779 | 0.1541±0.0400 | 0.6263±0.1349 | -5.814 | 0.004b | CBLN4 |
| 7 | cg19679123 | 0.4073±0.1601 | 0.8318±0.0875 | -4.030 | 0.016a | SMAD3 |
| 8 | cg14427009 | 0.2136±0.0373 | 0.6362±0.0527 | -11.327 |
<0.001c | PCDH17 |
| 9 | cg25993718 | 0.1236±0.0384 | 0.5085±0.1573 | -4.118 | 0.015a | CBLN4 |
| 10 | cg27423357 | 0.4072±0.0873 | 0.7177±0.1584 | -2.975 | 0.041a | FLT1 |
Pyrosequencing verification
In total, 10 candidate significantly differentially
methylated CpG sites of the HPV-16 group, including cg24575234,
cg20818778, cg14289461, cg06818777, cg08361126, cg06381931,
cg08976810, cg20881548, cg08264401 and cg25459558, screened using
the 850k methylation chip were verified by pyrosequencing. Among
these, cg25459558 was withdrawn from verification due to the
failure of primer design. The statistical comparison of the mean
value revealed that the other nine sites exhibited significant
differences (P<0.05), of which six sites exhibited highly
significant differences (P<0.01, Table IV). The statistical comparison of
means revealed that the most significantly different CpG sites
between the normal and HPV-16 samples cg24575234, cg14289461,
cg06818777, cg08361126, cg06381931, cg08976810, cg20881548 and
cg08264401, detected in the CHRM2, LAMA4, CHAD, ITGA8, COL11A1,
FGF10, IGF1 and TEK. Unexpectedly, the increasing mean value of the
CpG site, cg20818778, for the gene GNG4 was not statistically
significant (detailed statistical analysis data are contained in
Table IV).
 | Table IVVerification of different top 9
methylation CpG sites in the HPV-16 group using pyrosequencing. |
Table IV
Verification of different top 9
methylation CpG sites in the HPV-16 group using pyrosequencing.
| Sequence | Probe ID | Normal group | Cervical cancer
group HPV-16 | t-test | P-value | Gene |
|---|
| 1 | cg24575234 | 8.68±1.46 | 33.69±5.71 | 9.495 |
<0.001c | CHRM2 |
| 2 | cg20818778 | 14.92±0.603 |
28.17±1.33c | 1.916 | 0.092a | GNG4 |
| 3 | cg14289461 | 10.62±3.91 | 48.76±16.40 | 5.059 | 0.001b | LAMA4 |
| 4 | cg06818777 | 7.86±2.17 |
44.13±2.54c | 2.775 | 0.024a | CHAD |
| 5 | cg08361126 | 8.46±1.17 |
24.53±11.36c | 2.833 | 0.025a | ITGA8 |
| 6 | cg06381931 | 26.16±4.54 | 59.35±10.52 | 6.474 |
<0.001c | COL11A1 |
| 7 | cg08976810 | 9.39±1.68 | 52.08±12.63 | 7.492 |
<0.001c | FGF10 |
| 8 | cg20881548 | 36.35±4.50 | 60.61±10.68 | 4.684 | 0.002b | IGF1 |
| 9 | cg08264401 | 23.79±2.77 | 44.38±12.60 | 3.568 | 0.007b | TEK |
The mechanism of cervical cancer development remains
unclear, while individual differences are significant. Further
investigations of the methylation characteristics of a single gene
and a single locus as a biomarker for cancer screening will have a
high missed diagnosis rate. Multipoint joint detection is suggested
for the future improvement of cancer detection rate.
Discussion
Cervical cancer formation is affected by several
risk factors, including HPV infection. It has been reported that
HPV16 and HPV18 contribute to >70% of all cervical cancer cases
worldwide and are thus entitled as a ‘high-risk’ HPV genotype
(49-51).
In some studies, HPV-16/18 genotyping is used as a molecular marker
reflecting the underlying carcinogenic process. However, HPV
infection is self-limiting and regresses in some clinical cases
(30,52,53).
It is unclear whether HPV infection acts as a key determinant of
the progression to cervical cancer. Consequently, HPV positivity is
not a specific diagnostic indicator for cervical cancer or
diseases.
DNA methylation is one of the mechanisms that has
been closely related to the occurrence and development of cervical
cancer. The aberrant DNA methylation of human host cell genes or
HPV genomic DNA has been closely associated with the dysfunction of
various tumour suppressor genes during persistent high-risk HPV
(HR-HPV) infection and cervical carcinogenesis (54-56).
Studies have indicated that the tumour suppressor genes, p53 and
p73, demonstrate a higher degree of methylation in cervical cancer
samples than in normal samples (57,58).
The aberrant DNA methylation of CpG islands is comparatively rare
in normal cells, suggesting that the differentially methylated CpG
sites between cervical cancer and normal samples have the potential
to become reliable biomarkers of cervical cancer. In addition, DNA
hypermethylation has been associated with long-term HR-HPV
infection and is therefore considered a marker of cervical
intraepithelial neoplasia lesion severity and invasive cervical
cancer risk (59). However, the
high heterogeneity of those previously published data renders it
difficult to determine the appropriate methylation markers for
cervical cancer screening (60).
Additionally, the expression levels of E6 oncoprotein have a
different effect on the carcinogenic potential of HPV. For example,
the enhanced expression of the HPV-16 E6 oncogenes may trigger a
neoplastic transformation of squamous epithelial cells at the
uterine cervix (61). Thus, the
HPV promoter methylation profile could be an easy and measurable
biomarker for the examination of the high-risk HPV potential
carcinogenicity.
In the present study, the genome-wide methylation
level was evaluated by comparing HPV-16-positive or HPV-18-positive
cervical cancer cases with normal cervical tissues. The results of
the present study indicated that 106,378 and 70,744 sites
demonstrated differential expression in HPV-16 and HPV-18 cervical
cancer tissues as compared with normal cervical tissues,
respectively, indicating that the distribution of methylation sites
in cervical tissues varies greatly. It has been reported that
hypermethylation at CpG islands (CGIs) of genes acting as tumour
suppressors is a common mechanism involved in cancer occurrence
(62-64).
Other studies have also detected an apparently positive association
between the hypomethylation of proto-oncogenes and the progression
of cervical cancer. In the present study, 101,152 with higher
methylation levels and 5,226 with lower methylation levels CGIs in
HPV-16-positive cancer tissues than in normal cervical tissues were
identified. By contrast, 53,168 CGIs with increased methylation
levels and 17,576 CGIs with decreased methylation levels were
identified in HPV-18-positive cancer tissues compared with normal
cervical tissues. Genome-wide methylation level evaluation can
retrieve additional differential methylation sites that have not
been previously discovered.
Moreover, the differentially expressed methylation
genes were analysed through GO functional annotation. It has been
revealed that a number of methylated genes are closely associated
with HPV-positive cervical cancer cases, including SOX1, PAX1,
JAM3, EPB41L3, CADM1 and MAL (65,66).
For example, expression of SOX1 was shown to be associated with
early embryogenesis, central nervous system development, and neural
stem cell maintenance. Hypermethylated PAX1 has been detected in
cervical carcinoma (67). The
aforementioned methylated human gene biomarkers used in combination
may be clinically useful for the triage of women with HR-HPV
infections. The functional annotation data have previously
demonstrated that the highly enriched genes were mainly involved in
calcium binding, protein binding, cytoskeletal protein binding,
metal ion transmembrane transport activity and phosphotransferase
activity (68-71).
The results of cellular component analyses revealed that molecules
distributed in the cell periphery, plasma membrane, cell junction,
and cell membrane components were significantly enriched. Further
cluster analysis demonstrated that the differentially methylated
genes covered a variety of different functional communities,
indicating that there are many types of genes involved in the
regulation of the occurrence and progression of cervical cancer
(72-74).
Previous data indicate that a variety of cellular
pathways can be affected by the methylation status of specific
genes. KEGG pathway analysis in the present study revealed that
differentially methylated genes were mainly involved in focal
adhesion, regulation of actin cytoskeleton, and pathways in cancer.
Among these pathways, the most significant pathways were focal
adhesion and PI3K-AKT signalling pathways, which are a collection
of receptors and ligands on the plasma membrane associated with
intracellular and extracellular signalling pathways that regulate
cell growth and cell migration. Based on the KEGG pathway analysis
results, a total of nine genes, including CHRM2, GNG4, LAMA4, CHAD,
ITGA8, COL11A1, FGF10, IGF1 and TEK, associated with nine
significantly different CpG sites, cg24575234, cg20818778,
cg14289461, cg06818777, cg08361126, cg06381931, cg08976810,
cg20881548 and cg08264401, were screened from the focal adhesion
and PI3K-AKT signalling pathways of the HPV-16-positive group.
However, the pyrosequencing data of the present study indicated
that the increasing mean value of the CpG site, cg20818778, for the
gene GNG4 was not statistically significant. Thus, the most
significantly different CpG sites are cg24575234, cg14289461,
cg06818777, cg08361126, cg06381931, cg08976810, cg20881548 and
cg08264401, detected in the CHRM2, LAMA4, CHAD, ITGA8, COL11A1,
FGF10, IGF1 and TEK, indicating their possible diagnostic roles in
cervical carcinoma development.
Additionally, the 10 most significantly different
CpG sites of the HPV-18-positive group, cg03520644, cg25792518,
cg06958829, cg00172849, cg19707040, cg02501779, cg19679123,
cg14427009, cg25993718 and cg27423357 in COL11A1, CHAD, CHAD,
COL11A1, CTNNA2, CBLN4, SMAD3, PCDH17, CBLN4 and FLT1 were
selected, indicating their future applications as candidate
molecular markers of cervical cancer. Furthermore, the 10 selected
methylation significantly different sites were verified in
TCGA-CESC. All 10 sites exhibited significant differences
(P<0.05), and seven sites demonstrated highly significant
differences (P<0.01, Table
III).
Some of the genes screened in the present study have
already demonstrated potential functions in other diseases. Several
studies have revealed that CHRM2 is associated with the
pathophysiology of schizophrenia (75). GNG4 has been associated with immune
infiltration in the tumour microenvironment, which promotes tumour
cell migration and proliferation (76-78).
LAMA4 can regulate the proliferation and migration of gastric
cancer cells (47) and may be a
potential gastric cancer prognostic biomarker and therapeutic
target (79). ITGB8 silencing
inhibits the invasion and migration of lung cancer cells (80). Increased expression of COL11A1 has
been detected in several in various types of cancer, such as
ovarian, breast, pancreatic and colorectal cancer, and increased
levels of COL11A1 are often associated with poor survival,
chemoresistance and recurrence (48). Fgf10 induces migration and invasion
of pancreatic cancer cells (81).
CHAD, however, has been reported to be involved in confronting
hepatocellular carcinoma migration and proliferation and predicting
good survival (82). In addition,
PCDH17 methylation has been reported as a potential prognostic
biomarker in some cancer patient markers, including postoperative
renal cell carcinoma (83).
Validating whether these markers can be used as novel tools for
cervical cancer screening and investigating their role in normal
cervical and cervical cancer development will be the focus of our
future research.
In summary, the host cell gene methylation test may
be a promising method for cervical cancer screening. the Illumina
Human Methylation 850K BeadChip methylation chip was used for the
methylation site detection in HPV 16- and HPV 18-positive cervical
carcinoma and normal cervical tissues. The current findings
suggested that the methylation modification sites in cervical
carcinoma cells may be abnormal. Hypermethylation and
hypomethylation sites occur more frequently and are mainly enriched
in functional categories, including focal adhesion, regulation of
actin cytoskeleton, tumour growth, and pathways in cancer. The
eight most significantly different CpG sites, cg24575234,
cg14289461, cg06818777, cg08361126, cg06381931, cg08976810,
cg20881548 and cg08264401, were screened and verified from
HPV-16-positive samples and were associated with the CHRM2, LAMA4,
CHAD, ITGA8, COL11A1, FGF10, IGF1 and TEK genes. The 10 most
significantly different CpG sites of the HPV-18-positive group,
cg03520644, cg25792518, cg06958829, cg00172849, cg19707040,
cg02501779, cg19679123, cg14427009, cg25993718 and cg27423357,
which are located in COL11A1, CHAD, CHAD, COL11A1, CTNNA2, CBLN4,
SMAD3, PCDH17, CBLN4 and FLT1, were selected and verified in
TCGA-CESC. It is important to explore and develop DNA methylation
assays of improved sensitivity and specificity in order to
ameliorate the early detection of cervical cancer (84-86).
The findings of the present study may provide fundamental data for
the use of methylation biomarkers for cervical cancer diagnosis;
however, further research is required.
Supplementary Material
HPV Genotyping and grouping.
List of bisulfite PCR and sequencing
primers.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the innovation and
entrepreneurship talent project of Lanzhou (2015-RC-68),
distinguished professorship program of national research program on
prevention and control of major birth defects in reproductive
health (2017YFC1000900), special-funded program on national key
scientific instruments and equipment development (2016YFF0103800),
health and family planning commission research project of Jiangsu
province (H201619).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request. The Illumina 850K methylation chip analysis data have been
uploaded in the GEO public database repository (accession number:
GSE169622, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169622).
Authors' contributions
JQ and TC conceived and designed this study. YM and
CW were responsible for performing the experiments and writing the
first draft of the manuscript. JQ was responsible for revising the
first draft of the manuscript. MS and ML collected and analysed the
data. LL and YM interpreted the data of DNA methylation. JQ and TC
confirm the authenticity of all the raw data. All the authors
critically reviewed the original manuscript, edited and approved
the final version. All the authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki. Written informed consent was obtained
from all patients. The study was approved by the Medical Ethics
Examination of Lanzhou First People's Hospital, China
(2016-02).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xing B, Guo J, Sheng Y, Wu G and Zhao Y:
Human papillomavirus-negative cervical cancer: A comprehensive
review. Front Oncol. 10(606335)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Buskwofie A, David-West G and Clare CA: A
review of cervical cancer: Incidence and disparities. J Natl Med
Assoc. 112:229–232. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sosso SM, Tchouaket MCT, Fokam J, Simo RK,
Torimiro J, Tiga A, Lobe EE, Ambada G, Nange A, Semengue ENJ, et
al: Human immunodeficiency virus is a driven factor of human
papilloma virus among women: Evidence from a cross-sectional
analysis in Yaoundé, Cameroon. Virol J. 17(69)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mapanga W, Singh E, Feresu SA and
Girdler-Brown B: Treatment of pre- and confirmed cervical cancer in
HIV-seropositive women from developing countries: A systematic
review. Syst Rev. 9(79)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cegla P, Burchardt E, Roszak A,
Czepczynski R, Kubiak A and Cholewinski W: Influence of biological
parameters assessed in [18F]FDG PET/CT on overall survival in
cervical cancer patients. Clin Nucl Med. 44:860–863.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ding FN, Gao BH, Wu X, Gong CW, Wang WQ
and Zhang SM: miR-122-5p modulates the radiosensitivity of cervical
cancer cells by regulating cell division cycle 25A (CDC25A). FEBS
Open Bio. 9:1869–1879. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Song B, Ding C and Chen W, Sun H, Zhang M
and Chen W: Incidence and mortality of cervical cancer in China,
2013. Chin J Cancer Res. 29:471–476. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gu XY, Zheng RS, Sun KX, Zhang SW, Zeng
HM, Zou XN, Chen WQ and He J: Incidence and mort ality of cervical
cancer in China, 2014. Zhonghua Zhong Liu Za Zhi. 40:241–246.
2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
10
|
He R, Zhu B, Liu J, Zhang N, Zhang WH and
Mao Y: Women's cancers in China: A spatio-temporal epidemiology
analysis. BMC Womens Health. 21(116)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jee B, Yadav R, Pankaj S and Shahi SK:
Immunology of HPV-mediated cervical cancer: Current understanding.
Int Rev Immunol. 40:359–378. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Brianti P, De Flammineis E and Mercuri SR:
Review of HPV-related diseases and cancers. New Microbiol.
40:80–85. 2017.PubMed/NCBI
|
|
13
|
Tulay P and Serakinci N: The route to
HPV-associated neoplastic transformation: A review of the
literature. Crit Rev Eukaryot Gene Expr. 26:27–39. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen J: Signaling pathways in
HPV-associated cancers and therapeutic implications. Rev Med Virol.
25 (Suppl 1):S24–S53. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Lototskaja E, Sahharov O, Piirsoo M, Kala
M, Ustav M and Piirsoo A: Cyclic AMP-dependent protein kinase
exhibits antagonistic effects on the replication efficiency of
different HPV types. J Virol. 10(e0025121)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bhatt KH, Neller MA, Srihari S, Crooks P,
Lekieffre L, Aftab BT, Liu H, Smith C, Kenny L, Porceddu S and
Khanna R: Profiling HPV-16-specific T cell responses reveals broad
antigen reactivities in oropharyngeal cancer patients. J Exp Med.
217(e20200389)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bouvard V, Baan R, Straif K, Grosse Y,
Secretan B, Ghissassi FE, Benbrahim-Tallaa L, Guha N, Freeman C,
Galichet L, et al: A review of human carcinogens-Part B: Biological
agents. Lancet Oncol. 10:321–322. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tao G, Yaling G, Zhan G, Pu L and Miao H:
Human papillomavirus genotype distribution among HPV-positive women
in Sichuan province, Southwest China. Arch Virol. 163:65–72.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Castellsagué X: Natural history and
epidemiology of HPV infection and cervical cancer. Gynecol Oncol.
110 (3 Suppl 2):S4–S7. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Schellenbacher C, Roden RBS and Kirnbauer
R: Developments in L2-based human papillomavirus (HPV) vaccines.
Virus Res. 231:166–175. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lippert J, Bonlokke S, Utke A, Knudsen BR,
Sorensen BS, Steiniche T and Stougaard M: Targeted next generation
sequencing panel for HPV genotyping in cervical cancer. Exp Mol
Pathol. 118(104568)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Bordigoni A, Motte A, Tissot-Dupont H,
Colson P and Desnues C: Development and validation of a multiplex
qPCR assay for detection and relative quantification of HPV16 and
HPV18 E6 and E7 oncogenes. Sci Rep. 11(4039)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fan Z, Feng X, Zhang W, Li N, Zhang X and
Lin JM: Visual detection of high-risk HPV16 and HPV18 based on
loop-mediated isothermal amplification. Talanta.
217(121015)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Layman H, Rickert KW, Wilson S, Aksyuk AA,
Dunty JM, Natrakul D, Swaminathan N and DelNagro CJ: Development
and validation of a multiplex immunoassay for the simultaneous
quantification of type-specific IgG antibodies to E6/E7
oncoproteins of HPV16 and HPV18. PLoS One.
15(e0229672)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Peng S, Ferrall L, Gaillard S, Wang C, Chi
WY, Huang CH, Roden RBS, Wu TC, Chang YN and Hung CF: Development
of DNA vaccine targeting E6 and E7 proteins of human papillomavirus
16 (HPV16) and HPV18 for immunotherapy in combination with
recombinant vaccinia boost and PD-1 antibody. mBio.
12:e03224–e03220. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Berti FCB, Mathias C, Garcia LE, Gradia
DF, de Araujo Souza PS, Cipolla GA, de Oliveira JC and Malheiros D:
Comprehensive analysis of ceRNA networks in HPV16- and
HPV18-mediated cervical cancers reveals XIST as a pivotal competing
endogenous RNA. Biochim Biophys Acta Mol Basis Dis.
1867(166172)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hammer A, Rositch A, Qeadan F, Gravitt PE
and Blaakaer J: Age-specific prevalence of HPV16/18 genotypes in
cervical cancer: A systematic review and meta-analysis. Int J
Cancer. 138:2795–2803. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Luvero D, Lopez S, Bogani G, Raspagliesi F
and Angioli R: From the infection to the immunotherapy in cervical
cancer: Can we stop the natural course of the disease? Vaccines
(Basel). 8(597)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lin W, Niu Z, Zhang H, Kong Y, Wang Z,
Yang X and Yuan F: Imbalance of Th1/Th2 and Th17/Treg during the
development of uterine cervical cancer. Int J Clin Exp Pathol.
12:3604–3612. 2019.PubMed/NCBI
|
|
30
|
Ho GY, Bierman R, Beardsley L, Chang CJ
and Burk RD: Natural history of cervicovaginal papillomavirus
infection in young women. N Engl J Med. 338:423–428.
1998.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Bodily J and Laimins LA: Persistence of
human papillomavirus infection: Keys to malignant progression.
Trends Microbiol. 19:33–39. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Graham SV: The human papillomavirus
replication cycle, and its links to cancer progression: A
comprehensive review. Clin Sci (Lond). 131:2201–2221.
2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hebner CM and Laimins LA: Human
papillomaviruses: Basic mechanisms of pathogenesis and
oncogenicity. Rev Med Virol. 16:83–97. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Yoo SH, Ock CY, Keam B, Park SJ, Kim TM,
Kim JH, Jeon YK, Chung EJ, Kwon SK, Hah JH, et al: Poor prognostic
factors in human papillomavirus-positive head and neck cancer: Who
might not be candidates for de-escalation treatment? Korean J
Intern Med. 34:1313–1323. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Linde DS, Andersen MS, Mwaiselage JD,
Manongi R, Kjaer SK and Rasch V: Text messages to increase
attendance to follow-up cervical cancer screening appointments
among HPV-positive Tanzanian women (Connected2Care): Study protocol
for a randomised controlled trial. Trials. 18(555)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li X, Wu X, Li Y, Cui Y, Tian R, Singh N,
Ding M, Yang Y and Gao Y: Promoter hypermethylation of SOX11
promotes the progression of cervical cancer in vitro and
in vivo. Oncol Rep. 41:2351–2360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chen L, Qiu X, Zhang N, Wang Y, Wang M, Li
D, Wang L and Du Y: APOBEC-mediated genomic alterations link
immunity and viral infection during human papillomavirus-driven
cervical carcinogenesis. Biosci Trends. 11:383–388. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Lorincz AT: Cancer diagnostic classifiers
based on quantitative DNA methylation. Expert Rev Mol Diagn.
14:293–305. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yang S, Wu Y, Wang S, Xu P, Deng Y, Wang
M, Liu K, Tian T, Zhu Y, Li N, et al: HPV-related methylation-based
reclassification and risk stratification of cervical cancer. Mol
Oncol. 14:2124–2141. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fertey J, Hagmann J, Ruscheweyh HJ, Munk
C, Kjaer S, Huson D, Haedicke-Jarboui J, Stubenrauch F and Iftner
T: Methylation of CpG 5962 in L1 of the human papillomavirus 16
genome as a potential predictive marker for viral persistence: A
prospective large cohort study using cervical swab samples. Cancer
Med. 9:1058–1068. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang L, Hu D, Wang S, Zhang Y, Pang L,
Tao L and Jia W: Association between dense PAX1 promoter
methylation and HPV16 infection in cervical squamous epithelial
neoplasms of Xin Jiang Uyghur and Han women. Gene.
723(144142)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Franzen A, Vogt TJ, Muller T, Dietrich J,
Schrock A, Golletz C, Brossart P, Bootz F, Landsberg J, Kristiansen
G and Dietrich D: PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter
methylation is associated with HPV infection and transcriptional
repression in head and neck squamous cell carcinomas. Oncotarget.
9:641–650. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
El Aliani A, El-Abid H, El Mallali Y,
Attaleb M, Ennaji MM and El Mzibri M: Association between gene
promoter methylation and cervical cancer development: Global
distribution and a meta-analysis. Cancer Epidemiol Biomarkers Prev.
30:450–459. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Muñoz N, Bosch FX, de Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ:
Epidemiologic classification of human papillomavirus types
associated with cervical cancer. N Engl J Med. 348:518–527.
2003.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kishibuchi R, Kondo K, Soejima S, Tsuboi
M, Kajiura K, Kawakami Y, Kawakita N, Sawada T, Toba H, Yoshida M,
et al: DNA methylation of GHSR, GNG4, HOXD9 and SALL3 is a common
epigenetic alteration in thymic carcinoma. Int J Oncol. 56:315–326.
2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang M, Li C, Liu Y and Wang Z: Effect of
LAMA4 on prognosis and its correlation with immune infiltration in
gastric cancer. Biomed Res Int. 2021(6428873)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Nallanthighal S, Heiserman JP and Cheon
DJ: Collagen type XI alpha 1 (COL11A1): A novel biomarker and a key
player in cancer. Cancers (Basel). 13(935)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Xu HH, Wang K, Feng XJ, Dong SS, Lin A,
Zheng LZ and Yan WH: Prevalence of human papillomavirus genotypes
and relative risk of cervical cancer in China: A systematic review
and meta-analysis. Oncotarget. 9:15386–15397. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lin C, Franceschi S and Clifford GM: Human
papillomavirus types from infection to cancer in the anus,
according to sex and HIV status: A systematic review and
meta-analysis. Lancet Infect Dis. 18:198–206. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Malary M, Moosazadeh M, Hamzehgardeshi Z,
Afshari M, Moghaddasifar I and Afsharimoghaddam A: The prevalence
of cervical human papillomavirus infection and the most at-risk
genotypes among iranian healthy women: A systematic review and
meta-analysis. Int J Prev Med. 7(70)2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chan PK, Picconi MA, Cheung TH,
Giovannelli L and Park JS: Laboratory and clinical aspects of human
papillomavirus testing. Crit Rev Clin Lab Sci. 49:117–136.
2012.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kaliterna V and Barisic Z: Genital human
papillomavirus infections. Front Biosci (Landmark Ed).
23:1587–1611. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
54
|
Clarke MA, Wentzensen N, Mirabello L,
Ghosh A, Wacholder S, Harari A, Lorincz A, Schiffman M and Burk RD:
Human papillomavirus DNA methylation as a potential biomarker for
cervical cancer. Cancer Epidemiol Biomarkers Prev. 21:2125–2137.
2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Johannsen E and Lambert PF: Epigenetics of
human papillomaviruses. Virology. 445:205–212. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Szalmás A and Kónya J: Epigenetic
alterations in cervical carcinogenesis. Semin Cancer Biol.
19:144–152. 2009.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Jha AK, Sharma V, Nikbakht M, Jain V,
Sehgal A, Capalash N and Kaur J: A comparative analysis of
methylation status of tumor suppressor genes in paired biopsy and
serum samples from cervical cancer patients among north Indian
population. Genetika. 52:255–259. 2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
de la Cruz-Hernandez E, Perez-Cardenas E,
Contreras-Paredes A, Cantu D, Mohar A, Lizano M and Duenas-Gonzalez
A: The effects of DNA methylation and histone deacetylase
inhibitors on human papillomavirus early gene expression in
cervical cancer, an in vitro and clinical study. Virol J.
4(18)2007.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Lendvai Á, Johannes F, Grimm C, Eijsink
JJ, Wardenaar R, Volders HH, Klip HG, Hollema H, Jansen RC,
Schuuring E, et al: Genome-wide methylation profiling identifies
hypermethylated biomarkers in high-grade cervical intraepithelial
neoplasia. Epigenetics. 7:1268–1278. 2012.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Kelly H, Benavente Y, Pavon MA, De Sanjose
S, Mayaud P and Lorincz AT: Performance of DNA methylation assays
for detection of high-grade cervical intraepithelial neoplasia
(CIN2+): A systematic review and meta-analysis. Br J Cancer.
121:954–965. 2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Hoppe-Seyler K, Bossler F, Braun JA,
Herrmann AL and Hoppe-Seyler F: The HPV E6/E7 Oncogenes: Key
factors for viral carcinogenesis and therapeutic targets. Trends
Microbiol. 26:158–168. 2018.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Bachman KE, Park BH, Rhee I, Rajagopalan
H, Herman JG, Baylin SB, Kinzler KW and Vogelstein B: Histone
modifications and silencing prior to DNA methylation of a tumor
suppressor gene. Cancer Cell. 3:89–95. 2003.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Clark SJ and Melki J: DNA methylation and
gene silencing in cancer: Which is the guilty party? Oncogene.
21:5380–5387. 2002.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Tahara T, Shibata T, Yamashita H, Nakamura
M, Yoshioka D, Okubo M, Hirata I and Arisawa T: Chronic
nonsteroidal anti-inflammatory drug (NSAID) use suppresses multiple
CpG islands hyper methylation (CIHM) of tumor suppressor genes in
the human gastric mucosa. Cancer Sci. 100:1192–1197.
2009.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Clarke MA, Gradissimo A, Schiffman M, Lam
J, Sollecito CC, Fetterman B, Lorey T, Poitras N, Raine-Bennett TR,
Castle PE, et al: Human papillomavirus DNA methylation as a
biomarker for cervical precancer: Consistency across 12 genotypes
and potential impact on management of HPV-positive women. Clin
Cancer Res. 24:2194–2202. 2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Cuschieri K, Ronco G, Lorincz A, Smith L,
Ogilvie G, Mirabello L, Carozzi F, Cubie H, Wentzensen N, Snijders
P, et al: Eurogin roadmap 2017: Triage strategies for the
management of HPV-positive women in cervical screening programs.
Int J Cancer. 143:735–745. 2018.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Zhao Z, Zhang X, Zhao X, Cai J, Wu NY and
Wang J: SOX1 and PAX1 are hypermethylated in cervical
adenocarcinoma and associated with better prognosis. Biomed Res
Int. 2020(3981529)2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Meng M, Sang L and Wang X: S100 calcium
binding protein A11 (S100A11) promotes the proliferation, migration
and invasion of cervical cancer cells, and activates Wnt/β-catenin
signaling. Onco Targets Ther. 12:8675–8685. 2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Tian T, Li X, Hua Z, Ma J, Wu X, Liu Z,
Chen H and Cui Z: S100A7 promotes the migration, invasion and
metastasis of human cervical cancer cells through
epithelial-mesenchymal transition. Oncotarget. 8:24964–24977.
2017.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Chokchaichamnankit D, Watcharatanyatip K,
Subhasitanont P, Weeraphan C, Keeratichamroen S, Sritana N,
Kantathavorn N, Diskul-Na-Ayudthaya P, Saharat K, Chantaraamporn J,
et al: Urinary biomarkers for the diagnosis of cervical cancer by
quantitative label-free mass spectrometry analysis. Oncol Lett.
17:5453–5468. 2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Tomiyama N, Ikeda R, Nishizawa Y, Masuda
S, Tajitsu Y and Takeda Y: S100A16 up-regulates Oct4 and Nanog
expression in cancer stem-like cells of Yumoto human cervical
carcinoma cells. Oncol Lett. 15:9929–9933. 2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Lee HS, Yun JH, Jung J, Yang Y, Kim BJ,
Lee SJ, Yoon JH, Moon Y, Kim JM and Kwon YI: Identification of
differesntially-expressed genes by DNA methylation in cervical
cancer. Oncol Lett. 9:1691–1698. 2015.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Liu MY, Zhang H, Hu YJ, Chen YW and Zhao
XN: Identification of key genes associated with cervical cancer by
comprehensive analysis of transcriptome microarray and methylation
microarray. Oncol Lett. 12:473–478. 2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Ma X, Liu J, Wang H, Jiang Y, Wan Y, Xia Y
and Cheng W: Identification of crucial aberrantly methylated and
differentially expressed genes related to cervical cancer using an
integrated bioinformatics analysis. Bioscience Rep.
40(BSR20194365)2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Dean B and Scarr E: Possible involvement
of muscarinic receptors in psychiatric disorders: A focus on
schizophrenia and mood disorders. Curr Mol Med. 15:253–264.
2015.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Liang L, Huang J, Yao M, Li L, Jin XJ and
Cai XY: GNG4 promotes tumor progression in colorectal cancer. J
Oncol. 2021(9931984)2021.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Zhao H, Sheng D, Qian Z, Ye S, Chen J and
Tang Z: Identifying GNG4 might play an important role in colorectal
cancer TMB. Cancer Biomark. 32:435–450. 2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Wen S, Peng W, Chen Y, Du X, Xia J, Shen B
and Zhou G: Four differentially expressed genes can predict
prognosis and microenvironment immune infiltration in lung cancer:
A study based on data from the GEO. BMC Cancer.
22(193)2022.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Wang X, Hou Q and Zhou X: LAMA4 expression
is activated by zinc finger E-box-binding homeobox 1 and
independently predicts poor overall survival in gastric cancer.
Oncol Rep. 40:1725–1733. 2018.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Wang JF, Wang Y, Zhang SW, Chen YY, Qiu Y,
Duan SY, Li BP and Chen JQ: Expression and prognostic analysis of
integrins in gastric cancer. J Oncol. 2020(8862228)2020.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Itoh N and Ohta H: Fgf10: A
paracrine-signaling molecule in development, disease, and
regenerative medicine. Curr Mol Med. 14:504–509. 2014.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Deng X, Wei W, Huang N, Shi Y, Huang M,
Yan Y, Li D, Yi J and Wang X: Tumor repressor gene chondroadherin
oppose migration and proliferation in hepatocellular carcinoma and
predicts a good survival. Oncotarget. 8:60270–60279.
2017.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Lin YL, Wang YP, Li HZ and Zhang X:
Aberrant promoter methylation of PCDH17 (Protocadherin 17) in serum
and its clinical significance in renal cell carcinoma. Med Sci
Monit. 23:3318–3323. 2017.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Xu W, Xu M, Wang L, Zhou W, Xiang R, Shi
Y, Zhang Y and Piao Y: Integrative analysis of DNA methylation and
gene expression identified cervical cancer-specific diagnostic
biomarkers. Signal Transduct Target Ther. 4(55)2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Bhat S, Kabekkodu SP, Varghese VK,
Chakrabarty S, Mallya SP, Rotti H, Pandey D, Kushtagi P and
Satyamoorthy K: Aberrant gene-specific DNA methylation signature
analysis in cervical cancer. Tumour Biol.
39(1010428317694573)2017.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Varghese VK, Shukla V, Kabekkodu SP,
Pandey D and Satyamoorthy K: DNA methylation regulated microRNAs in
human cervical cancer. Mol Carcinog. 57:370–382. 2018.PubMed/NCBI View Article : Google Scholar
|