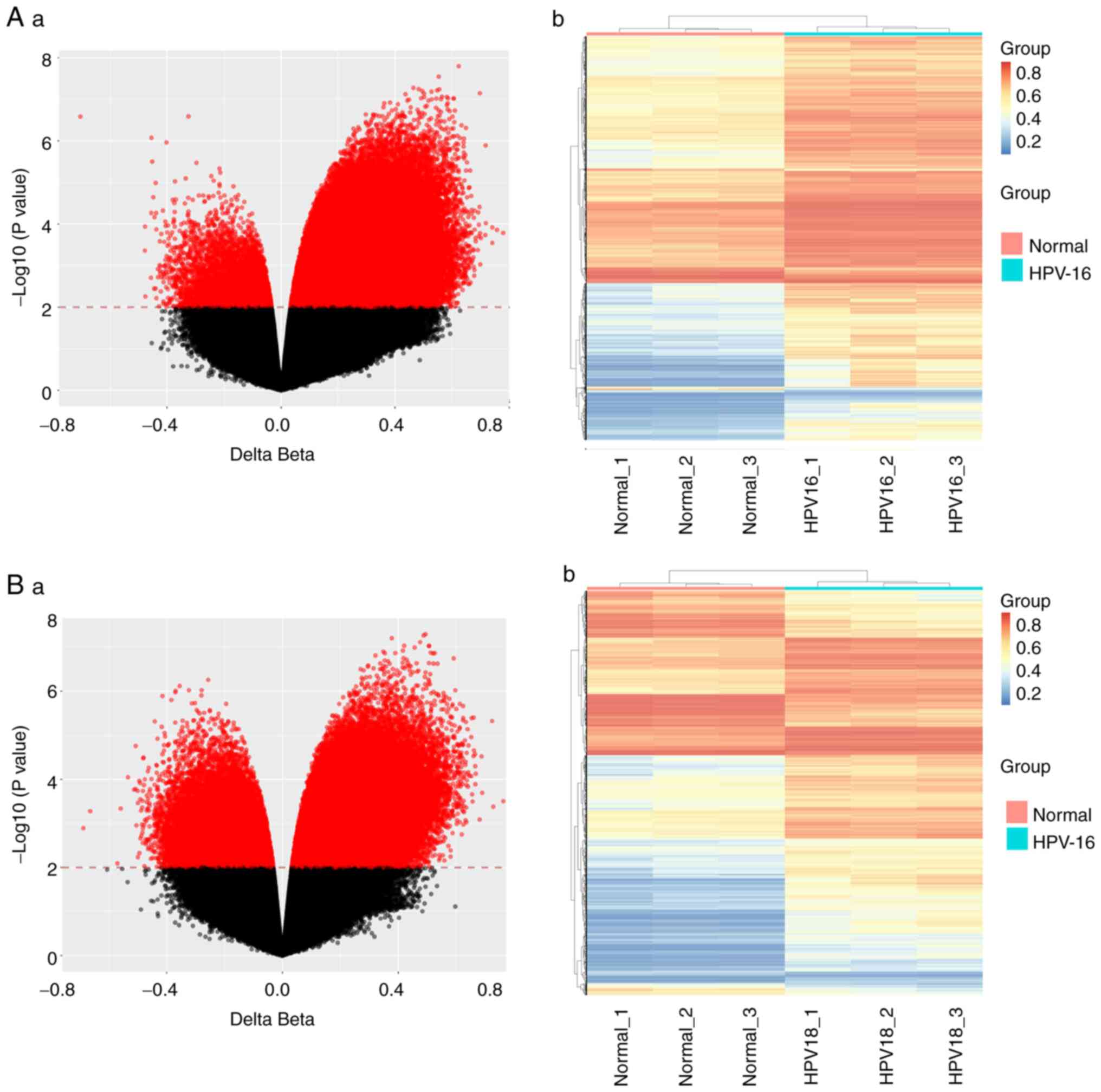
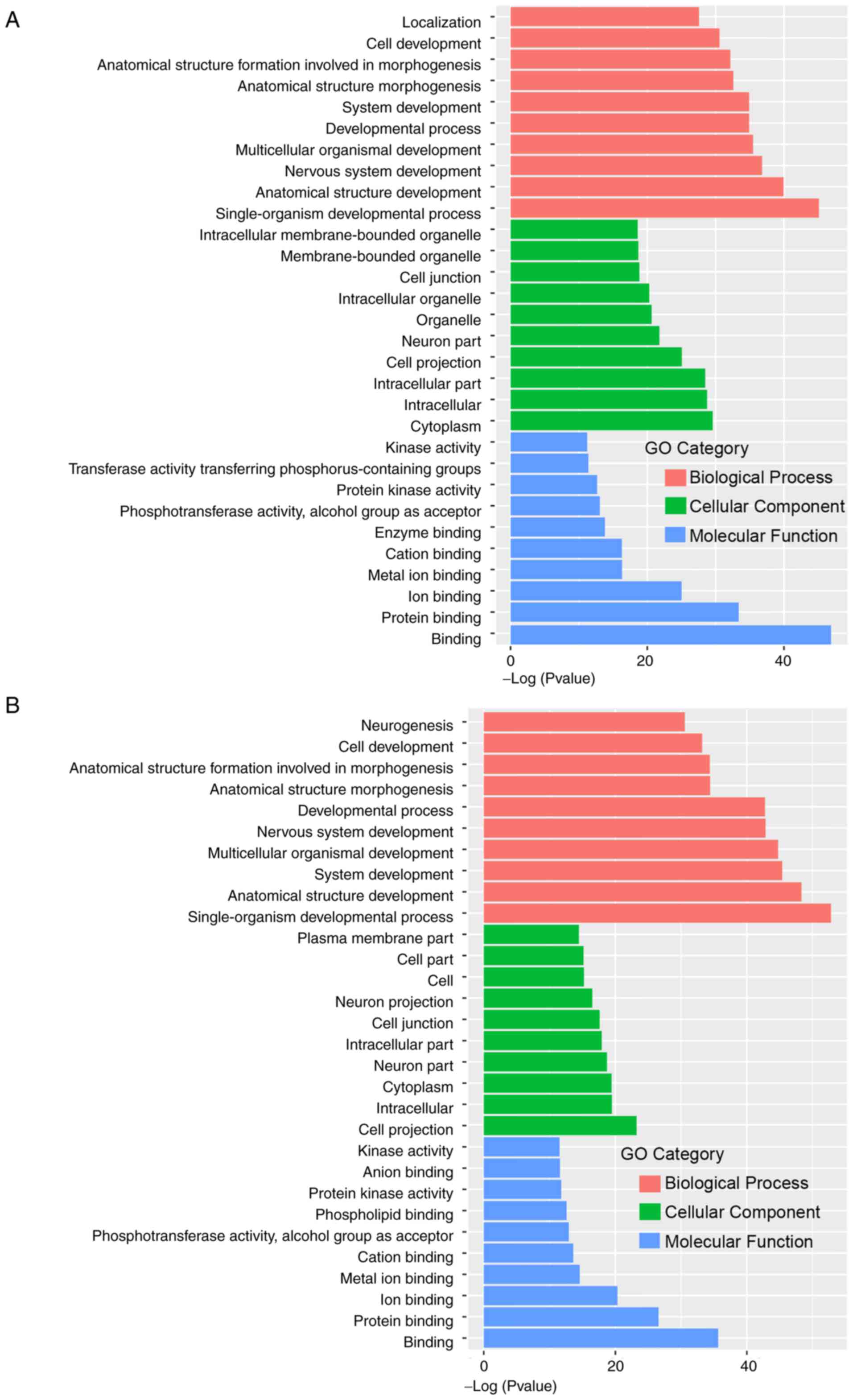
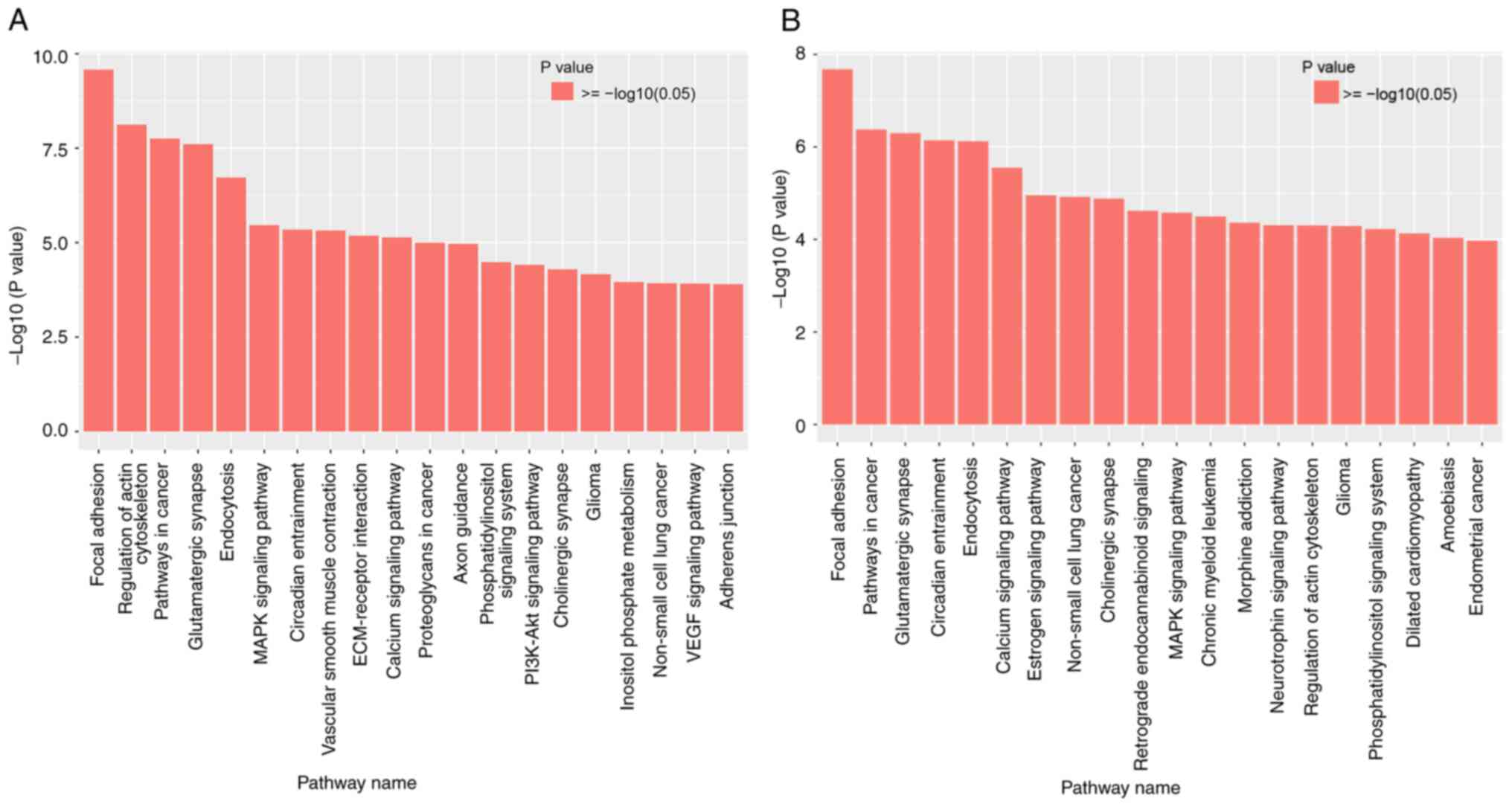
|
1
|
Xing B, Guo J, Sheng Y, Wu G and Zhao Y:
Human papillomavirus-negative cervical cancer: A comprehensive
review. Front Oncol. 10(606335)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Buskwofie A, David-West G and Clare CA: A
review of cervical cancer: Incidence and disparities. J Natl Med
Assoc. 112:229–232. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sosso SM, Tchouaket MCT, Fokam J, Simo RK,
Torimiro J, Tiga A, Lobe EE, Ambada G, Nange A, Semengue ENJ, et
al: Human immunodeficiency virus is a driven factor of human
papilloma virus among women: Evidence from a cross-sectional
analysis in Yaoundé, Cameroon. Virol J. 17(69)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mapanga W, Singh E, Feresu SA and
Girdler-Brown B: Treatment of pre- and confirmed cervical cancer in
HIV-seropositive women from developing countries: A systematic
review. Syst Rev. 9(79)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cegla P, Burchardt E, Roszak A,
Czepczynski R, Kubiak A and Cholewinski W: Influence of biological
parameters assessed in [18F]FDG PET/CT on overall survival in
cervical cancer patients. Clin Nucl Med. 44:860–863.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ding FN, Gao BH, Wu X, Gong CW, Wang WQ
and Zhang SM: miR-122-5p modulates the radiosensitivity of cervical
cancer cells by regulating cell division cycle 25A (CDC25A). FEBS
Open Bio. 9:1869–1879. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Song B, Ding C and Chen W, Sun H, Zhang M
and Chen W: Incidence and mortality of cervical cancer in China,
2013. Chin J Cancer Res. 29:471–476. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gu XY, Zheng RS, Sun KX, Zhang SW, Zeng
HM, Zou XN, Chen WQ and He J: Incidence and mort ality of cervical
cancer in China, 2014. Zhonghua Zhong Liu Za Zhi. 40:241–246.
2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
10
|
He R, Zhu B, Liu J, Zhang N, Zhang WH and
Mao Y: Women's cancers in China: A spatio-temporal epidemiology
analysis. BMC Womens Health. 21(116)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jee B, Yadav R, Pankaj S and Shahi SK:
Immunology of HPV-mediated cervical cancer: Current understanding.
Int Rev Immunol. 40:359–378. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Brianti P, De Flammineis E and Mercuri SR:
Review of HPV-related diseases and cancers. New Microbiol.
40:80–85. 2017.PubMed/NCBI
|
|
13
|
Tulay P and Serakinci N: The route to
HPV-associated neoplastic transformation: A review of the
literature. Crit Rev Eukaryot Gene Expr. 26:27–39. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen J: Signaling pathways in
HPV-associated cancers and therapeutic implications. Rev Med Virol.
25 (Suppl 1):S24–S53. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Lototskaja E, Sahharov O, Piirsoo M, Kala
M, Ustav M and Piirsoo A: Cyclic AMP-dependent protein kinase
exhibits antagonistic effects on the replication efficiency of
different HPV types. J Virol. 10(e0025121)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bhatt KH, Neller MA, Srihari S, Crooks P,
Lekieffre L, Aftab BT, Liu H, Smith C, Kenny L, Porceddu S and
Khanna R: Profiling HPV-16-specific T cell responses reveals broad
antigen reactivities in oropharyngeal cancer patients. J Exp Med.
217(e20200389)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bouvard V, Baan R, Straif K, Grosse Y,
Secretan B, Ghissassi FE, Benbrahim-Tallaa L, Guha N, Freeman C,
Galichet L, et al: A review of human carcinogens-Part B: Biological
agents. Lancet Oncol. 10:321–322. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tao G, Yaling G, Zhan G, Pu L and Miao H:
Human papillomavirus genotype distribution among HPV-positive women
in Sichuan province, Southwest China. Arch Virol. 163:65–72.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Castellsagué X: Natural history and
epidemiology of HPV infection and cervical cancer. Gynecol Oncol.
110 (3 Suppl 2):S4–S7. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Schellenbacher C, Roden RBS and Kirnbauer
R: Developments in L2-based human papillomavirus (HPV) vaccines.
Virus Res. 231:166–175. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lippert J, Bonlokke S, Utke A, Knudsen BR,
Sorensen BS, Steiniche T and Stougaard M: Targeted next generation
sequencing panel for HPV genotyping in cervical cancer. Exp Mol
Pathol. 118(104568)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Bordigoni A, Motte A, Tissot-Dupont H,
Colson P and Desnues C: Development and validation of a multiplex
qPCR assay for detection and relative quantification of HPV16 and
HPV18 E6 and E7 oncogenes. Sci Rep. 11(4039)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fan Z, Feng X, Zhang W, Li N, Zhang X and
Lin JM: Visual detection of high-risk HPV16 and HPV18 based on
loop-mediated isothermal amplification. Talanta.
217(121015)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Layman H, Rickert KW, Wilson S, Aksyuk AA,
Dunty JM, Natrakul D, Swaminathan N and DelNagro CJ: Development
and validation of a multiplex immunoassay for the simultaneous
quantification of type-specific IgG antibodies to E6/E7
oncoproteins of HPV16 and HPV18. PLoS One.
15(e0229672)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Peng S, Ferrall L, Gaillard S, Wang C, Chi
WY, Huang CH, Roden RBS, Wu TC, Chang YN and Hung CF: Development
of DNA vaccine targeting E6 and E7 proteins of human papillomavirus
16 (HPV16) and HPV18 for immunotherapy in combination with
recombinant vaccinia boost and PD-1 antibody. mBio.
12:e03224–e03220. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Berti FCB, Mathias C, Garcia LE, Gradia
DF, de Araujo Souza PS, Cipolla GA, de Oliveira JC and Malheiros D:
Comprehensive analysis of ceRNA networks in HPV16- and
HPV18-mediated cervical cancers reveals XIST as a pivotal competing
endogenous RNA. Biochim Biophys Acta Mol Basis Dis.
1867(166172)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hammer A, Rositch A, Qeadan F, Gravitt PE
and Blaakaer J: Age-specific prevalence of HPV16/18 genotypes in
cervical cancer: A systematic review and meta-analysis. Int J
Cancer. 138:2795–2803. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Luvero D, Lopez S, Bogani G, Raspagliesi F
and Angioli R: From the infection to the immunotherapy in cervical
cancer: Can we stop the natural course of the disease? Vaccines
(Basel). 8(597)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lin W, Niu Z, Zhang H, Kong Y, Wang Z,
Yang X and Yuan F: Imbalance of Th1/Th2 and Th17/Treg during the
development of uterine cervical cancer. Int J Clin Exp Pathol.
12:3604–3612. 2019.PubMed/NCBI
|
|
30
|
Ho GY, Bierman R, Beardsley L, Chang CJ
and Burk RD: Natural history of cervicovaginal papillomavirus
infection in young women. N Engl J Med. 338:423–428.
1998.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Bodily J and Laimins LA: Persistence of
human papillomavirus infection: Keys to malignant progression.
Trends Microbiol. 19:33–39. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Graham SV: The human papillomavirus
replication cycle, and its links to cancer progression: A
comprehensive review. Clin Sci (Lond). 131:2201–2221.
2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hebner CM and Laimins LA: Human
papillomaviruses: Basic mechanisms of pathogenesis and
oncogenicity. Rev Med Virol. 16:83–97. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Yoo SH, Ock CY, Keam B, Park SJ, Kim TM,
Kim JH, Jeon YK, Chung EJ, Kwon SK, Hah JH, et al: Poor prognostic
factors in human papillomavirus-positive head and neck cancer: Who
might not be candidates for de-escalation treatment? Korean J
Intern Med. 34:1313–1323. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Linde DS, Andersen MS, Mwaiselage JD,
Manongi R, Kjaer SK and Rasch V: Text messages to increase
attendance to follow-up cervical cancer screening appointments
among HPV-positive Tanzanian women (Connected2Care): Study protocol
for a randomised controlled trial. Trials. 18(555)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li X, Wu X, Li Y, Cui Y, Tian R, Singh N,
Ding M, Yang Y and Gao Y: Promoter hypermethylation of SOX11
promotes the progression of cervical cancer in vitro and
in vivo. Oncol Rep. 41:2351–2360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chen L, Qiu X, Zhang N, Wang Y, Wang M, Li
D, Wang L and Du Y: APOBEC-mediated genomic alterations link
immunity and viral infection during human papillomavirus-driven
cervical carcinogenesis. Biosci Trends. 11:383–388. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Lorincz AT: Cancer diagnostic classifiers
based on quantitative DNA methylation. Expert Rev Mol Diagn.
14:293–305. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yang S, Wu Y, Wang S, Xu P, Deng Y, Wang
M, Liu K, Tian T, Zhu Y, Li N, et al: HPV-related methylation-based
reclassification and risk stratification of cervical cancer. Mol
Oncol. 14:2124–2141. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fertey J, Hagmann J, Ruscheweyh HJ, Munk
C, Kjaer S, Huson D, Haedicke-Jarboui J, Stubenrauch F and Iftner
T: Methylation of CpG 5962 in L1 of the human papillomavirus 16
genome as a potential predictive marker for viral persistence: A
prospective large cohort study using cervical swab samples. Cancer
Med. 9:1058–1068. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang L, Hu D, Wang S, Zhang Y, Pang L,
Tao L and Jia W: Association between dense PAX1 promoter
methylation and HPV16 infection in cervical squamous epithelial
neoplasms of Xin Jiang Uyghur and Han women. Gene.
723(144142)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Franzen A, Vogt TJ, Muller T, Dietrich J,
Schrock A, Golletz C, Brossart P, Bootz F, Landsberg J, Kristiansen
G and Dietrich D: PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter
methylation is associated with HPV infection and transcriptional
repression in head and neck squamous cell carcinomas. Oncotarget.
9:641–650. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
El Aliani A, El-Abid H, El Mallali Y,
Attaleb M, Ennaji MM and El Mzibri M: Association between gene
promoter methylation and cervical cancer development: Global
distribution and a meta-analysis. Cancer Epidemiol Biomarkers Prev.
30:450–459. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Muñoz N, Bosch FX, de Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ:
Epidemiologic classification of human papillomavirus types
associated with cervical cancer. N Engl J Med. 348:518–527.
2003.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kishibuchi R, Kondo K, Soejima S, Tsuboi
M, Kajiura K, Kawakami Y, Kawakita N, Sawada T, Toba H, Yoshida M,
et al: DNA methylation of GHSR, GNG4, HOXD9 and SALL3 is a common
epigenetic alteration in thymic carcinoma. Int J Oncol. 56:315–326.
2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang M, Li C, Liu Y and Wang Z: Effect of
LAMA4 on prognosis and its correlation with immune infiltration in
gastric cancer. Biomed Res Int. 2021(6428873)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Nallanthighal S, Heiserman JP and Cheon
DJ: Collagen type XI alpha 1 (COL11A1): A novel biomarker and a key
player in cancer. Cancers (Basel). 13(935)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Xu HH, Wang K, Feng XJ, Dong SS, Lin A,
Zheng LZ and Yan WH: Prevalence of human papillomavirus genotypes
and relative risk of cervical cancer in China: A systematic review
and meta-analysis. Oncotarget. 9:15386–15397. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lin C, Franceschi S and Clifford GM: Human
papillomavirus types from infection to cancer in the anus,
according to sex and HIV status: A systematic review and
meta-analysis. Lancet Infect Dis. 18:198–206. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Malary M, Moosazadeh M, Hamzehgardeshi Z,
Afshari M, Moghaddasifar I and Afsharimoghaddam A: The prevalence
of cervical human papillomavirus infection and the most at-risk
genotypes among iranian healthy women: A systematic review and
meta-analysis. Int J Prev Med. 7(70)2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chan PK, Picconi MA, Cheung TH,
Giovannelli L and Park JS: Laboratory and clinical aspects of human
papillomavirus testing. Crit Rev Clin Lab Sci. 49:117–136.
2012.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kaliterna V and Barisic Z: Genital human
papillomavirus infections. Front Biosci (Landmark Ed).
23:1587–1611. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
54
|
Clarke MA, Wentzensen N, Mirabello L,
Ghosh A, Wacholder S, Harari A, Lorincz A, Schiffman M and Burk RD:
Human papillomavirus DNA methylation as a potential biomarker for
cervical cancer. Cancer Epidemiol Biomarkers Prev. 21:2125–2137.
2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Johannsen E and Lambert PF: Epigenetics of
human papillomaviruses. Virology. 445:205–212. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Szalmás A and Kónya J: Epigenetic
alterations in cervical carcinogenesis. Semin Cancer Biol.
19:144–152. 2009.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Jha AK, Sharma V, Nikbakht M, Jain V,
Sehgal A, Capalash N and Kaur J: A comparative analysis of
methylation status of tumor suppressor genes in paired biopsy and
serum samples from cervical cancer patients among north Indian
population. Genetika. 52:255–259. 2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
de la Cruz-Hernandez E, Perez-Cardenas E,
Contreras-Paredes A, Cantu D, Mohar A, Lizano M and Duenas-Gonzalez
A: The effects of DNA methylation and histone deacetylase
inhibitors on human papillomavirus early gene expression in
cervical cancer, an in vitro and clinical study. Virol J.
4(18)2007.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Lendvai Á, Johannes F, Grimm C, Eijsink
JJ, Wardenaar R, Volders HH, Klip HG, Hollema H, Jansen RC,
Schuuring E, et al: Genome-wide methylation profiling identifies
hypermethylated biomarkers in high-grade cervical intraepithelial
neoplasia. Epigenetics. 7:1268–1278. 2012.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Kelly H, Benavente Y, Pavon MA, De Sanjose
S, Mayaud P and Lorincz AT: Performance of DNA methylation assays
for detection of high-grade cervical intraepithelial neoplasia
(CIN2+): A systematic review and meta-analysis. Br J Cancer.
121:954–965. 2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Hoppe-Seyler K, Bossler F, Braun JA,
Herrmann AL and Hoppe-Seyler F: The HPV E6/E7 Oncogenes: Key
factors for viral carcinogenesis and therapeutic targets. Trends
Microbiol. 26:158–168. 2018.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Bachman KE, Park BH, Rhee I, Rajagopalan
H, Herman JG, Baylin SB, Kinzler KW and Vogelstein B: Histone
modifications and silencing prior to DNA methylation of a tumor
suppressor gene. Cancer Cell. 3:89–95. 2003.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Clark SJ and Melki J: DNA methylation and
gene silencing in cancer: Which is the guilty party? Oncogene.
21:5380–5387. 2002.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Tahara T, Shibata T, Yamashita H, Nakamura
M, Yoshioka D, Okubo M, Hirata I and Arisawa T: Chronic
nonsteroidal anti-inflammatory drug (NSAID) use suppresses multiple
CpG islands hyper methylation (CIHM) of tumor suppressor genes in
the human gastric mucosa. Cancer Sci. 100:1192–1197.
2009.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Clarke MA, Gradissimo A, Schiffman M, Lam
J, Sollecito CC, Fetterman B, Lorey T, Poitras N, Raine-Bennett TR,
Castle PE, et al: Human papillomavirus DNA methylation as a
biomarker for cervical precancer: Consistency across 12 genotypes
and potential impact on management of HPV-positive women. Clin
Cancer Res. 24:2194–2202. 2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Cuschieri K, Ronco G, Lorincz A, Smith L,
Ogilvie G, Mirabello L, Carozzi F, Cubie H, Wentzensen N, Snijders
P, et al: Eurogin roadmap 2017: Triage strategies for the
management of HPV-positive women in cervical screening programs.
Int J Cancer. 143:735–745. 2018.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Zhao Z, Zhang X, Zhao X, Cai J, Wu NY and
Wang J: SOX1 and PAX1 are hypermethylated in cervical
adenocarcinoma and associated with better prognosis. Biomed Res
Int. 2020(3981529)2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Meng M, Sang L and Wang X: S100 calcium
binding protein A11 (S100A11) promotes the proliferation, migration
and invasion of cervical cancer cells, and activates Wnt/β-catenin
signaling. Onco Targets Ther. 12:8675–8685. 2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Tian T, Li X, Hua Z, Ma J, Wu X, Liu Z,
Chen H and Cui Z: S100A7 promotes the migration, invasion and
metastasis of human cervical cancer cells through
epithelial-mesenchymal transition. Oncotarget. 8:24964–24977.
2017.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Chokchaichamnankit D, Watcharatanyatip K,
Subhasitanont P, Weeraphan C, Keeratichamroen S, Sritana N,
Kantathavorn N, Diskul-Na-Ayudthaya P, Saharat K, Chantaraamporn J,
et al: Urinary biomarkers for the diagnosis of cervical cancer by
quantitative label-free mass spectrometry analysis. Oncol Lett.
17:5453–5468. 2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Tomiyama N, Ikeda R, Nishizawa Y, Masuda
S, Tajitsu Y and Takeda Y: S100A16 up-regulates Oct4 and Nanog
expression in cancer stem-like cells of Yumoto human cervical
carcinoma cells. Oncol Lett. 15:9929–9933. 2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Lee HS, Yun JH, Jung J, Yang Y, Kim BJ,
Lee SJ, Yoon JH, Moon Y, Kim JM and Kwon YI: Identification of
differesntially-expressed genes by DNA methylation in cervical
cancer. Oncol Lett. 9:1691–1698. 2015.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Liu MY, Zhang H, Hu YJ, Chen YW and Zhao
XN: Identification of key genes associated with cervical cancer by
comprehensive analysis of transcriptome microarray and methylation
microarray. Oncol Lett. 12:473–478. 2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Ma X, Liu J, Wang H, Jiang Y, Wan Y, Xia Y
and Cheng W: Identification of crucial aberrantly methylated and
differentially expressed genes related to cervical cancer using an
integrated bioinformatics analysis. Bioscience Rep.
40(BSR20194365)2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Dean B and Scarr E: Possible involvement
of muscarinic receptors in psychiatric disorders: A focus on
schizophrenia and mood disorders. Curr Mol Med. 15:253–264.
2015.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Liang L, Huang J, Yao M, Li L, Jin XJ and
Cai XY: GNG4 promotes tumor progression in colorectal cancer. J
Oncol. 2021(9931984)2021.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Zhao H, Sheng D, Qian Z, Ye S, Chen J and
Tang Z: Identifying GNG4 might play an important role in colorectal
cancer TMB. Cancer Biomark. 32:435–450. 2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Wen S, Peng W, Chen Y, Du X, Xia J, Shen B
and Zhou G: Four differentially expressed genes can predict
prognosis and microenvironment immune infiltration in lung cancer:
A study based on data from the GEO. BMC Cancer.
22(193)2022.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Wang X, Hou Q and Zhou X: LAMA4 expression
is activated by zinc finger E-box-binding homeobox 1 and
independently predicts poor overall survival in gastric cancer.
Oncol Rep. 40:1725–1733. 2018.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Wang JF, Wang Y, Zhang SW, Chen YY, Qiu Y,
Duan SY, Li BP and Chen JQ: Expression and prognostic analysis of
integrins in gastric cancer. J Oncol. 2020(8862228)2020.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Itoh N and Ohta H: Fgf10: A
paracrine-signaling molecule in development, disease, and
regenerative medicine. Curr Mol Med. 14:504–509. 2014.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Deng X, Wei W, Huang N, Shi Y, Huang M,
Yan Y, Li D, Yi J and Wang X: Tumor repressor gene chondroadherin
oppose migration and proliferation in hepatocellular carcinoma and
predicts a good survival. Oncotarget. 8:60270–60279.
2017.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Lin YL, Wang YP, Li HZ and Zhang X:
Aberrant promoter methylation of PCDH17 (Protocadherin 17) in serum
and its clinical significance in renal cell carcinoma. Med Sci
Monit. 23:3318–3323. 2017.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Xu W, Xu M, Wang L, Zhou W, Xiang R, Shi
Y, Zhang Y and Piao Y: Integrative analysis of DNA methylation and
gene expression identified cervical cancer-specific diagnostic
biomarkers. Signal Transduct Target Ther. 4(55)2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Bhat S, Kabekkodu SP, Varghese VK,
Chakrabarty S, Mallya SP, Rotti H, Pandey D, Kushtagi P and
Satyamoorthy K: Aberrant gene-specific DNA methylation signature
analysis in cervical cancer. Tumour Biol.
39(1010428317694573)2017.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Varghese VK, Shukla V, Kabekkodu SP,
Pandey D and Satyamoorthy K: DNA methylation regulated microRNAs in
human cervical cancer. Mol Carcinog. 57:370–382. 2018.PubMed/NCBI View Article : Google Scholar
|