Introduction
Nasopharyngeal carcinoma (NPC) is an epithelial
carcinoma arising from the nasopharyngeal mucosal lining, with
>70% of new cases in east and southeast Asia. Although NPC is a
relatively rare disease in Northern China (~1-5 per 100,000
individuals per year) (1),
incidence remains high in endemic regions of China (2).
Knowledge on the etiology and pathogenesis of NPC is
underdeveloped. Multiple factors, including Epstein-Barr virus
(EBV) infection, host genetics, and environmental factors, such as
oral hygiene, have been suggested to contribute to the development
of NPC (3). Previous
epidemiological studies identified oral hygiene as a potential risk
factor for NPC (3-5).
EBV is an enveloped herpes virus with double-stranded DNA that only
infects humans. EBV DNA is frequently detected in saliva, throat
washing, gingival crevicular fluid, and nasopharyngeal epithelium
(6). Poor oral health can increase
the risk of NPC by stimulating EBV replication, and nurturing the
overgrowth of oral bacteria (7).
Since oral health status is affected by the oral microbial
equilibrium and activities, a link between NPC-risk and the oral
bacterial status has been hypothesized by researchers.
Porphyromonas gingivalis (P. gingivalis), a keystone
pathogen in chronic periodontitis (8), was also found to coexist with EBV, a
potential causative agent of NPC, in individuals with periodontal
disease (6). Thus, the combined
presence of EBV and periodontopathic bacteria could increase the
risk of developing periodontitis (6,9,10).
However, whether P. gingivalis is present in
NPCs in a non-endemic area of China, has yet to be
investigated.
Thus, to the best of our knowledge, for the first
time, the presence of P. gingivalis was retrospectively
evaluated by immunohistochemistry (IHC) and nested PCR, and
determined its possible association with NPC. The overall survival
(OS) of patients with P. gingivalis-positive NPC was
directly compared with that of patients with EBV-positive and P.
gingivalis/EBV-negative NPC, and the prognostic significance of
different infection status were assessed.
Materials and methods
Samples and clinical data
Formalin-fixed paraffin-embedded tissue (FFPE)
specimens from patients with primary NPC who were diagnosed at
Luoyang Central Hospital Affiliated to Zhengzhou University and the
First Affiliated Hospital of Henan University of Science and
Technology (Luoyang, China) between January 2011 and July 2017 were
collected. All specimens were reviewed by a single pathologist
under a light microscope (Eclipse 80i; Nikon Corporation) at the
Department of Pathology, the First Affiliated Hospital of Henan
University of Science and Technology following hematoxylin and
eosin (H&E) staining. Histological classification was
re-evaluated according to the current World Health Organization
(WHO) classification (3). NPC are
grouped into keratinizing squamous, non-keratinizing and basaloid
squamous. Non-keratinizing NPC can be divided into differentiated
and undifferentiated tumors (3).
Patients with nasopharyngeal tumors other than the WHO types, those
with poor quality, and those without sufficient sample available
for investigation were excluded from the study. Clinical
information associated with each sample was recorded. The present
study was approved (approval no. 2021-03-B053) by the Ethics
Committee of the First Affiliated Hospital of Henan University of
Science and Technology (Luoyang, China).
P. gingivalis detection and
identification
P. gingivalis was detected using IHC and
nested PCR.
IHC
Primary NPC FFPE tissues were used for the IHC
analysis of P. gingivalis using polyclonal rabbit anti-whole
cell P. gingivalis 33277 antibody (a generous gift from Dr
Richard Lamont) (11). This
antibody does not react with human cells or with other bacteria at
dilutions of 1:500 or greater (a dilution of 1:1,000 was used with
NPC tissue sections). Pre-immune rabbit IgG was used as a negative
control. IHC was performed as previously described (12). The sections were evaluated by two
pathologists under a light microscope (Eclipse 80i; Nikon
Corporation) after IHC staining. Staining intensity was classified
using a numerical scale, as previously described by the authors
(12). In the present study, IHC
scores ≥2 were categorized as P. gingivalis-positive.
DNA extraction
Genomic DNA from FFPE tissue was extracted using the
QIAamp DNA FFPE Tissue kit (cat. no. 56404, Qiagen, Inc.) according
to the manufacturer's instructions. The quantity and purity of the
DNA were accessed by NanoDrop 2000 (Thermo Fisher Scientific, Inc.)
at 260/280 nm (ratios of 1.8-2.0 favorable results).
Designation and synthesis of
primers
The universal bacterial primer pair, 27F/1492R
(forward, 5'-AGAGTTTGATCCTGGCTCAG-3' and reverse,
5'-ACGGCTACCTTGTTACGACTT-3') and the P. gingivalis specific
primer pair, 404F/R (forward, 5'-AGGCAGCTTGCCATACTGCG-3' and
reverse, 5'-ACTGTTAGCAACTACCGATGT-3'), were used as previously
described (13-15).
The primer pair 27F/1492R was used for the first round of PCR
amplification, generating a full-length 16S rDNA product. P.
gingivalis specific primers 404F/R, targeting the internal
sequence of 16S rDNA, were used to detect P. gingivalis in
the second round of PCR amplification. To increase sensitivity,
nested PCR was performed based on the sequence of the 16S rRNA
fragment of P. gingivalis genomic DNA. The expected size of
the amplification product by the inner primer pair was 404 bp in
length. Oligonucleotide primers were synthesized by Genewiz,
Inc.
Nested PCR
The nested PCR assay included two rounds of
consecutive PCR amplifications and was performed as described
herein. Briefly, the first round of amplification contained the
outer primer pair (27F/1429R) and was performed in a reaction
volume of 50 µl consisting of 2 µl of 50 ng DNA and 48 µl of
reaction mixture containing 2X Taq Plus Master Mix (cat. no.
P212-AA; Vazyme Biotech Co., Ltd.), and 10 pmol of each primer. The
reaction was performed under the following thermocycling
conditions: 94˚C for 10 min, 25 PCR cycles (94˚C for 30 sec, 60˚C
for 30 sec, and 72˚C for 60 sec). The final cycle included
extension for 5 min at 72˚C. Then, 1 µl of the reaction products
was transferred into a new tube and diluted 1:100 with
nuclease-free water. Subsequently, 2 µl of this dilution was used
as a template for the second-round reaction. The second-round
reaction mixture contained 10 pmol of each of the inner pair
primers 404F/R and the same Taq polymerase system as used in the
first round. Samples were amplified for 30 cycles under the same
conditions reported for the first round of amplification, except
that elongation was performed at 72˚C for 30 sec in this round of
amplification.
Identification
The amplification products obtained by nested PCR
were run on 2% agarose gel, using a horizontal electrophoresis
system (DYCP-32C; Beijing Liuyi Biotechnology Co., Ltd.), then
visualized on ChemiDoc™ XRS+ (Bio-Rad Laboratories, Inc.). The size
of the product was estimated by comparison with DL 2000 DNA markers
(Takara Bio, Inc.), and DNA bands close to the expected size (based
on the PCR product obtained from the amplification of positive
control and DNA markers) were identified as P. gingivalis
positive. The products of positive sample were sent to Genewiz,
Inc. for DNA sequencing.
Positive and negative controls were included for
each batch of amplification. DNA extracted from the American Type
Culture Collection (cat. no. 33277) cultures (from the authors'
laboratory) served as a positive control, and a tube containing
distilled water in place of the DNA template was used as a negative
control. Nested PCR was performed blinded to the results obtained
by IHC.
DNA sequencing and reads analysis
The products of nested PCR were subjected to DNA
sequencing in both (forward and reverse) directions with 404F/R
primers by Genewiz, Inc. The obtained map results were analyzed by
Chromas 2.22 (Technelysium Pty. Ltd.) and sequencing reads were
analyzed via BLAST search in NCBI (https://blast. ncbi.nlm.nih.gov/). Bacterial species
were identified if subjects showed the lowest expectation
(E) value in the list of BLAST results.
EBV detection
Reverse transcription-quantitative PCR was performed
using the following primer sets: EBV forward,
5'-CCTGGTCATCCTTTGCCA-3'; and EBV reverse,
5'-TGCTTCGTTATAGCCGTAGT-3' (8),
using SYBR Master Mix (cat. no. Q111-02-AA; Vazyme Biotech Co.,
Ltd.) in a Bio-Rad CFX96™ real time system (Bio-Rad Laboratories,
Inc.). The amplification reaction was performed in a total volume
of 20 µl containing 2X AceQ qPCR SYBR Master Mix (10 µl), 10 µM
forward and reverse primers (1 µl), and 20 ng genomic DNA (2 µl)
and distilled water (7 µl). The thermocycling conditions were as
follows: 10 sec at 95˚C and 40 cycles of 5 sec at 95˚C and 30 sec
at 60˚C. Post-PCR melting curves confirmed the specificity of
single-target amplification.
Statistical analysis
All statistical analyses were performed using SPSS
Statistics 19.0 software (IBM Corp.). Cohen's Kappa coefficient was
used to evaluate the concordance between IHC and nested PCR. All
patients were linked to data from a mortality registry up to May
18, 2021. The primary endpoint was OS, measured using the duration
from the date of diagnosis to the end of follow-up or the date of
death by any cause. Kaplan-Meier methodology and the log-rank test
were performed to determine survival differences among groups.
Chi-square or Fisher's exact test was used to
compare categorical variables between patients with different P.
gingivalis and EBV infection statuses, and to determine the
associations between P. gingivalis status and clinical
characteristics. P<0.05 was considered to indicate a
statistically significant difference.
Results
Clinical and histological features of
NPC
A total of 58 patients with NPC diagnosed from
January 2011 to July 2017 were identified, in the two hospitals.
Among these, six patients were excluded because their samples
presented histology other than a WHO type, and seven were excluded
due to poor quality or having insufficient sample for
investigation. Thus, a total of 45 subjects were included in the
present study. A total of 30 (66.7%) patients were male and 15
(33.3%) patients were female. All tumors were classified as
non-keratinizing undifferentiated NPC (WHO Type-III), and no other
type cases were identified. Unfortunately, tumor staging and other
information were unknown for most patients at the time of
diagnosis.
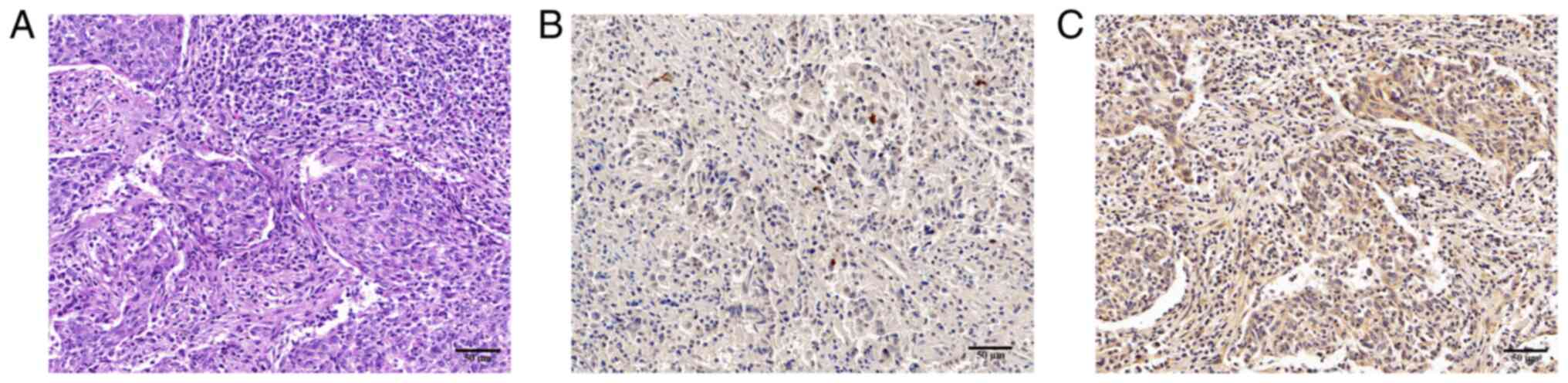
Detection of P. gingivalis by IHC
Among the 45 samples, 26 (57.8%) cases were P.
gingivalis-positive (P. gingivalis+), and 19
(42.2%) cases were P. gingivalis-negative (P.
gingivalis-). P. gingivalis expression was
detected as dark brown staining, which was primarily localized to
the cytoplasm of epithelial cells (Fig. 1).
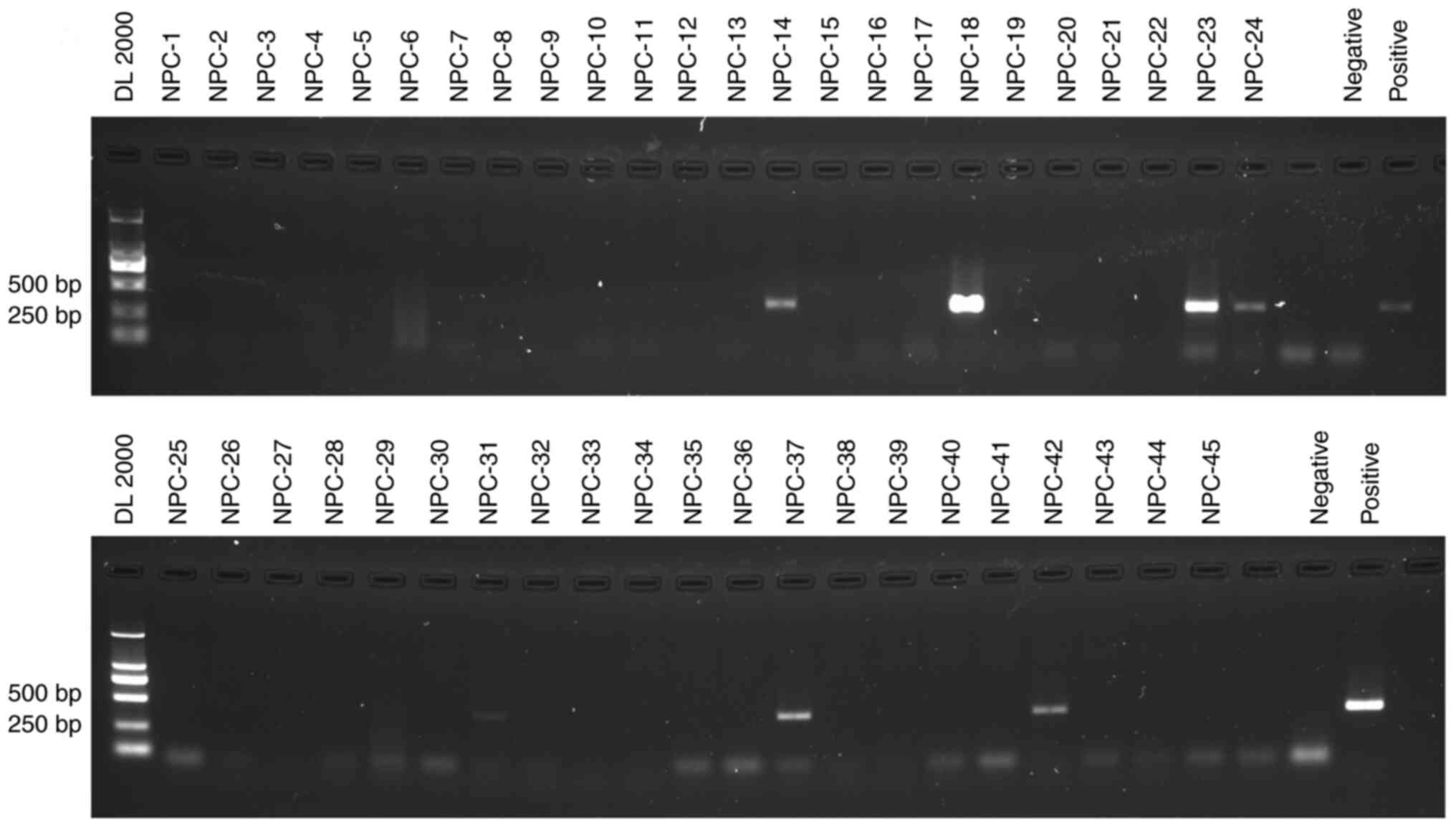
Detection and identification of P.
gingivalis using nested PCR and DNA sequencing
DNA extracted from the 45 FFPE tissues was examined
by single-step PCR, nested PCR, and the products were confirmed by
Sanger sequencing. P. gingivalis DNA was not detected by
single-step PCR (Fig. S2). After
two rounds of amplification, seven (15.6%) samples produced a clear
and expected 404 bp band, which was specific for P.
gingivalis DNA (Fig. 2). Since
nested PCR represents a highly sensitive method to detect low copy
numbers of P. gingivalis DNA, it is possible that the nested
amplification of a common bacterium could result in a
false-positive signal. Therefore, the direct sequencing map was
verified on Chromas and the sequencing reads were analyzed by
BLAST. All 14 sequencing maps from the seven products corresponded
specifically to single reads (Fig.
S1). Sequencing of the nested PCR products and BLAST analysis
confirmed the presence of P. gingivalis in DNA from NPC
tissue (Table I). This indicated
that the results obtained by P. gingivalis PCR results were
unlikely to be due to a PCR artifact; thus, P. gingivalis is
present in NPC tumor tissues. In summary, the identification of
P. gingivalis DNA from tumor tissues and the results for
P. gingivalis+ in FFPE tissues by IHC validate
the presence of P. gingivalis in tissues of NPC.
 | Table IAge and ID of specimens and sequence
identity of nested PCR products to Porphyromonas gingivalis
ATCC 33277 16S rDNA sequence by NCBI BLAST. Sequencing data are
included in Fig. S1. |
Table I
Age and ID of specimens and sequence
identity of nested PCR products to Porphyromonas gingivalis
ATCC 33277 16S rDNA sequence by NCBI BLAST. Sequencing data are
included in Fig. S1.
| Subject ID | Age | PCR primers | Sequencing
primers | Identity, % | E-value |
|---|
| NPC-14 | 49 | 27F/1492R;
404F/R | 404F/R | 100, 100 | 1e-171, 8e-173 |
| NPC-18 | 70 | 27F/1492R;
404F/R | 404F/R | 100, 100 | 7e-168, 5e-180 |
| NPC-23 | 51 | 27F/1492R;
404F/R | 404F/R | 100, 100 | 6e-174, 1e-176 |
| NPC-24 | 61 | 27F/1492R;
404F/R | 404F/R | 99, 99 | 5e-170, 1e-175 |
| NPC-31 | 59 | 27F/1492R;
404F/R | 404F/R | 99, 99 | 2e-169, 3e-172 |
| NPC-37 | 32 | 27F/1492R;
404F/R | 404F/R | 100, 100 | 3e-172, 5e-175 |
| NPC-42 | 61 | 27F/1492R;
404F/R | 404F/R | 99, 100 | 2e-174, 5e-175 |
Comparison of different techniques for
the detection of P. gingivalis
Nested PCR was able to detect P.
gingivalis-specific DNA in FFPE tissues from seven patients
with NPC, of which specimens from five patients were histologically
classified as P. gingivalis+. Two P.
gingivalis+ specimens examined by nested PCR were
revealed to be negative by IHC. Nested PCR failed to detect P.
gingivalis DNA from the FFPE tissues of 21 patients with NPC
that were categorized as P. gingivalis+ based on
the results of IHC. The concordance rate was 48.9% (kappa=0.422;
P<0.001) between nested PCR and IHC (Table II). There was agreement between
these two methods for the detection of P. gingivalis in FFPE
tissues from patients with NPC.
 | Table IIHistological examination and nested
PCR for the detection of Porphyromonas gingivalis in FFPE
samples from patients with NPC. |
Table II
Histological examination and nested
PCR for the detection of Porphyromonas gingivalis in FFPE
samples from patients with NPC.
| | Porphyromonas
gingivalis results | |
|---|
| Patients (no.) | Histology | Nested PCR | Kappa | P-value |
|---|
| 5 | + | + | 0.422 | <0.001 |
| 21 | + | - | | |
| 2 | - | + | | |
| 17 | - | - | | |
| Total=45 | 26 | 7 | | |
| Positive rate
(%) | 57.8 | 15.6 | | |
Association between P. gingivalis
infection and the clinicopathological status of patients with
NPC
An association was identified between P.
gingivalis infection and the clinical features of patients with
NPC (Table III). The presence of
P. gingivalis was not significantly associated with age, EBV
status, or prognosis in terms of both DNA and expression level. NPC
was found to predominantly affect male (30 males vs. 15 females),
while the P. gingivalis-positive rate of female patients
with NPC was significantly higher than that of male patients with
NPC (80.0% vs. 46.7%, P<0.05) in terms of the expression level,
but not based on DNA. Furthermore, there were twice as numerous
male patients with NPC compared with female patients with NPC,
which is consistent with previous studies from high-incidence areas
(1,16). The relationship between P.
gingivalis infection and the sex of patients with NPC was not
consistent with the results of nested PCR and IHC.
 | Table IIIClinicopathological features of
patients with nasopharyngeal carcinoma. |
Table III
Clinicopathological features of
patients with nasopharyngeal carcinoma.
| | Porphyromonas
gingivalis positive cases (%) |
|---|
| Clinicopathological
features | Nested PCR |
Immunohistochemistry |
|---|
| Sex | | |
|
Male
(30) | 3 (10.0) | 14
(46.7)a |
|
Female
(15) | 4 (26.7) | 12
(80.0)a |
| Age, years | | |
|
≥50(28) | 5 (17.9) | 15 (53.6) |
|
<50(17) | 2 (11.8) | 11 (64.7) |
| Histologic
classification | | |
|
Keratinizing
(0) | 0 (0) | 0 (0) |
|
Non-keratinizing
(45) | 7 (15.6) | 26 (57.8) |
| EBV status | | |
|
Positive
(40) | 7 (17.5) | 24 (60.0) |
|
Negative
(5) | 0 (0) | 2 (40.0) |
| Prognosis | | |
|
Alive
(20) | 1 (5.0) | 10 (50.0) |
|
Died
(25) | 6 (24.0) | 16 (64.0) |
P. gingivalis and EBV status
EBV-positive NPCs were predominant in the current
study. Among the 45 specimens, 40 (88.9%) possessed EBV-positive
(EBV+) tumors, seven of which (15.6%) exhibited EBV and
P. gingivalis co-infection. Thus, based on DNA status, the
samples were classified into three groups: i) EBV-/P.
gingivalis- (5 of 45 patients, 11.1%); ii)
EBV+/P. gingivalis- (33 of 45
patients, 73.3%); and iii) EBV+/P.
gingivalis+ (7 of 45 patients, 15.6%). The clinical
characteristics of patients are shown in Table IV.
 | Table IVCharacteristics of patients with
different infection statuses. |
Table IV
Characteristics of patients with
different infection statuses.
| | Number of patients
(%) |
|---|
| Clinicopathological
features |
Pg-/EBV- |
Pg-/EBV+ |
Pg+/EBV+ | χ2 | P-value |
|---|
| Sex | | | | 3.947 | 0.160 |
|
Male
(30) | 5(100) | 22 (66.7) | 3 (42.9) | | |
|
Female
(15) | 0 (0) | 11(33.3) | 4 (57.1) | | |
| Age, years | | | | 30.705 | <0.001 |
|
≥50(28) | 2 (40.0) | 21 (63.6) | 5 (71.4) | | |
|
<50(17) | 3 (60.0) | 12 (36.4) | 2 (28.6) | | |
| Histologic
classification | | | | | |
|
Keratinizing
(0) | 0 | 0 | 0 | | 0 |
|
Non-keratinizing
(45) | 5(100) | 33(100) | 7(100) | | |
| Prognosis | | | | 0.896 | 0.793 |
|
Alive
(20) | 3 (60.0) | 16 (48.5) | 1 (14.3) | | |
|
Died
(25) | 2 (40.0) | 17 (51.5) | 6 (85.7) | | |
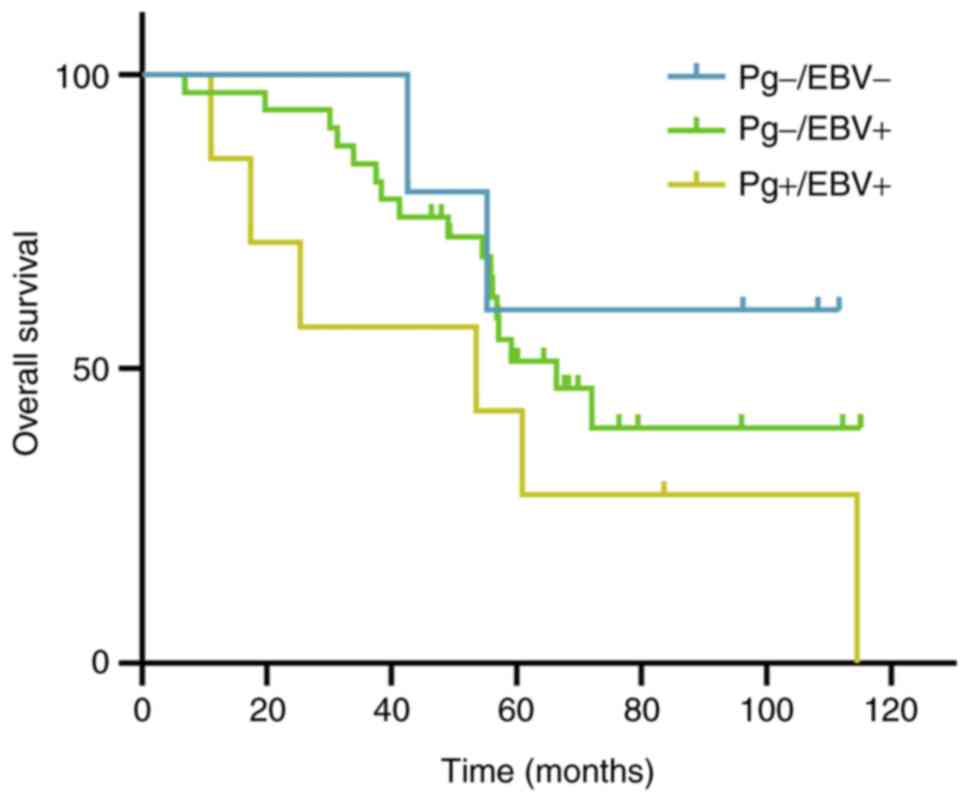
Survival analysis
A total of 45 patients were followed up for survival
analysis over a period of 115 months. The median follow-up period
was 60.9 months for all groups (range, 6.9-115.1 months). Most
patients had a minimum of 5 years follow-up; after treatment for
<60 months (46.3-59.6 months), there were only five survivors.
Due to the low patient number, the survival rate exceeded 40% in
two groups and the median survival time could not be calculated in
the other group. In the present study, the mean survival time among
the three groups was compared. Patients in the
EBV+/P. gingivalis- group, which
contained the highest number of patients, presented a shorter mean
time compared with those with EBV-/P.
gingivalis- NPC (74.8±6.7 vs. 86.5±13.8). Patients
with EBV+/P. gingivalis+ tended to
have the shortest mean survival time (56.8 months). However, there
were no significant differences among the three groups (P=0.255;
Table IV). Furthermore, the
5-year OS rate in patients with EBV-/P.
gingivalis- NPC was higher than that in patients
with EBV+/P. gingivalis+ NPC (60.0%
vs. 42.9%), although the number of specimens was too low for any
statistically meaningful analysis. The 5-year OS rate differed
between P. gingivalis positive (whether EBV infected or not)
and P. gingivalis negative patients with NPC, the difference
was not significant (14.3% vs. 50%, P=0.120). A Kaplan-Meier curve
for OS is shown in Figs. 3 and
S3.
Discussion
P. gingivalis is one of the most common
bacterial pathogens in human periodontitis. In the present study,
for the first time, the presence of P. gingivalis in FFPE
tissues from patients with NPC was retrospectively investigated and
confirmed using two complementary approaches. P. gingivalis
was detected in 26 (57.8%) and seven (15.6%) of 45 NPC tumor
tissues by IHC and nested PCR, respectively; and the presence of
P. gingivalis was confirmed by DNA sequencing in NPC FFPE
tissues. Moreover, P. gingivalis DNA and EBV DNA were found
to co-exist in the tumor tissues of patients with NPC.
NPC is not endemic within Central China, although
EBV was predominant among the NPC cases (88.9%). A significant
association between the presence of P. gingivalis and
infection with EBV has also been reported (17). To the best of our knowledge, the
presence of P. gingivalis and infection with EBV has not
been reported in NPC. Furthermore, no differences in the clinical
characteristics of patients with P. gingivalis-positive NPC
and those with P. gingivalis-negative NPC were observed. The
co-infection of P. gingivalis with EBV may affect the OS of
patients with NPC; however, a larger sample size is needed for a
statistically meaningful analysis.
Among the 45 cases, no EBV negative and P.
gingivalis positive cases were reported. This is a very
interesting question that leads the authors to think all possible
distribution patterns of P. gingivalis and EBV. There were
cases being reported with negative EBV DNA but positive P.
gingivalis in the patients with chronic periodontitis (8). However, to the best of our knowledge,
it was not found in NPC so far. Considering the regional and
pathogenic characteristics of NPC, it could be hypothesized that
even if there were such cases in clinical practice, it would be
very rare.
Nested PCR with 27F/1429R and 404F/R primer pairs
successfully identified P. gingivalis DNA in FFPE tissues
from 7 out of 45 NPC patients. False-positive results with nested
PCR were unlikely in the present study for the following reasons:
First, negative controls performed in parallel with the samples
during the two rounds of amplification revealed no detectable or
specific band (404 bp). Second, the specificity of the 404F/R
primers that were used for the second round of amplification of
nested PCR has been widely investigated (13-15).
The products of nested PCR appeared to be specific for P.
gingivalis, which was confirmed by DNA sequencing. Finally,
nested PCR has higher specificity compared with single-step PCR.
P. gingivalis DNA was not detected using 404F/R primers by
single-step PCR. Consequently, use of nested PCR was found to be
more efficient for the reliable detection of P. gingivalis
DNA in FFPE tissues.
Although the presence of P. gingivalis in NPC
was verified in the present study, several questions remain
unanswered, for example, the mechanism by which P.
gingivalis infects the nasopharynx.
The nasopharynx is a tubular space that represents a
transitional area between the nasal cavities and the oropharynx.
This region is suitable for the growth of anaerobic bacteria during
infection, although limited bacteria are present under normal
healthy conditions (18). The
mucosal epithelium in the nasopharynx, which possesses a small
crypt epithelium similar to the oropharynx, consists of a special
type of stratified squamous epithelium that is typically observed
in the respiratory tract (19,20).
Furthermore, oral pathogens can translocate to remote body organs
via the local or oral blood circulation, or pass through the
gastrointestinal tract (21).
Since P. gingivalis is a gram-negative anaerobic pathogen
that can penetrate and invade oral epithelial and endothelial
cells, and the nasopharynx is histologically similar and in close
proximity to the oropharynx, infection by P. gingivalis
arising from the oral cavity is highly plausible when the
environment of the human nasopharynx changes. Previous studies have
reported the occurrence of anaerobes in the nasopharynx during
respiratory infection (18,22).
Alternatively, when sneezing and covering the mouth, due to
increased local pressure, it is likely that bacteria in the mouth,
oropharynx and other parts of the mouth may enter the nasopharynx
where is under relatively little pressure. It appears that P.
gingivalis reaches the nasopharynx by direct mucosal dispersion
or through the flow of saliva from the oral cavity upon
swallowing.
Another question is whether P. gingivalis
positivity affects the development of NPC. Mounting evidence
suggests there is a relationship between P. gingivalis
infection and the development of certain cancers (23-25).
Persistent exposure to P. gingivalis may induce tumor-like
characteristic changes in oral epithelial cells and promote
tumorigenesis (26). By contrast,
no studies have investigated the relationship between P.
gingivalis and NPC carcinogenesis, and the mechanism remains
unknown. The existence of P. gingivalis-positive patients
with NPC in the present study demonstrated that P.
gingivalis following infection may constantly colonize the
nasopharynx. This explains why P. gingivalis DNA and EBV DNA
were found to co-exist in seven specimens of NPC in the present
study. Persistent EBV infection in epithelial cells could induce
progressive genomic changes, which promote the clonal evolution of
NPC (3). P. gingivalis and
other anaerobic bacteria can reactivate EBV infection via the
production of butyric acid; this may contribute to the progression
of EBV-related diseases (27,28).
These findings suggested that P. gingivalis may accelerate
the replication of EBV in EBV-related diseases (27).
Although P. gingivalis-positive cases were
identified, this finding suggested that P. gingivalis is not
etiologically linked to NPC carcinogenesis in Central China.
Further studies are needed to define the influence of P.
gingivalis on NPC.
A third question relates to the discrepancy between
the overexpression of P. gingivalis antigen and P.
gingivalis DNA-positive NPC. IHC was able to identify a
specific protein associated with P. gingivalis, and the
results may indicate localization within the tissues; while PCR
detected nucleic acid of P. gingivalis within the tissue,
regardless of the localization. Several FFPE samples that were
positive for P. gingivalis were identified upon IHC
staining; of these, only five were detected as P.
gingivalis-positive following nested PCR. Certain investigators
have suggested that the formalin fixation process may result in the
formation of crosslinks between proteins and nucleic acids, which
is a challenge for DNA detection methods such as PCR. The lower
detection rate of PCR may be resultant from the fixation of our
tissues. For another, the PCR samples were picked from a small
portion of the sliced tissues while antibody in IHC could cover the
whole area of the slide. In fact, according to the present study,
nested PCR is more sensitive to detect P. gingivalis in the
nasopharynx compared with routine PCR. Two FFPE cases that were
P. gingivalis-positive by nested PCR were revealed to be
P. gingivalis-negative by IHC, with a score of 1. This may
be attributed to the authors' relatively strict IHC scoring
standard, at least partially. The IHC positive standard used in the
current study was similar to that previously used in studies on
esophageal cancer (12). In
addition, the present PCR samples were only from a small portion of
sliced tissues while antibody in IHC could cover the whole area of
the slide. This could be a major reason for the discrepancy
observed in the study. Notably, these two different assays have
their own emphasis on detecting organisms. Nevertheless, despite
the different results from specific samples, the statistical
results of these two methods showed the same trends of the presence
of P. gingivalis in FFPE tissues from patients with NPC.
Although the mean survival time and survival rate
differed between P. gingivalis-positive and P.
gingivalis-negative patients with NPC, the differences were not
significant. When the influence of EBV infection was considered,
the mean survival time and 5-year survival rate among patients with
P. gingivalis-/EBV-, P.
gingivalis-/EBV+ and P.
gingivalis+/EBV+ were also found to
differ; however, the differences were not significant. Therefore,
there was less power to yield meaningful outcomes in this
situation, and additional studies are needed. In general, high
expression levels of EBV encoding region expression, or
EBV-positivity, are associated with non-keratinizing carcinoma and
a favorable prognosis for patients with NPC (29); however, in the current study,
EBV-negative patients with NPC tended to live longer than
EBV-positive patients. It is considered that the EBV-negative group
in the present study was too small to allow meaningful comparisons
to be made.
There are several limitations to the present study.
First, it was a retrospective study; tumor staging and EBV status
were obtained when the patients were diagnosed, and information on
smoking habits and alcohol use were incomplete or absent. This
limits the potential to evaluate the outcome data. Second, it is
difficult to estimate the prognosis for patients with P.
gingivalis-related NPC due to the small number of identified
patients with NPC in Henan, although data were collected from two
cohorts. Certain clinicopathological subgroups obtained too few
patients for analysis with adequate statistical power. For example,
more females with NPC had P. gingivalis infection (by IHC),
and patients with EBV+/P. gingivalis+
had worse outcomes in the present study. The reason and
significance underlying these findings are unclear. Last, also the
most important, the consistence of the two methods used in the
present study are not favorable enough. The reasons were
aforementioned. An interesting phenomenon was identified in the
current study; P. gingivalis was revealed to preferably
infect female patients with NPC than male. Given the relatively
small sample size in the present study, the authors are uncertain
about the probable reason, which could be an interesting topic in
future investigation. Further studies are required to establish
P. gingivalis as a co-etiologic factor of NPC. Reducing
infection and maintaining oral hygiene could be potential
strategies to decrease the risk of NPC.
In conclusion, the present study of P.
gingivalis in a low-incidence population confirmed the presence
of P. gingivalis in NPC tumor tissues. It is proposed by the
authors that P. gingivalis is not etiologically relevant to
NPC in central China, despite its coinfection with EBV-the most
common causal factor for NPC.
Supplementary Material
Sequencing of Porphyromonas
gingivalis 16S rDNA nested PCR products from NPC formalin-fixed
paraffin-embedded tissues. (A) Partial of sequencing map from
NPC-18 sequenced in forward direction with 404-F primers. (B)
Nested PCR product sequences for each NPC subjects. NPC,
nasopharyngeal carcinoma.
Single-step PCR products detecting
Porphyromonas gingivalis from formalin-fixed
paraffin-embedded tissues for all subjects run on agarose gel with
DL 2000 as DNA marker, including negative and positive controls.
(A) Products examined from NPC-1 to NPC-12, positive and negative.
(B) Products examined from NPC-13 to NPC-22. (C) Products examined
from NPC-23 to NPC-45. NPC, nasopharyngeal carcinoma.
Kaplan-Meier curve showing the 5-year
overall survival for P. gingivalis positive and P.
gingivalis negative patients with nasopharyngeal carcinoma. Pg,
Porphyromonas gingivalis; +, positive; -, negative.
Acknowledgements
The authors would like to thank Professors Jianqiang
Mi and Lan Chen from Department of Pathology, the First Affiliated
Hospital of Henan University of Science and Technology for their
assistance while preparing and slicing paraffin blocks; Kelei Guo
from Medical Record Department of the First Affiliated Hospital of
Henan University of Science and Technology for clinical data
collection; and Dr Linlin Shi from Henan Key Laboratory of Cancer
Epigenetics; Cancer Hospital, The First Affiliated Hospital
(College of Clinical Medicine) of Henan University of Science and
Technology for manuscript editing.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81972571), the Special
Project in Social Development of Luoyang (Medical and Health
Services Key Program; grant no. 2101038A).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
BG, YW and JG contributed to the experimental
studies. YQ and SG contributed to the study design, supervision of
experiments, and manuscript review. JH and LM contributed to the
collection of samples, the acquisition of clinical data, and
patient follow-ups. BG and YW conceived of the study and prepared
the manuscript. All authors read and approved the final manuscript.
BG and SG confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved (approval no.
2021-03-B053) by the Ethics Committee of the First Affiliated
Hospital of Henan University of Science (Luoyang, China). There was
an informed consent waiver from the Ethics Committee of the First
Affiliated Hospital of Henan University of Science.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tsao SW, Yip YL, Tsang CM, Pang PS, Lau
VMY, Zhang G and Lo KW: Etiological factors of nasopharyngeal
carcinoma. Oral Oncol. 50:330–338. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen YP, Chan A, Le QT, Blanchard P, Sun Y
and Ma J: Nasopharyngeal carcinoma. Lancet. 394:64–80.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Pihlstrom BL, Michalowicz BS and Johnson
NW: Periodontal diseases. Lancet. 366:1809–1820. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xu FH, Xiong D, Xu YF, Cao SM, Xue WQ, Qin
HD, Liu WS, Cao JY, Zhang Y, Feng QS, et al: An epidemiological and
molecular study of the relationship between smoking, risk of
nasopharyngeal carcinoma, and Epstein-Barr virus activation. J Natl
Cancer Inst. 104:1396–1410. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kato A, Imai K, Ochiai K and Ogata Y:
Prevalence and quantitative analysis of Epstein-Barr virus DNA and
Porphyromonas gingivalis associated with Japanese chronic
periodontitis patients. Clin Oral Investig. 19:1605–1610.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Turkoz FP, Celenkoglu G, Dogu GG, Kalender
ME, Coskun U, Alkis N, Ozkan M, Turk HM and Arslan UY: Risk factors
of nasopharyngeal carcinoma in Turkey-an epidemiological survey of
the anatolian society of medical oncology. Asian Pac J Cancer Prev.
12:3017–3021. 2011.PubMed/NCBI
|
|
8
|
Hajishengallis G and Lamont RJ: Dancing
with the stars: How choreographed bacterial interactions dictate
nososymbiocity and give rise to keystone pathogens, accessory
pathogens, and pathobionts. Trends Microbiol. 24:477–489.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kato A, Imai K, Sato H and Ogata Y:
Prevalence of Epstein-Barr virus DNA and Porphyromonas gingivalis
in Japanese peri-implantitis patients. BMC Oral Health.
17(148)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kato A, Imai K, Ochiai K and Ogata Y:
Higher prevalence of Epstein-Barr virus DNA in deeper periodontal
pockets of chronic periodontitis in Japanese patients. PLoS One.
8(e71990)2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yilmaz O, Watanabe K and Lamont RJ:
Involvement of integrins in fimbriae-mediated binding and invasion
by Porphyromonas gingivalis. Cell Microbiol. 4:305–314.
2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gao S, Li S, Ma Z, Liang S, Shan T, Zhang
M, Zhu X, Zhang P, Liu G, Zhou F, et al: Presence of Porphyromonas
gingivalis in esophagus and its association with the
clinicopathological characteristics and survival in patients with
esophageal cancer. Infect Agent Cancer. 11(3)2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Dougherty WJ, Bae KS, Watkins BJ and
Baumgartner JC: Black-pigmented bacteria in coronal and apical
segments of infected root canals. J Endod. 24:356–358.
1998.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fouad AF, Barry J, Caimano M, Clawson M,
Zhu Q, Carver R, Hazlett K and Radolf JD: PCR-based identification
of bacteria associated with endodontic infections. J Clin
Microbiol. 40:3223–3231. 2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Siqueira JF Jr, Rôças IN, Alves FR and
Santos KR: Selected endodontic pathogens in the apical third of
infected root canals: A molecular investigation. J Endod.
30:638–643. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lo KW, To KF and Huang DP: Focus on
nasopharyngeal carcinoma. Cancer Cell. 5:423–428. 2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Slots J, Kamma JJ and Sugar C: The
herpesvirus-Porphyromonas gingivalis-periodontitis axis. J
Periodontal Res. 38:318–323. 2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Könönen E, Syrjänen R, Takala A and
Jousimies-Somer H: . Nasopharyngeal carriage of anaerobes during
health and acute otitis media by two years of age. Diagn Microbiol
Infect Dis. 46:167–172. 2003.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bryant WS: The transition of the ciliated
epithelium of the nose into the squamous epithelium of the pharynx.
J Anat Physiol. 50:172–176. 1916.PubMed/NCBI
|
|
20
|
Kano M, Kondo S, Wakisaka N, Moriyama-Kita
M, Nakanishi Y, Endo K, Murono S, Nakamura H and Yoshizaki T: The
influence of human papillomavirus on nasopharyngeal carcinoma in
Japan. Auris Nasus Larynx. 44:327–332. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Mohammed H, Varoni EM, Cochis A, Cordaro
M, Gallenzi P, Patini R, Staderini E, Lajolo C, Rimondini L and
Rocchetti V: Oral dysbiosis in pancreatic cancer and liver
cirrhosis: A review of the literature. Biomedicines.
6(115)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Haraldsson G, Holbrook WP and Könönen E:
Clonal similarity of salivary and nasopharyngeal Fusobacterium
nucleatum in infants with acute otitis media experience. J Med
Microbiol. 53:161–165. 2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Atanasova KR and Yilmaz O: Looking in the
Porphyromonas gingivalis cabinet of curiosities: The microbium, the
host and cancer association. Mol Oral Microbiol. 29:55–66.
2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Olsen I and Yilmaz Ö: Possible role of
Porphyromonas gingivalis in orodigestive cancers. J Oral Microbiol.
11(1563410)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Irfan M, Delgado RZR and Frias-Lopez J:
The oral microbiome and cancer. Front Immunol.
11(591088)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Geng F, Liu J, Guo Y, Li C, Wang H, Wang
H, Zhao H and Pan Y: Persistent exposure to Porphyromonas
gingivalis promotes proliferative and invasion capabilities, and
tumorigenic properties of human immortalized oral epithelial cells.
Front Cell Infect Microbiol. 7(57)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Imai K, Inoue H, Tamura M, Cueno ME, Inoue
H, Takeichi O, Kusama K, Saito I and Ochiai K: The periodontal
pathogen Porphyromonas gingivalis induces the Epstein-Barr virus
lytic switch transactivator ZEBRA by histone modification.
Biochimie. 94:839–846. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Makino K, Takeichi O, Imai K, Inoue H,
Hatori K, Himi K, Saito I, Ochiai K and Ogiso B: Porphyromonas
endodontalis reactivates latent Epstein-Barr virus. Int Endod J.
51:1410–1419. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ke K, Wang H, Fu S, Zhang Z, Duan L, Liu D
and Ye J: Epstein-barr virus-encoded RNAs as a survival predictor
in nasopharyngeal carcinoma. Chin Med J (Engl). 127:294–299.
2014.PubMed/NCBI
|