Introduction
Liver cancer is a prevalent form of malignant tumor
and the deadliest malignant tumor of the digestive system. In
total, ~85% of these cases are of hepatocellular carcinoma (HCC).
China accounts for nearly 50% of all newly diagnosed HCC cases and
51% of all deaths worldwide (1).
Surgery is considered the first choice of treatment, which,
however, is accompanied by high rates of recurrence and metastasis,
very low 5-year survival rate, and extremely poor prognosis
(2). Despite advancements in
immunotherapy and targeted therapies over the past decade, patients
suffering from liver cancer still present a 5-year survival rate
<20% (3). Consequently, the
mechanisms underlying the prognosis of HCC shall be well elucidated
and biomarkers with strong sensitivity and specificity shall be
identified to contributed to a valuable regime for HCC diagnosis
and treatment.
Dysfunction of cell cycle regulators is a
significant event in carcinogenesis and progression.
Cyclin-dependent kinase inhibitor 3 (CDKN3) is part of a family of
bispecific protein phosphatases located on human chromosome 14 and
is a cell cycle protein-dependent kinase inhibitor that functions
well in the cell cycle regulation (4). In previous studies, CDKN3 was
identified to remarkably affect the progression of several types of
cancers, including rectal and cervical cancer (5,6).
CDKN3 has been found to be expressed in HCC tissues, and its
overexpression drives HCC cell proliferation. Therefore, CDKN3 can
be regarded as an oncogene in HCC (7). Existing research have not well
elucidated the specific mechanisms underlying the role of CDKN3 in
HCC development. On these accounts, the present study holds the
primary objective of delving deeper into the mechanism of CDKN3 in
HCC.
Materials and methods
Materials
Among the HCC specimens and normal paraneoplastic
tissues from the Department of Pathology of Longhua Central
Hospital from January 2022 to December 2023, 16 cases with complete
medical records and comprehensive clinical information were
selected. The cases were not treated with radiotherapy or
chemotherapy, and all pathological indices were reevaluated by two
pathologists who reached a unanimous opinion. The present study
adhered to the declaration of Helsinki, and approval (approval no.
(2024-097-01) was obtained of the Ethics Committee of Longhua
Central Hospital (Shenzhen, China). Written informed consent was
acquired by all participants.
Inclusion criteria were as follows: i) Patients with
liver cancer who underwent surgery at Longhua Central Hospital; ii)
age ≥18 years old; iii) the specimen contains both cancerous and
control adjacent or normal liver tissue; iv) complete clinical
information; and v) the patient did not receive any adjuvant
therapy such as chemotherapy or radiotherapy before surgery.
Exclusion criteria were as follows: i) age <18 years old; ii)
the specimen does not contain corresponding adjacent cancerous
tissue or normal liver tissue; iii) received adjuvant therapy such
as chemotherapy or radiotherapy before surgery; and iv) clinical
information is incomplete.
The cancer genome atlas (TCGA)
database
General information of patients with liver HCC
(LIHC) was downloaded from the TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga)
and data on 371 patients with LIHC were obtained, including 245
male patients and 117 female patients. There were 27 patients aged
21-40, 140 patients aged 41-60, 181 patients aged 61-80, and 10
patients aged >81. There were 54 patients with stage I tumors,
173 patients with stage II tumors, 118 patients with stage III
tumors, and 12 patients with stage IV tumors.
The Assistant for Clinical
Bioinformatics database
The Assistant for Clinical Bioinformatics database
(https://www.aclbi.com/static/index.html) is a platform
that integrates information from multiple databases (8). Currently, it contains sample
information for all 33 tumors from TCGA database, seven
pediatric/hematologic tumors from the target database, tumor
samples from the Gene Expression Omnibus (GEO) database, non-tumor
sample information, cell line data from the cancer cell line
encyclopedia database, and sample information of 24 tumors from the
international cancer genome consortium database. The analysis steps
were as follows: i) pan-cancer analysis; ii) select sample: HCC;
iii) select gene: CDKN3; and iv) expression.
SangerBox database
The SangerBox database (http://sangerbox.com/home.html) is a web-based
platform (9) that offers
interactive graphical analysis tools, integrates multiple databases
and conducts fast batch processing of these data, remarkably
weakening the difficulty for users to obtain data and improving the
efficiency of data processing in the analysis of raw information.
The steps used were as follows: i) pan-cancer analysis; ii)
prognostic cancer gene expression analysis; iii) input gene: CDKN3;
iv) sample source selection: all samples; v) data source: TCGA +
GTEx; vi) survival data: overall survival (OS), progression-free
interval (PFI), disease-free interval (DFI) and disease-specific
survival (DSS).
STRING database
STRING (https://string-db.org/) is a search tool used for
analyzing biological gene or protein interactions. The interaction
data hosted on the database were sourced from high-throughput
experimental data, automated text mining, computer genome
prediction, and data from other databases, which has the largest
coverage of species and the largest amount of information on
interactions among numerous protein interaction databases currently
available, including a biological database containing proven and
predictable proteins and protein interactions (10). In the present study, the search
conditions were set as follows: i) select the protein by its name;
ii) enter CDKN3 in the protein names; and iii) select Homo
sapiens as the organism.
Gene expression profiling interactive
analysis (GEPIA) database
The GEPIA database (http://gepia.cancer-pku.cn/) comprises both the TCGA
cancer database and the GTEx normal tissue database (11). The screening criteria for the
differential expression analysis were as follows: i) expression
DIY, expression on box plots; ii) gene, CDKN3; iii) dataset
selection, HCC; iv) matched normal data, TCGA normal and GETx data.
The settings of the conditions for survival analysis were as
follows: i) dataset selection: HCC; ii) methods: OS; iii) group
cut-off: median.
Kaplan-Meier plotter database
Microarray and RNA-seq data derived from GEO, EGA
and TCGA databases were utilized for the construction of the
database. A total of 54,675 genes were subjected to meta-analysis
by integrating gene expression information with clinical prognostic
information, and survival-related molecular markers were identified
and validated (12,13). The steps used were as follows: i)
start KM Plotter for liver cancer; ii) input the target gene:
CDKN3; and iii) draw a Kaplan-Meier plot.
Genomic data commons (GDC)
database
GDC database (https://gdc.cancer.gov/) was developed by the National
Cancer Institute, consolidating data from multiple cancer
databases, offering unified storage, management, and visualization
while facilitating global sharing with cancer genomics
researchers.
Immunohistochemistry
For 16 samples of HCC tissues and normal tissues
adjacent to the cancer, paraffin-embedded 4-µm thick tissue
sections were received from each sample and stained (100˚C, 15 min)
with the Roche Ventana BenchMark XT immunohistochemistry system
using multimer technology. The samples were heat-repaired using
EDTA (pH 9.0). The slides then underwent 30 min of incubation with
rabbit anti-human CDKN3 antibodies (1:200; cat. no. abs115945;
Absin) at 37˚C, and another 32 min of incubation with horseradish
peroxidase-labeled secondary antibodies [1:100, cat. no. K20716;
Roche Diagnostics (Shanghai) Co., Ltd.] at 37˚C in succession.
Tissue slices then underwent color development using
diaminobenzidine and hematoxylin re-staining. Positive CDKN3 was
localized in the perinuclear region and cytoplasm, with yellowish
to tan coloration observed under a light microscope. Based on this
staining, the percentage of positive cells for CDKN3 protein
expression in tumor cells was assessed.
The proportion of positive cells to the total number
of cells is ≤10%, 11-25%, 26-50% and >50%, respectively, rated
as 0, 1, 2 and 3 points. At the same time, the degree of staining
of positive cells was observed and no staining, light yellow,
brownish yellow, and yellow brown were rated as 0, 1, 2, and 3
points, respectively. The final staining score for each slice is
obtained by multiplying the positive cell percentage score with the
positive cell staining degree score. A final score of 0-2 indicates
no expression, while a score of 3-9 indicates positive
expression.
Statistical analysis
Data analysis and processing relied on R software
(version 4.2.2; https://cran.r-project.org/). The mRNA expression
levels were converted to expression log2(TPM+1).
Measurements with normal distribution presented in the format of
the mean ± standard deviation (SD), and an independent sample
t-test served for the between-group variance comparison.
Measurements failing to obey a normal distribution presented as M
(P25, P75), and the Mann-Whitney U test assisted in the relevant
comparison. Counting data presented as the number of samples or
percentages, comparison between groups relied on the χ2
test, and grading information comparison relied on the rank-sum
test. A receiver operating characteristic (ROC) curve was plotted
for analyzing the diagnostic value of CDKN3 in patients. The
survival of high- and low-CDKN3 expression groups were analyzed
using the Kaplan-Meier survival curves followed by the log-rank
test. Univariate and multivariate Cox regression analyses together
assisted in ascertaining risk factors affecting the poor prognosis
of HCC. P<0.05 was considered to indicate a statistically
significant difference.
Results
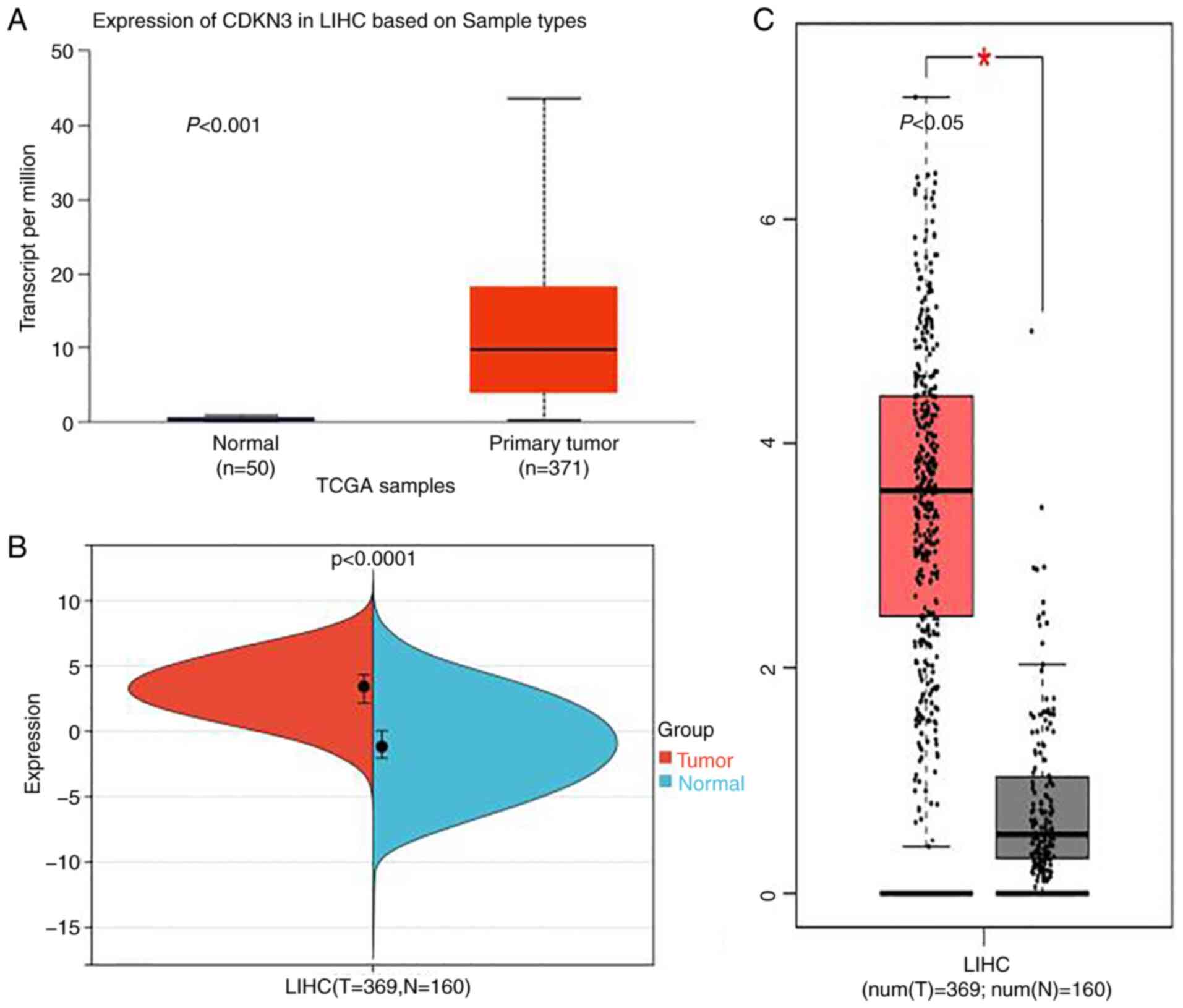
CDKN3 gene expression analysis
According to the analysis results of the CDKN3
expression in HCC via the SangerBox, UALCAN and GEPIA databases,
tumor tissue presented significantly higher CDKN3 expression versus
normal tissues, and the difference exhibited a statistical
significance (P<0.05) (Fig.
1A-C).
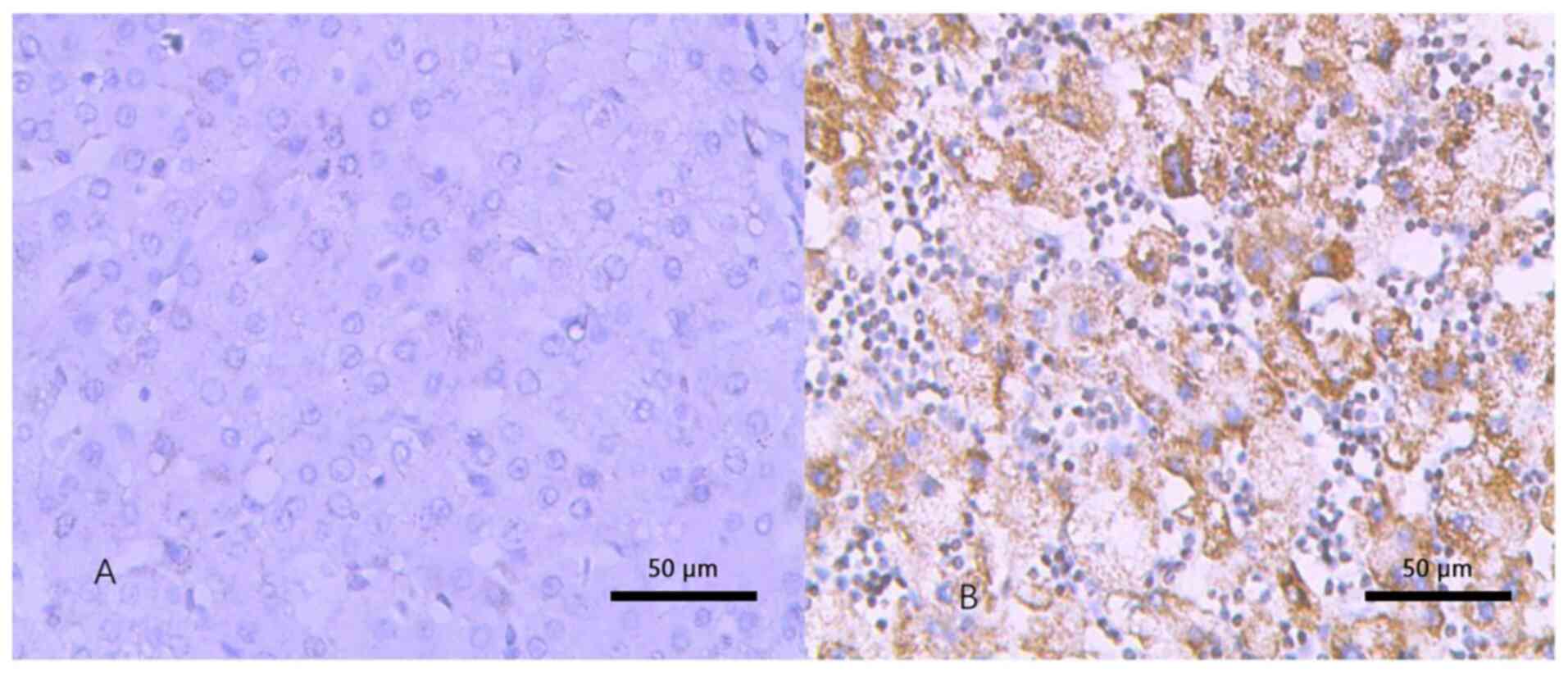
Immunohistochemistry
Immunohistochemical analysis suggested that CDKN3
was localized in the perinuclear region and cytoplasm of the cells
and presented a high expression in HCC tissues versus normal
tissues (P<0.05) (Fig. 2).
Immunohistochemical scoring was performed on the expression of
CDKN3 in 16 cases of liver cancer tissues, and the expression of
CDKN3 in corresponding normal tissues was also evaluated. After
statistical analysis, the positive expression rate of CDKN3 in LIHC
was 93.75%, while the positive expression rate of CDKN3 in normal
liver tissues was 18.75%, and the difference was statistically
significant (P<0.05) (Table
I).
 | Table IComparison of immunohistochemical
positivity rates between tumor tissue and normal tissue
(P<0.05). |
Table I
Comparison of immunohistochemical
positivity rates between tumor tissue and normal tissue
(P<0.05).
| | Positive (%) | Negative (%) |
|---|
| Normal | 18.75 (3/16) | 81.25 (13/16) |
| Tumor | 93.75 (15/16) | 6.25 (1/16) |
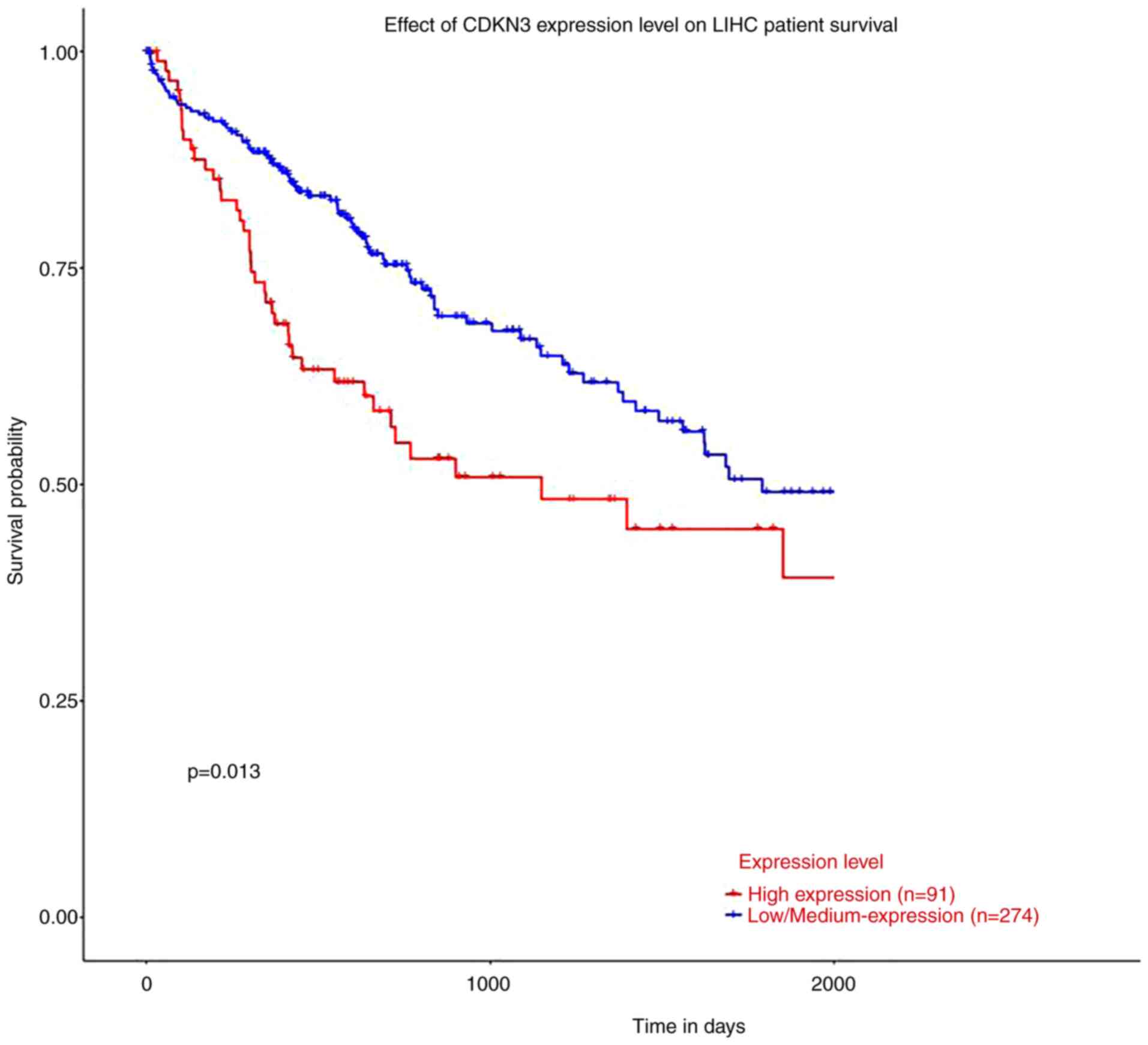
CDKN3 at different expression
levels
According to the analysis results of how CDKN3
expression affected the OS of patients with HCC by the UALCAN
database, patients in the low-expression group presented
considerably extended survival time versus the high-expression
group and P<0.05 (Fig. 3).
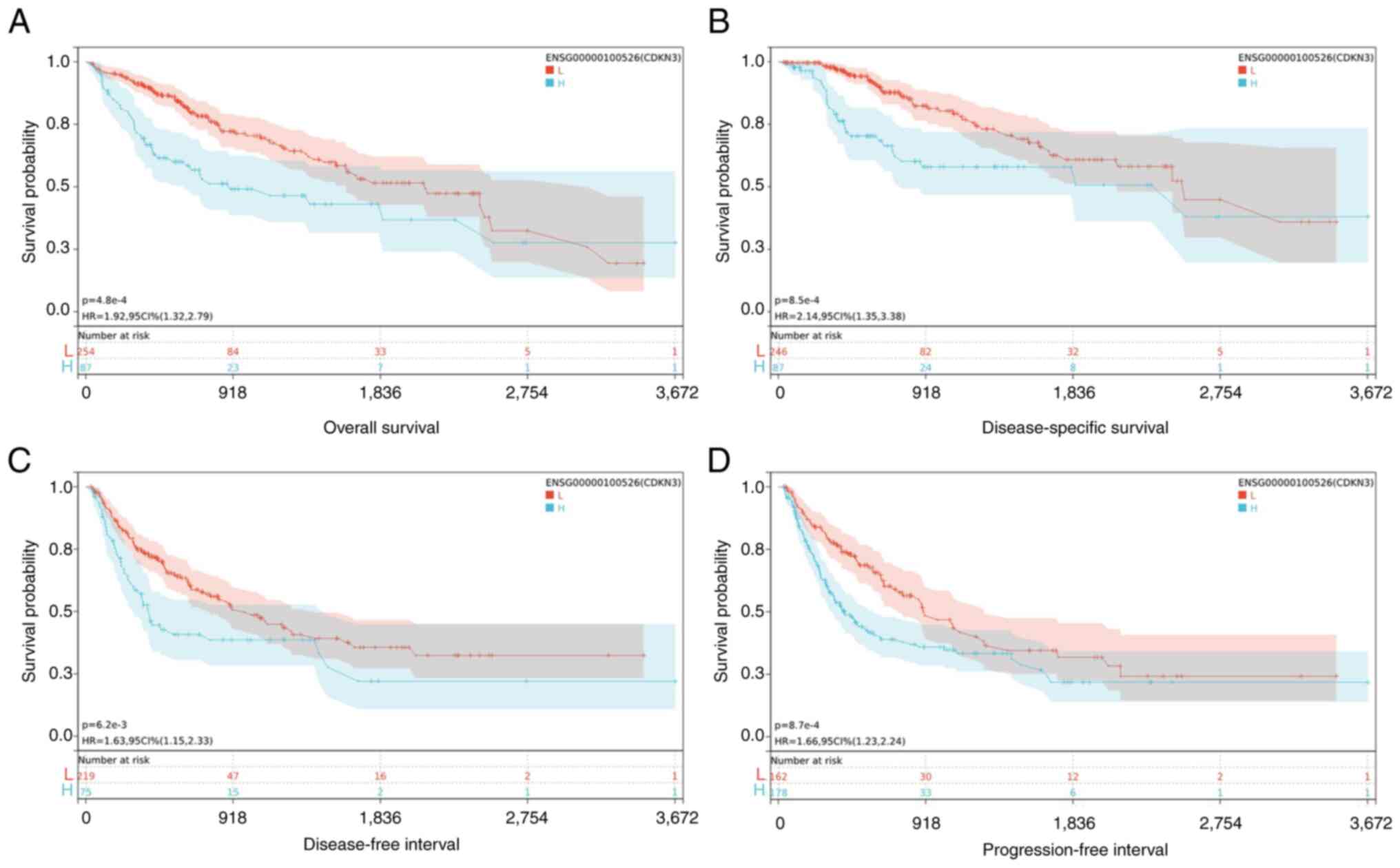
The tumor prognosis included OS, PFI, DFI and DSS,
and their correlation with CDKN3 was measured by utilizing the
SangerBox database. The specific method was as follows: the optimal
cut-off values for CDKN3 were calculated by virtue of the ‘maxstat’
package in R (maximally selected rank statistics with some P-value
approximations version: 0.7-25), with the minimum grouping of
samples >25% and the maximum grouping <75%. The optimal
cut-off values were calculated as -9.9658 (OS), -5.0116 (PFI),
-5.0116 (DFI) and -9.9658 (DSS). Based on these values, patients
fell into high- and low-expression groups, and the log-rank test
method was employed for a deeper interpretation of their difference
in prognosis by virtue of the survfit function of the ‘survival’
package in R. The low-expression group showed more favorable
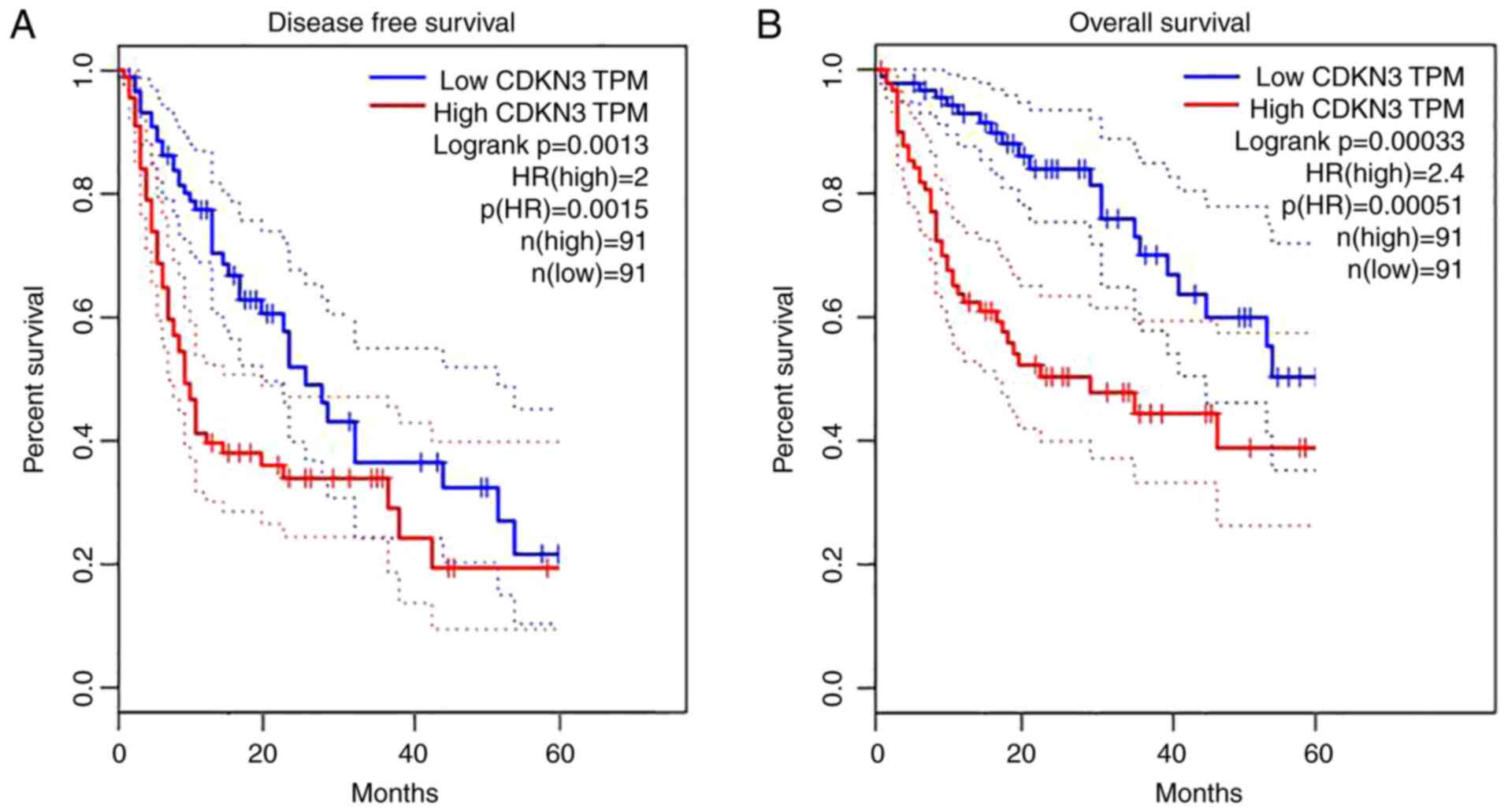
prognosis versus the high-expression group (P<0.05) (Fig. 4).
Moreover, the prognosis of the two expression groups
in patients with HCC was compared using the GEPIA database. The
high-expression group exhibited considerably better DFS and OS
versus the low-expression group (P<0.05) (Fig. 5).
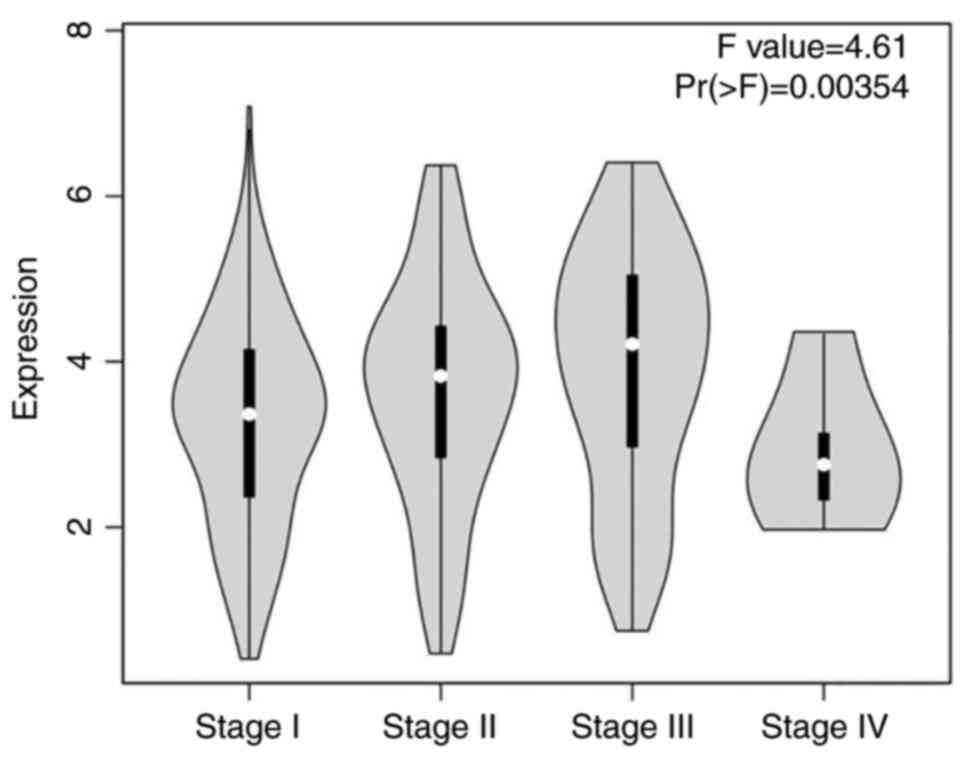
According to the analysis on the linkage between
CDKN3 expression and HCC tumor stage grading using the GEPIA
database, the expression level of CDKN3 underwent an obvious
elevation in higher tumor stage (P<0.05). However, in stage IV,
because patients with tumors at this stage were in the advanced
stage, fewer specimens could be obtained by surgical resection.
Because this led to an insufficient number of specimens, the
statistical data may have been skewed. However, based on the
samples obtained, the levels of CDKN3 in grade I, II and III
patients increased progressively with grading (Fig. 6).
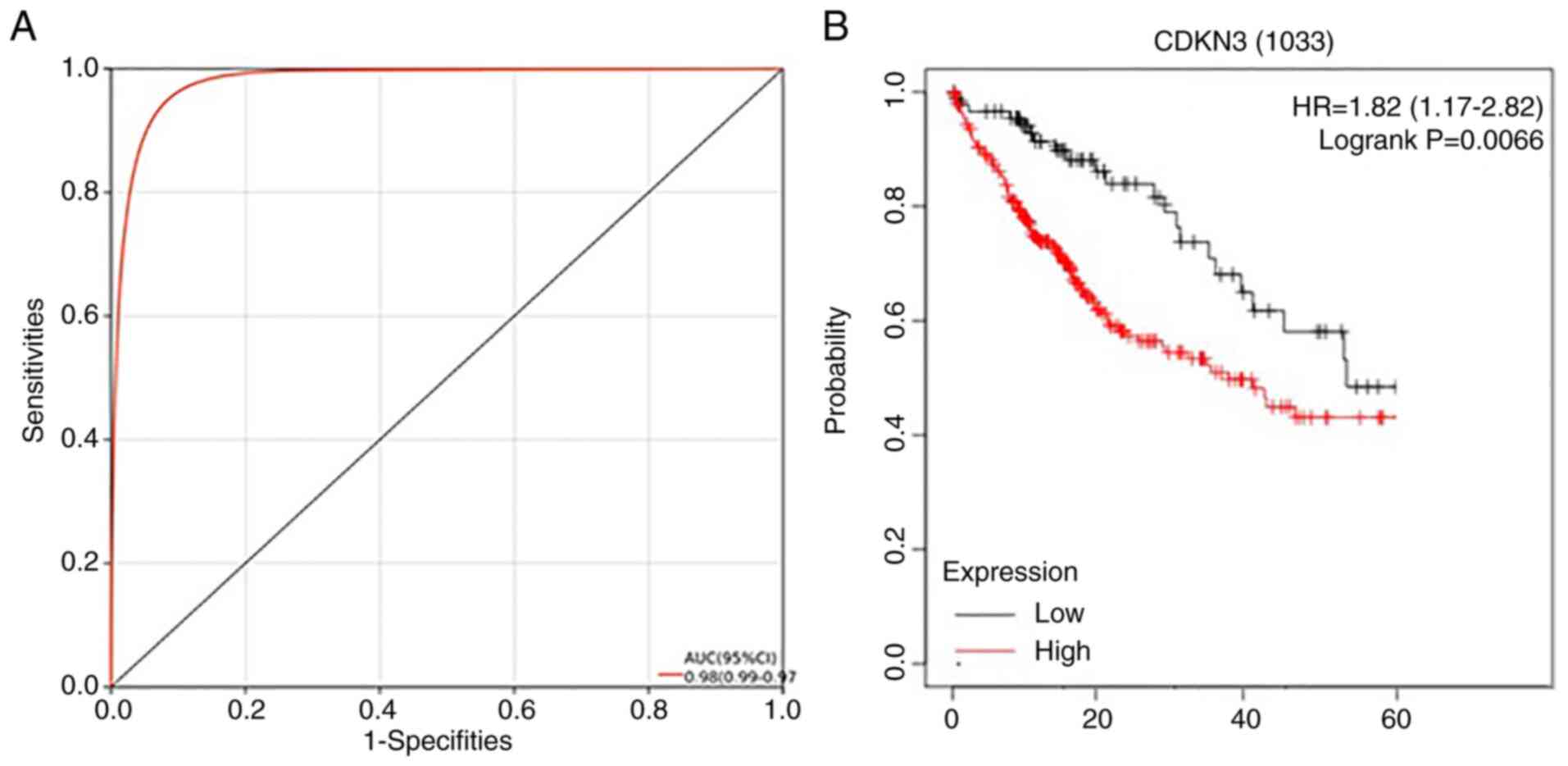
ROC curves for the diagnosis of HCC by
CDKN3
The Kaplan-Meier analysis was conducted on specific
genes for the examination of their prognostic values. The different
quartiles of gene expression were taken as the criteria to classify
patients into two cohorts, with the relevant comparison results
illustrated in the Kaplan-Meier survival plot, from which we
obtained the hazard ratio (HR), 95% confidence interval (CI) and
log-rank P-values. Collectively, CDKN3 had a favorable diagnostic
value for HCC, with an area under curve (AUC) of 0.98 and 95% CI of
0.99-0.97. CDKN3 could also well predict the poor prognosis of
patients with HCC (P<0.05), proving its statistical significance
(Fig. 7).
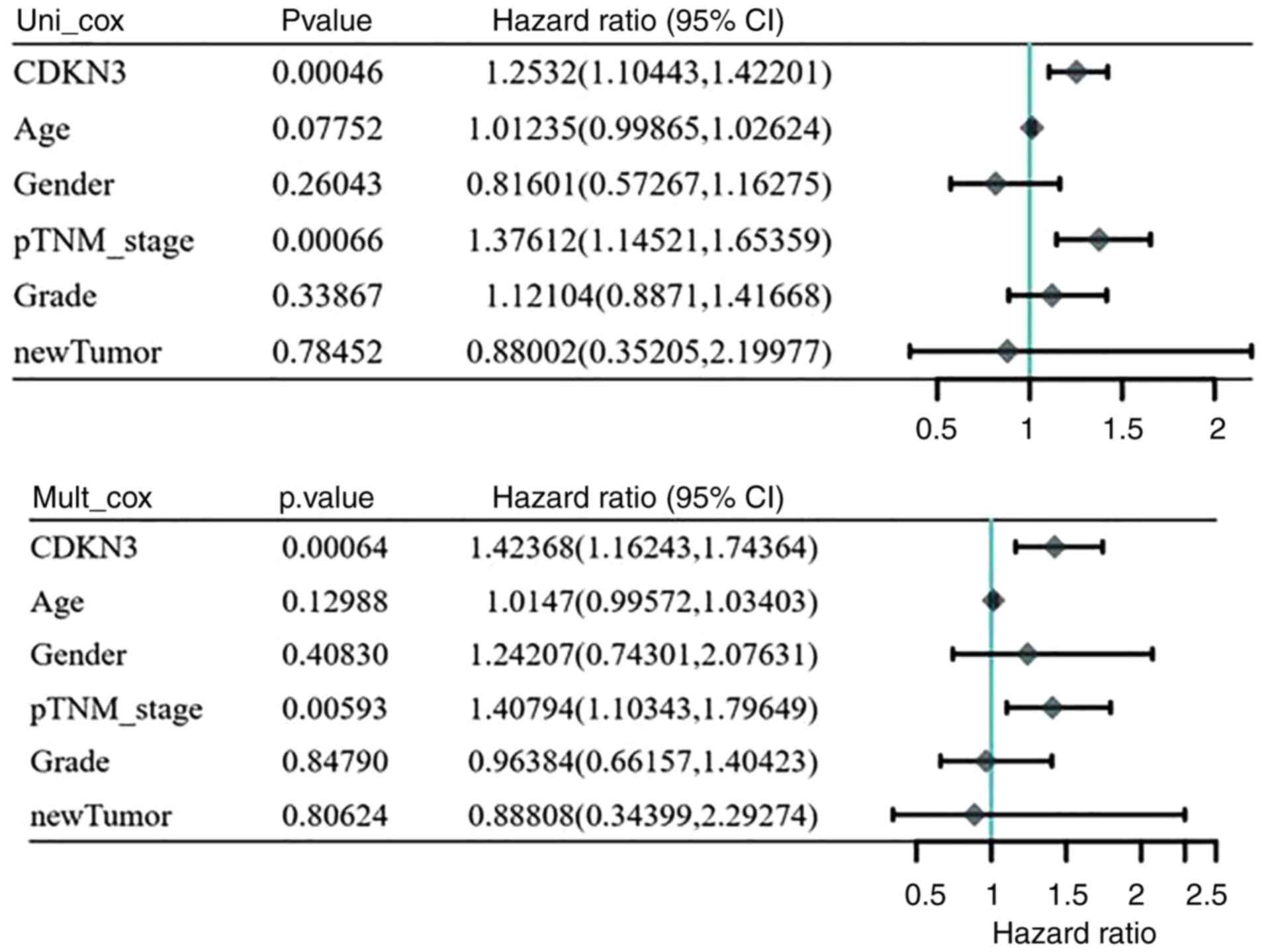
Univariate and multivariate
analysis
One-way multifactorial analysis was conducted using
Assistant for Clinical Bioinformatics. Univariate and
multifactorial analyses revealed the association between prognosis
and CDKN3 expression and tumor-node-metastasis staging for patients
with HCC (P<0.05) (Fig. 8).
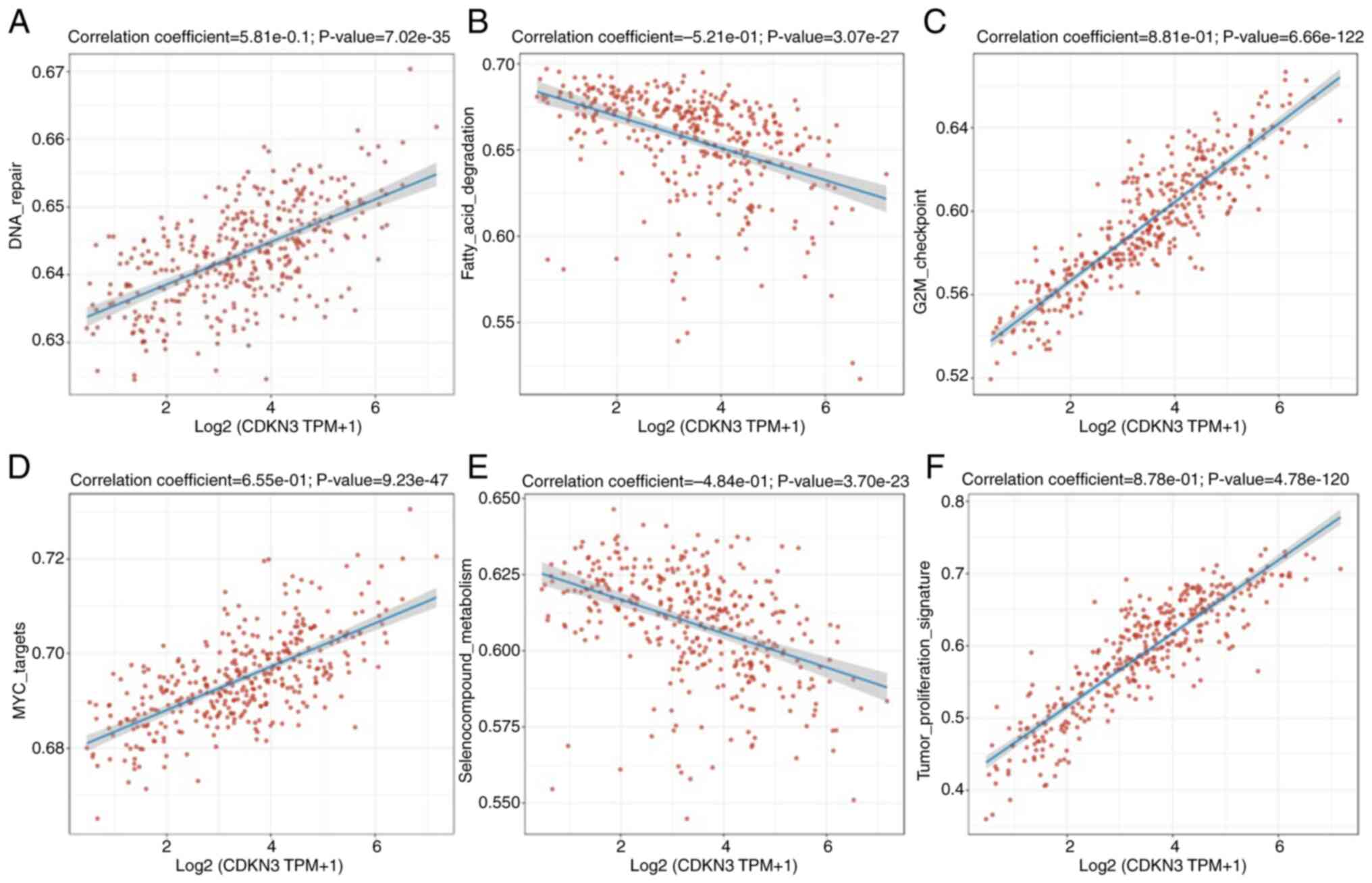
CDKN3 and pathway correlation
RNA-seq data and the respective clinical information
for HCC were obtained from the Genomic Data Commons data portal
(https://portal.gdc.com). The gene set variation
analysis package in R was utilized for analyzing the genes
contained in relevant pathways. Lastly, the gene collection
analysis was conducted based on pathway scores using Spearman
correlation. All of the analysis relied on R (version 4.0.3). The
present study demonstrated the participation of CDKN3 in several
biological processes, including DNA repair (0.581), fatty acid
degradation (0.521), G2/M checkpoint regulation (0.881), MYC
targets (0.655), seleno-compound metabolism (0.484), tumor
proliferation signature (0.878) and other pathways (P<0.05)
(Fig. 9A-F).
Gene enrichment analysis
Numerous functional nodes that exhibit a conceptual
overlapping phenomenon will be generated upon the direct annotation
of a set of genes; hence the redundant analysis dose does not
benefit the further examination. Therefore, the obtained functional
nodes must be filtered and screened, thereby obtaining a larger
number of meaningful functional information. The most commonly used
methods are based on Gene Ontology (GO) and Kyoto Encyclopedia of
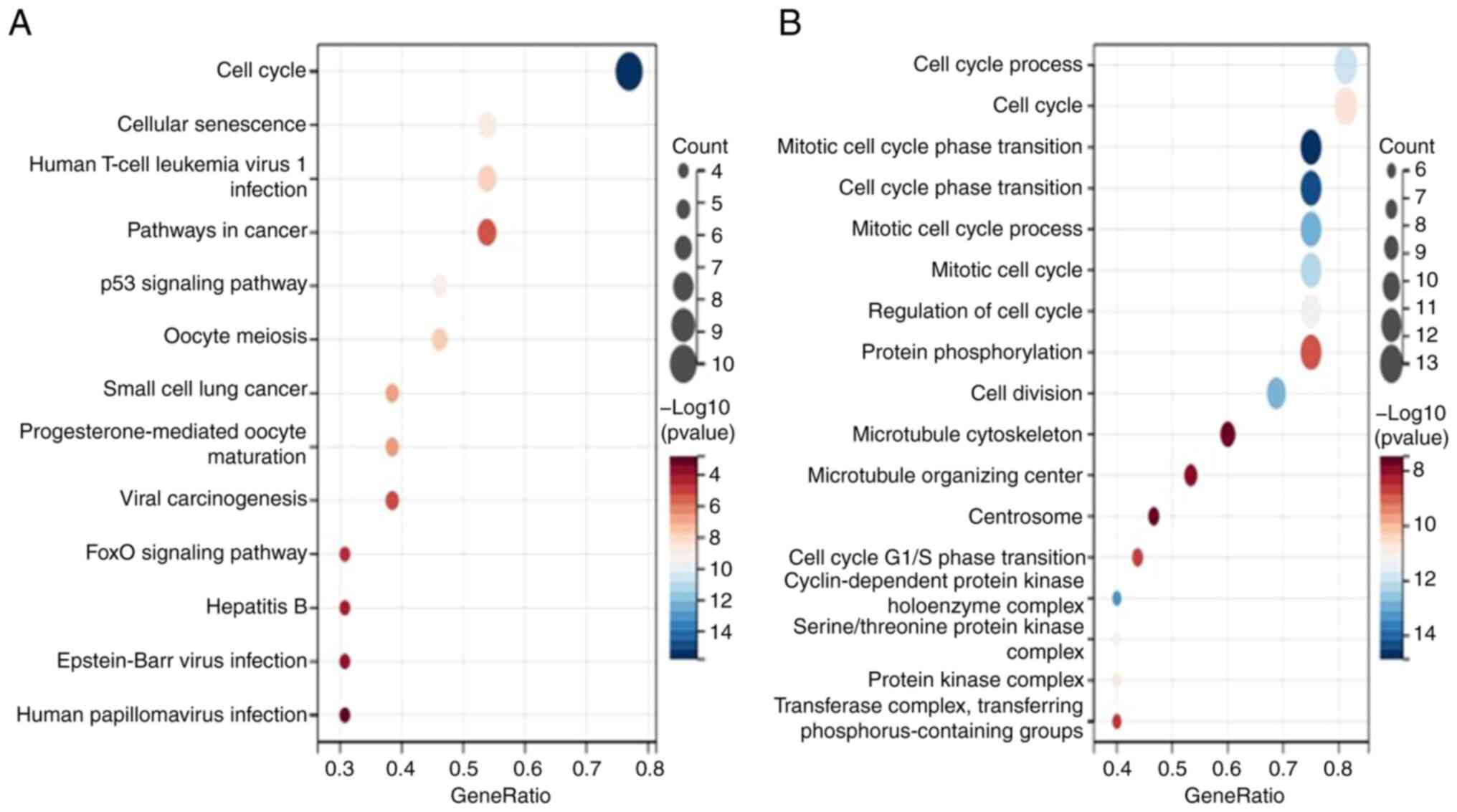
Genes and Genomes (KEGG) enrichment analyses. According to KEGG
analysis, CDKN3 is associated with the cell cycle, p53 signaling
pathway, cellular senescence, human T-cell leukemia virus 1
infection, oocyte meiosis, small-cell lung cancer,
progesterone-mediated oocyte maturation, cancer pathway, viral
oncogenesis, FOXO signaling pathway, hepatitis B, EB virus
infection, human papillomavirus infection, and other pathways; the
cell cycle pathway was the most dominant pathway. GO analysis
indicated that mitotic cell cycle phase transition, cell cycle
phase transition, cell cycle protein-dependent protein kinase
holoenzyme complex, mitotic cell cycle process, cell division, cell
cycle process, mitotic cell cycle, serine/threonine protein kinase
complex, protein kinase complex, cell cycle regulation, cell cycle,
cell cycle protein-dependent protein kinase activity, transferase
complexes, transfer of phosphorus-containing groups, protein
phosphorylation, cell cycle G1/S phase transition, microtubule
organizing centers, microtubule cytoskeleton, centrosome, and other
pathways were related, wherein the mitotic cell cycle phase
transition and cell cycle pathways were the dominant pathways
(Fig. 10).
Protein-protein interaction network of
CDKN3 in HCC
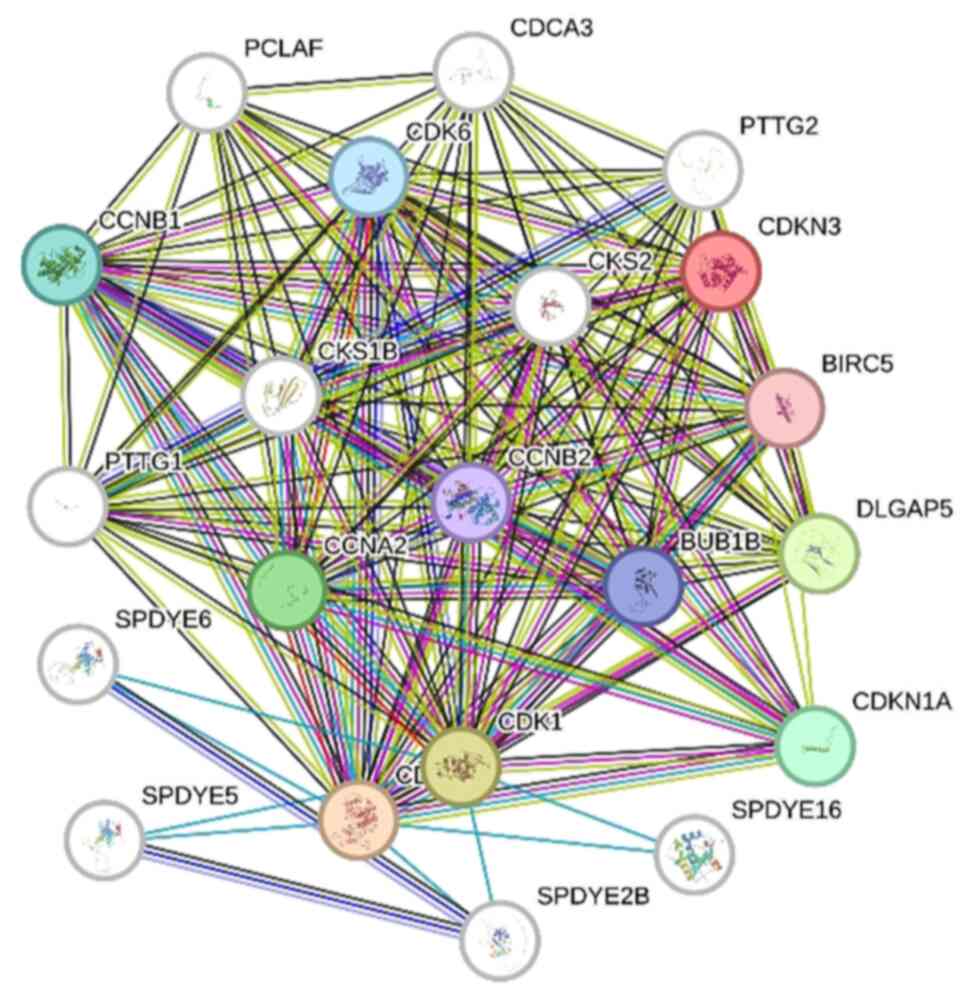
Proteins usually perform their function by
interacting with other proteins. By studying the interaction
networks between proteins, the functional regulation and signaling
processes of proteins within the cell can be better understood. The
interactions of CDKN3 with other proteins were analyzed using the
STRING database, in which multiple interacting proteins were found
to be centered on CDKN3, such as PTTG2, BIRC5, CKS2, CCNB2 and
CCNA2, as shown in Fig. 11.
Discussion
HCC is the second most common cause of
cancer-related deaths and ranks 6th in incidence, with rates
currently increasing in China (14). Advances in surgical procedures,
chemotherapy and targeted immunotherapy obviously prolong the life
span of patients with HCC. Additionally, the widespread adoption of
multidisciplinary comprehensive treatment protocols has improved
patient outcomes. Despite these developments, the prevention and
control of HCC remain challenging (15). On these accounts, efforts shall be
made to identify new HCC markers and potential therapeutic targets,
and to investigate their molecular mechanisms in the pathogenesis
of HCC to prevent and control the disease.
In the present study, CDKN3 presented a high
expression in HCC tumor tissues, which was further validated by
immunohistochemistry, conforming to previous findings (16-18).
The value of CDKN3 in diagnosing HCC and determining its prognosis
was well recognized here. According to Fig. 9, CDKN3 expression in HCC is mainly
related to DNA repair, fatty acid degradation, G2M checkpoint, MYC
target, selenium compound metabolism, tumor proliferation markers
and other pathways. According to unifactorial and multifactorial
analyses, CDKN3 significantly marks worse prognosis of patients
with HCC. As demonstrated in Fig.
10, the function of CDKN3 in HCC was closely correlated with
cell cycle-related pathways, as demonstrated previously in other
studies (7). The CDKN3 gene
interacts with multiple pathways related to the cell cycle, such as
DNA repair and G2/M checkpoint, in liver cancer to regulate tumor
growth, metastasis and drug resistance. Research has shown that the
KAP protein encoded by CDKN3 can inhibit the activity of CDK2,
thereby affecting the normal progression of the cell cycle. In
terms of DNA repair, CDKN3 may interfere with the function of CDK2,
weaken the cell's ability to repair DNA damage, increase genomic
instability, and promote tumor occurrence and progression. In G2/M
checkpoint regulation, CDKN3 may hinder the transition of cells
from G2 phase to M phase by inhibiting the activity of CDK1,
leading to cell cycle arrest or abnormal division, which may be
related to the invasiveness and metastatic ability of tumor cells.
In addition, overexpression of CDKN3 may lead to tumor cell
resistance to chemotherapy drugs by affecting the expression of
cell cycle related proteins. However, the specific molecular
mechanisms by which CDKN3 regulates tumor growth, metastasis, and
drug resistance through these pathways have not been fully
elucidated, and further research is needed to reveal its detailed
functional network. Thus, CDKN3 affects HCC development by
controlling the cell cycle.
Cell cycle dysregulation leads to uncontrolled and
abnormal cell proliferation, during which cell cycle-related
proteins associated with tumorigenesis undergo aberrant changes
(19). CDKN3 can well control the
cell cycle by acting as a dual-specificity protein phosphatase
dephosphorylating cell cycle protein dependent kinase (20). Hence, CDKN3 can negatively regulate
cell cycle progression, thus crucially impacting the development of
various tumors (4,21-22).
In summary, the role of CDKN3 gene in LIHC has
received widespread attention in recent years. Research has shown
that CDKN3 plays an important role in the occurrence and
development of liver cancer by regulating the cell cycle
progression. The protein KAP (CDK2 Associated Protein) encoded by
CDKN3 can inhibit the activity of cell cycle dependent kinase,
thereby affecting cell proliferation. In liver cancer, CDKN3 often
exhibits abnormal expression, and its overexpression is closely
related to the invasiveness, metastatic ability, and poor prognosis
of the tumor. Therefore, CDKN3 is considered a potential
therapeutic target. By targeting and inhibiting the expression or
function of CDKN3, it may effectively suppress the proliferation of
liver cancer cells and induce their apoptosis, providing a new
approach for precise treatment of liver cancer.
From the perspective of clinical translational
value, the study of CDKN3 has important practical significance.
Firstly, the expression level of CDKN3 may serve as a biomarker for
the diagnosis and prognosis of liver cancer, helping clinicians
identify high-risk patients earlier and develop personalized
treatment plans. Secondly, the development of small molecule
inhibitors or gene editing technologies targeting CDKN3 may provide
new drug options for the treatment of liver cancer. In addition,
the study of the interaction between CDKN3 and other signaling
pathways (such as PI3K/AKT, p53) may reveal multi-target treatment
strategies for liver cancer, thereby improving treatment efficacy
and reducing the occurrence of drug resistance.
However, there are still some limitations in current
research on CDKN3. Firstly, the present study is retrospective, and
information bias may be caused by incomplete records (such as
missing clinical parameters, interrupted follow-up) or improper
sample preservation (such as degradation of immunohistochemical
specimens). It is also possible that differences in
immunohistochemistry between different batches (such as antibody
clone numbers, staining processes) may lead to measurement errors
in CDKN3 expression. Secondly, the specific mechanism of action of
CDKN3 in liver cancer has not been fully elucidated, especially the
differential expression and functional heterogeneity in different
subtypes of liver cancer still need further exploration. Thirdly,
existing research is mostly based on cell experiments and animal
models, lacking support from large-scale clinical data, which
limits the clinical translation process of CDKN3 as a therapeutic
target. In addition, the safety and efficacy of CDKN3 inhibitors
still need to be validated through rigorous clinical trials.
Although immunohistochemistry was used to validate the expression
of CDKN3 in tumor samples, the present study mainly relied on
bioinformatics tools for data analysis; In the future, the authors
will further validate the role of CDKN3 in LIHC through western
blotting and PCR methods.
Although the present study has numerous limitations,
it still has some innovation: combining clinical sample analysis,
public database mining, and experimental verification to confirm
the role of CDKN3 in LIHC; Multi-level research from expression
characteristics, clinical significance to molecular mechanisms; A
new standard for CDKN3 as a prognostic biomarker was proposed, and
the optimal critical value was determined. CDKN3 was also proposed
as a potential biomarker for immunotherapy, demonstrating the
association between CDKN3 and tumor microenvironment.
Future research directions should focus on the
following aspects: firstly, in-depth analysis of the molecular
mechanism of CDKN3 in liver cancer, especially its interaction with
other key signaling pathways; Secondly, conducting multi-center and
large-scale clinical studies to verify the reliability of CDKN3 as
a biomarker and therapeutic target; The third is to develop
efficient and low toxicity CDKN3 targeted drugs, and explore their
combined effects with other treatment methods such as immunotherapy
and radiotherapy. Through interdisciplinary collaboration and the
application of cutting-edge technologies, CDKN3 is expected to
become an important breakthrough point in the field of liver cancer
treatment.
In summary, the present study has laid a theoretical
foundation for the pathogenesis and immunotherapy research of LIHC.
However, there are also some shortcomings: differences in sample
sources, sequencing platforms, and batches in public databases may
lead to biased results and affect the comparability of CDKN3
expression. Lack of functional validation: Bioinformatics only
provides relevant conclusions and lacks experimental validation of
the protein level and specific molecular mechanism of CDKN3; The
molecular characteristics of subtypes of liver cancer (such as
HBV/HCV related and non-alcoholic fatty liver cancer) are
different, but some datasets are not clearly stratified, which
limits the generalization of conclusions Although the co-expression
network or pathway enrichment of CDKN3 can be predicted, its
upstream regulation (such as methylation, miRNA) and downstream
effects in liver cancer still require experimental exploration.
In addition, the present study also has certain
limitations, such as not being able to fully control for all
potential confounding factors, such as the patient's genetic
background, environmental exposure, comorbidities and treatment
history. These factors may affect the association between CDKN3
expression and liver cancer progression, leading to biased results.
In future research, it will be attempted to collect treatment
response data and information on co-existing diseases in patients,
conduct supplementary analysis on existing molecular marker data
and concurrently improve the analysis method to control as numerous
known confounding factors as possible.
Supplementary Material
Details of pathological section
(diagnosis: liver cancer).
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XL conducted experiments, wrote and edited the
manuscript. KC wrote the original draft and collected data. JL
designed the study, performed data curation and project
administration. XL and KC confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study adhered to the declaration of
Helsinki, and approval (approval no. (2024-097-01) was obtained of
the Ethics Committee of Longhua Central Hospital (Shenzhen, China).
Written informed consent was acquired by all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shi JF, Cao M, Wang Y, Bai FZ, Lei L, Peng
J, Feletto E, Canfell K, Qu C and Chen W: Is it possible to halve
the incidence of liver cancer in China by 2050? Int J Cancer.
148:1051–1065. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Xia Y, Li J, Liu G, Wang K, Qian G, Lu Z,
Yang T, Yan Z, Lei Z, Si A, et al: Long-term effects of repeat
hepatectomy vs percutaneous radiofrequency ablation among patients
with recurrent hepatocellular carcinoma: A randomized clinical
trial. JAMA Oncol. 6:255–263. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ding J and Wen Z: Survival improvement and
prognosis for hepatocellular carcinoma: Analysis of the SEER
database. BMC Cancer. 21(1157)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhang C, Shen Q, Gao M, Li J and Pang B:
The role of cyclin dependent kinase inhibitor 3 (CDKN3) in
promoting human tumors: Literature review and pan-cancer analysis.
Heliyon. 10(e26061)2024.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Barrón EV, Roman-Bassaure E,
Sánchez-Sandoval AL, Espinosa AM, Guardado-Estrada M, Medina I,
Juárez E, Alfaro A, Bermúdez M, Zamora R, et al: CDKN3 mRNA as a
biomarker for survival and therapeutic target in cervical cancer.
PLoS One. 10(e0137397)2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li WH, Zhang L and Wu YH: CDKN3 regulates
cisplatin resistance to colorectal cancer through TIPE1. Eur Rev
Med Pharmacol Sci. 24:3614–3623. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dai W, Miao H, Fang S, Fang T, Chen N and
Li M: CDKN3 expression is negatively associated with pathological
tumor stage and CDKN3 inhibition promotes cell survival in
hepatocellular carcinoma. Mol Med Rep. 14:1509–1514.
2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Huang R, Guo L, Chen C, Xiang Y, Li G,
Zheng J, Wu Y, Yuan X, Zhou J, Gao W and Xiang S: System analysis
identifies UBE2C as a novel oncogene target for adrenocortical
carcinoma. PLoS One. 18(e0289418)2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shi T, Hu Z, Tian L and Yang Y: Pan-cancer
landscape of CENPO and its underlying mechanism in LUAD. Respir
Res. 24(113)2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Szklarczyk D, Kirsch R, Koutrouli M,
Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT,
Pyysalo S, et al: The STRING database in 2023: Protein-protein
association networks and functional enrichment analyses for any
sequenced genome of interest. Nucleic Acids Res. 51:D638–D646.
2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gao J, Yang S, Xie G, Pan J and Zhu F:
Integrating network pharmacology and experimental verification to
explore the pharmacological mechanisms of Aloin against gastric
cancer. Drug Des Devel The. 16:1947–1961. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hu J, Qiu D, Yu A, Hu J, Deng H, Li H, Yi
Z, Chen J and Zu X: YTHDF1 Is a potential pan-cancer biomarker for
prognosis and immunotherapy. Front Oncol. 11(607224)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hou W, Kong L, Hou Z and Ji H: CD44 is a
prognostic biomarker and correlated with immune infiltrates in
gastric cancer. BMC Med Genomics. 15(225)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xie D, Shi J, Zhou J, Fan J and Gao Q:
Clinical practice guidelines and real-life practice in
hepatocellular carcinoma: A Chinese perspective. Clin Mol Hepatol.
29:206–216. 2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
An L, Zeng HM, Zheng RS, Zhang SW, Sun KX,
Zou XN, Chen R, Wang SM, Gu XY, Wei WW and He J: Liver cancer
epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi. 41:721–727.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mori J, Sawada T, Baba T, Hayakawa A,
Kanemoto Y, Nishimura K, Amano R, Siril YJ, Okada M, Kurokawa T and
Kato S: Identification of cell cycle-associated and -unassociated
regulators for expression of a hepatocellular carcinoma oncogene
cyclin-dependent kinase inhibitor 3. Biochem Biophys Res Commun.
625:46–52. 2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Deng X, Ma J, Zhou W, Yuan Y, Wang B and
Meng X: GID2 interacts with CDKN3 and regulates pancreatic cancer
growth and apoptosis. Lab Invest. 103(100122)2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gao C, Fan X, Liu Y, Han Y, Liu S, Li H,
Zhang Q, Wang Y and Xue F: Comprehensive analysis reveals the
potential roles of CDKN3 in pancancer and verification in
endometrial cancer. Int J Gen Med. 16:5817–5839. 2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ghafouri-Fard S, Khoshbakht T, Hussen BM,
Dong P, Gassler N, Taheri M, Baniahmad A and Dilmaghani NA: A
review on the role of cyclin dependent kinases in cancers. Cancer
Cell Int. 22(325)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yu C, Cao H, He X, Sun P, Feng Y, Chen L
and Gong H: Cyclin-dependent kinase inhibitor 3 (CDKN3) plays a
critical role in prostate cancer via regulating cell cycle and DNA
replication signaling. Biomed Pharmacother. 96:1109–1118.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang J, Zhang Y and Zhang J:
N6-Methyladenosine modified circ-NAB1 modulates cell cycle and
epithelial-mesenchymal transition via CDKN3 in endometrial cancer.
Cell Mol Biol (Noisy-le-grand). 70:161–169. 2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Al Sharie AH, Abu Zahra AM, El-Elimat T,
Darweesh RF, Al-Khaldi AK, Abu Mousa BM, Amer MSB, Al Zu'bi YO,
Al-Kammash K, Abu Lil A, et al: Cyclin dependent kinase inhibitor 3
(CDKN3) upregulation is associated with unfavorable prognosis in
clear cell renal cell carcinoma and shapes tumor immune
microenvironment: A bioinformatics analysis. Medicine (Baltimore).
102(e35004)2023.PubMed/NCBI View Article : Google Scholar
|