|
1
|
Atun R and Cavalli F: The global fight
against cancer: Challenges and opportunities. Lancet. 391:412–413.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jariwala N, Rajasekaran D, Srivastava J,
Gredler R, Akiel MA, Robertson CL, Emdad L, Fisher PB and Sarkar D:
Role of the staphylococcal nuclease and tudor domain containing 1
in oncogenesis (Review). Int J Oncol. 46:465–473. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cui X, Zhang X, Liu M, Zhao C, Zhang N,
Ren Y, Su C, Zhang W, Sun X, He J, et al: A pan-cancer analysis of
the oncogenic role of staphylococcal nuclease domain-containing
protein 1 (SND1) in human tumors. Genomics. 112:3958–3967.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang Y, Wang X, Cui X, Zhuo Y, Li H, Ha C,
Xin L, Ren Y, Zhang W, Sun X, et al: Oncoprotein SND1 hijacks
nascent MHC-I heavy chain to ER-associated degradation, leading to
impaired CD8+ T cell response in tumor. Sci Adv.
6(eaba5412)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Dhiman G, Srivastava N, Goyal M, Rakha E,
Lothion-Roy J, Mongan NP, Miftakhova RR, Khaiboullina SF, Rizvanov
AA and Baranwalet M: Metadherin: A therapeutic target in multiple
cancers. Front Oncol. 9(349)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Manna D and Sarkar D: Multifunctional role
of astrocyte elevated gene-1 (AEG-1) in cancer: Focus on drug
resistance. Cancers. 13(1792)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Guo F, Wan L, Zheng A, Stanevich V, Wei Y,
Satyshur KA, Shen M, Lee W, Kang Y and Xing Y: Structural insights
into the tumor-promoting function of the MTDH-SND1 complex. Cell
Rep. 8:1704–1713. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Davis E, Ermi AG and Sarkar D: Astrocyte
elevated gene-1/Metadherin (AEG-1/MTDH): A promising molecular
marker and therapeutic target for hepatocellular carcinoma. Cancers
(Basel). 17(1375)2025.PubMed/NCBI View Article : Google Scholar
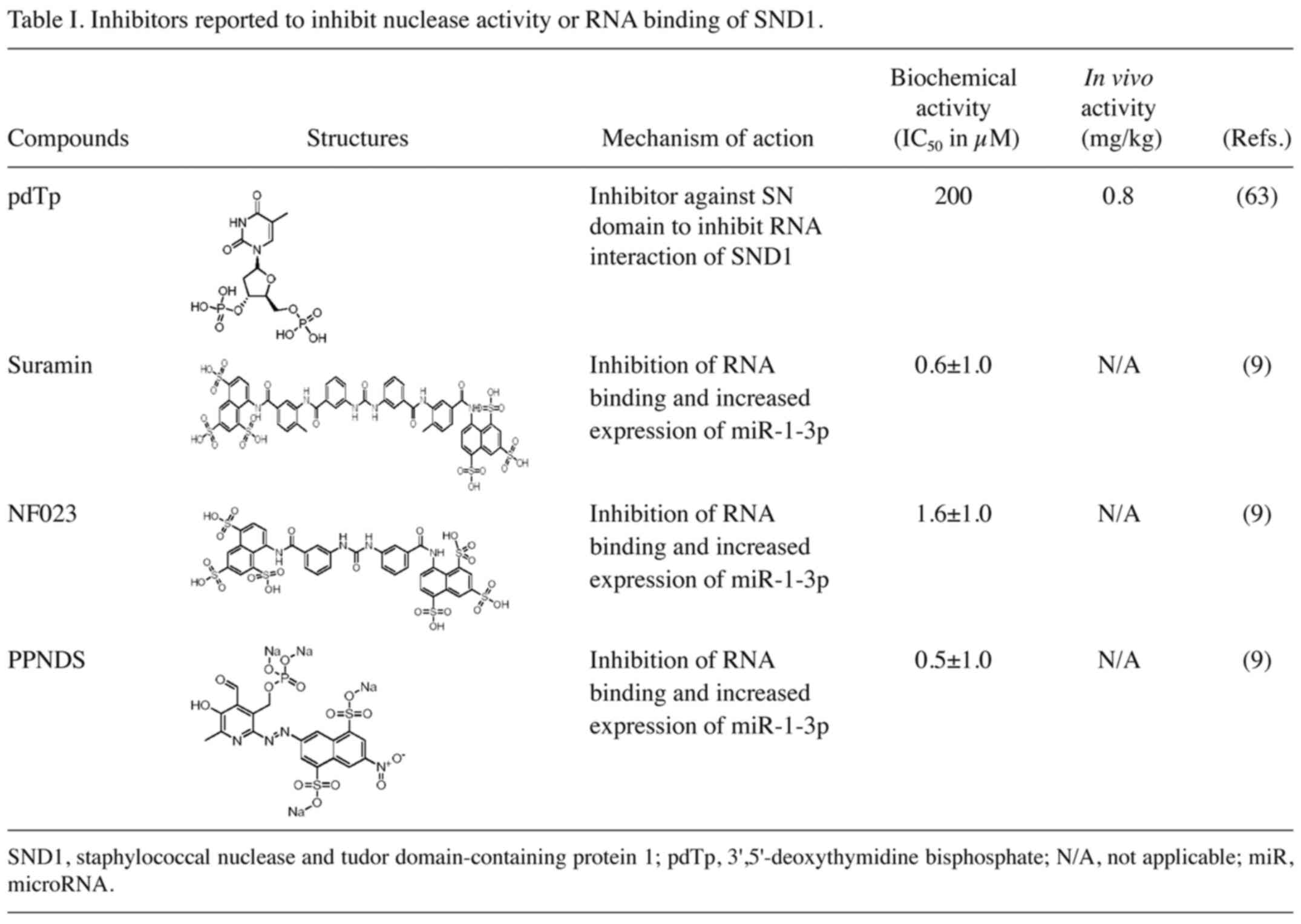
|
|
9
|
Lehmusvaara S, Haikarainen T, Saarikettu
J, Nieto GM and Silvennoinen O: Inhibition of RNA binding in snd1
increases the levels of mir-1-3p and sensitizes cancer cells to
navitoclax. Cancers (Basel). 14(3100)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
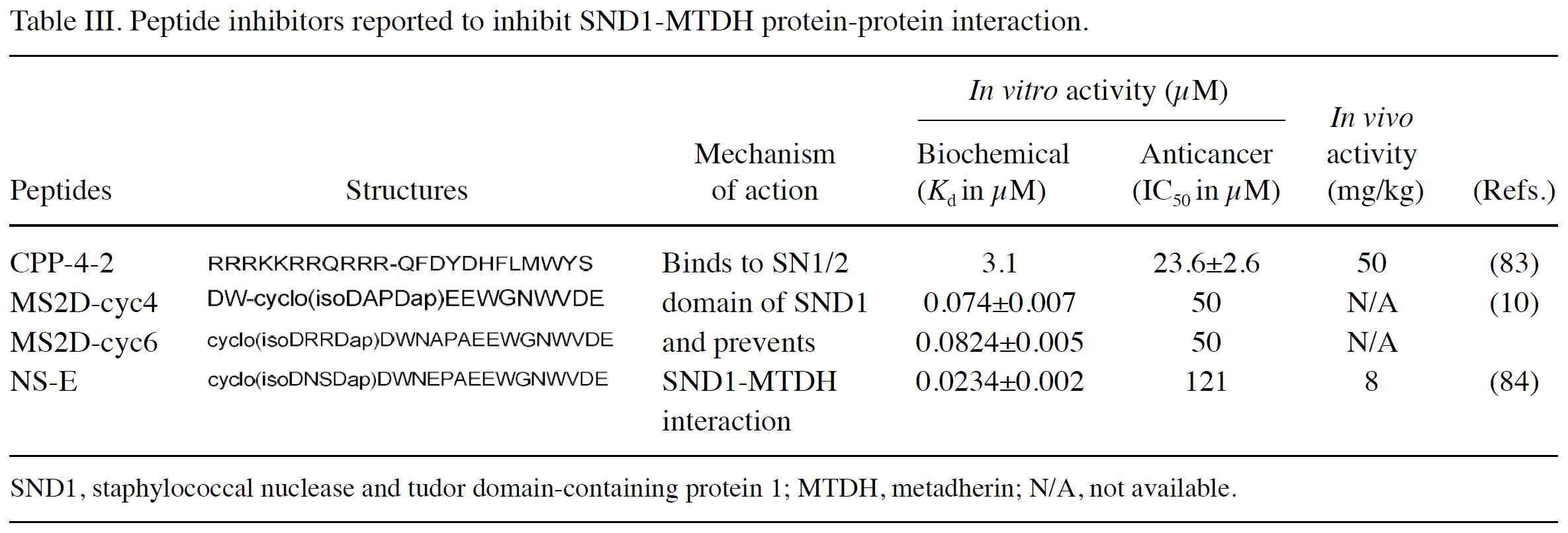
Chen H, Zhan M, Liu J, Liu Z, Shen M, Yang
F, Kang Y, Yin F and Li Z: Structure-based design, optimization,
and evaluation of potent stabilized peptide inhibitors disrupting
MTDH and SND1 interaction. J Med Chem. 65:12188–12199.
2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
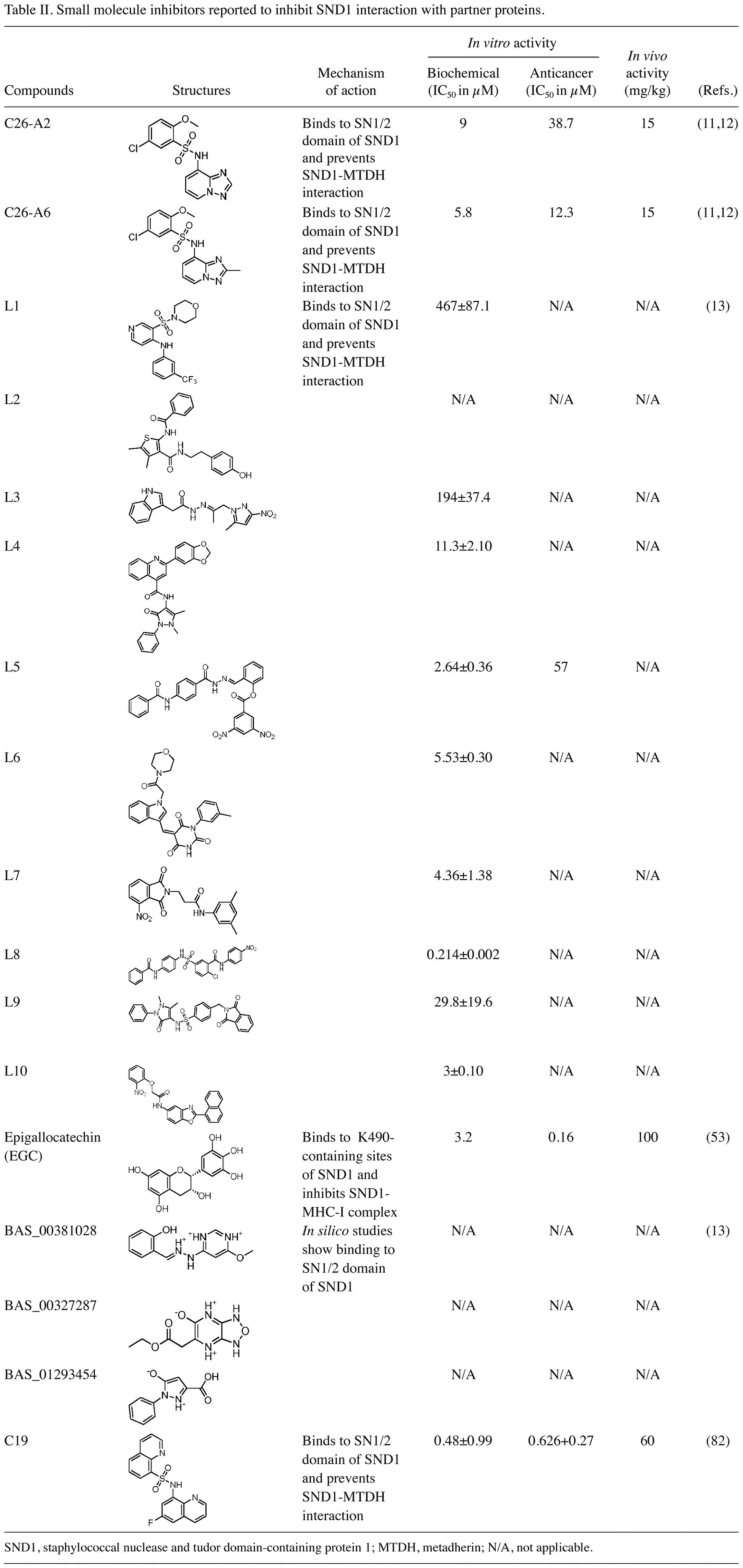
Shen M, Wei Y, Kim H, Wan L, Jiang YZ,
Hang X, Raba M, Remiszewski S, Rowicki M, Wu CG, et al:
Small-molecule inhibitors that disrupt the MTDH-SND1 complex
suppress breast cancer progression and metastasis. Nat Cancer.
3:43–59. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shen M, Smith HA, Wei Y, Jiang YZ, Zhao S,
Wang N, Rowicki M, Tang Y, Hang X, Wu S, et al: Pharmacological
disruption of the MTDH-SND1 complex enhances tumor antigen
presentation and synergizes with anti-PD-1 therapy in metastatic
breast cancer. Nat Cancer. 3:60–74. 2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xu Y, Guo X, Yan D, Dang X, Guo L, Jia T
and Wang Q: Molecular dynamics simulation-driven focused virtual
screening and experimental validation of inhibitors for MTDH-SND1
protein-protein interaction. J Chem Inf Model. 63:3614–3627.
2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Almansour NM: Cheminformatics and
biomolecular dynamics studies towards the discovery of
anti-staphylococcal nuclease domain-containing 1 (SND1) inhibitors
to treat metastatic breast cancer. Saudi Pharm J.
31(101751)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li CL, Yang WZ, Chen YP and Yuan HS:
Structural and functional insights into human Tudor-SN, a key
component linking RNA interference and editing. Nucleic Acids Res.
36:3579–3589. 2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Callebaut I and Mornon JP: The human
EBNA-2 coactivator p100: Multidomain organization and relationship
to the staphylococcal nuclease fold and to the tudor protein
involved in Drosophila melanogaster development. Biochem J.
321:125–132. 1997.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Theobald DL, Mitton-Fry RM and Wuttke DS:
Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys
Biomol Struct. 32:115–133. 2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ying M and Chen D: Tudor domain-containing
proteins of Drosophila melanogaster. Dev Growth Differ. 54:32–43.
2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Leverson JD, Koskinen PJ, Orrico FC,
Rainio EM, Jalkanen KJ, Dash AB, Eisenman RN and Ness SA: Pim-1
kinase and p100 cooperate to enhance c-Myb activity. Mol Cell.
2:417–425. 1998.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liang S, Zhu C, Suo C, Wei H, Yu Y, Gu X,
Chen L, Yuan M, Shen S, Li S, et al: Mitochondrion-localized SND1
promotes mitophagy and liver cancer progression through PGAM5.
Front Oncol. 12(857968)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wright T, Wang Y and Bedford MT: The role
of the PRMT5-SND1 axis in hepatocellular carcinoma. Epigenomes.
5(2)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Quintana AM, Liu F, O'Rourke JP and Ness
SA: Identification and regulation of c-Myb target genes in MCF-7
cells. BMC Cancer. 11(30)2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zeng Q, Liu CH, Wu D, Jiang W, Zhang N and
Tang H: LncRNA and circRNA in patients with non-alcoholic fatty
liver disease: A systematic review. Biomolecules.
13(560)2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yankey A, Oh M, Lee BL, Desai TK and
Somarowthu S: A novel partnership between lncTCF7 and SND1
regulates the expression of the TCF7 gene via recruitment of the
SWI/SNF complex. Sci Rep. 14(19384)2024.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ochoa B, Chico Y and Martínez MJ: Insights
into SND1 oncogene promoter regulation. Front Oncol.
1(606)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hu YZ, Hu ZL, Liao TY, Li Y and Pan YL:
LncRNA SND1-IT1 facilitates TGF-β1-induced
epithelial-to-mesenchymal transition via miR-124/COL4A1 axis in
gastric cancer. Cell Death Discov. 8(73)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev. 15:321–333. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ganesan H, Nandy SK, Banerjee A, Pathak S,
Zhang H and Sun XF: RNA-interference-mediated miR-122-based gene
regulation in colon cancer, a structural in silico analysis. Int J
Mol Sci. 23(15257)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ahmed EA, Rajendran P and Scherthan H: The
microRNA-202 as a diagnostic biomarker and a potential tumor
suppressor. Int J Mol Sci. 23(5870)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang Y, Dong L, Wan F, Chen F, Liu D, Chen
D and Long J: MiR-9-3p regulates the biological functions and drug
resistance of gemcitabine-treated breast cancer cells and affects
tumor growth through targeting MTDH. Cell Death Dis.
12(861)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Banerjee S, Kalyani-Yabalooru SR and
Karunagaran D: Identification of mRNA and non-coding RNA hubs using
network analysis in organ tropism regulated triple negative breast
cancer metastasis. Comput Biol Med. 127(104076)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Levy DE and Lee CK: What does stat3 do? J
Clin Investig. 109:1143–1148. 2002.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tsuchiya N, Ochiai M, Nakashima K, Ubagai
T, Sugimura T and Nakagama H: SND1, a component of RNA-induced
silencing complex, is up-regulated in human colon cancers and
implicated in early stage colon carcinogenesis. Cancer Res.
67:9568–9576. 2007.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bromberg J: Stat proteins and oncogenesis.
J Clin Investig. 109:1139–1142. 2002.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kennell J and Cadigan KM: APC and
beta-catenin degradation. Adv Exp Med Biol. 656:1–12.
2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chidambaranathan-Reghupaty S, Mendoza R,
Fisher PB and Sarkar D: The multifaceted oncogene SND1 in cancer:
Focus on hepatocellular carcinoma. Hepatoma Res.
4(32)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Gao X, Shi X, Fu X, Ge L, Zhang Y, Su C,
Yang X, Silvennoinen O, Yao Z, He J, et al: Human tudor
staphylococcal nuclease (Tudor-SN) protein modulates the kinetics
of AGTR1-3' UTR granule formation. FEBS Lett. 588:2154–2161.
2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wu J, Jiang Y, Zhang Q, Mao X, Wu T, Hao
M, Zhang S, Meng Y, Wan X, Qiu L and Han J: KDM6A-SND1 interaction
maintains genomic stability by protecting the nascent DNA and
contributes to cancer chemoresistance. Nucleic Acids Res.
52:7665–7686. 2024.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhang H, Gao M, Zhao W and Yu L: The
chromatin architectural regulator SND1 mediates metastasis in
triple-negative breast cancer by promoting CDH1 gene methylation.
Breast Cancer Res. 25(129)2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lyko F: The DNA methyltransferase family:
A versatile toolkit for epigenetic regulation. Nat Rev Genetics.
19:81–92. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gao X, Yan F, Lin J, Gao L, Lu XL, Wei SC,
Shen N, Pang JX, Ning QY, Komeno Y, et al: AML1/ETO cooperates with
HIF1α to promote leukemogenesis through DNMT3a transactivation.
Leukemia. 29:1730–1740. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kim G, Kim JY, Lim SC, Lee KY, Kim O and
Choi HS: SUV39H1/DNMT3A-dependent methylation of the RB1 promoter
stimulates PIN1 expression and melanoma development. FASEB J.
32:5647–5660. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yu L, Xu J, Liu J, Zhang H, Sun C, Wang Q,
Shi C, Zhou X, Hua D, Luo W, et al: The novel chromatin
architectural regulator SND1 promotes glioma proliferation and
invasion and predicts the prognosis of patients. Neuro Oncol.
21:742–754. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kim SK and Cho SW: The evasion mechanisms
of cancer immunity and drug intervention in the tumor
microenvironment. Front Pharmacol. 13(868695)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Garrido F, Aptsiauri N, Doorduijn EM, Lora
AM and van Hall T: The urgent need to recover MHC class I in
cancers for effective immunotherapy. Curr Opin Immunol. 39:44–51.
2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Seliger B, Cabrera T, Garrido F and
Ferrone S: HLA class I antigen abnormalities and immune escape by
malignant cells. Semin Cancer Biol. 12:3–13. 2002.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Gabathuler R, Reid G, Kolaitis G, Driscoll
J and Jefferies WA: Comparison of cell lines deficient in antigen
presentation reveals a functional role for TAP-1 alone in antigen
processing. J Exp Med. 180:1415–1425. 1994.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Qin Z, Harders C, Cao X, Huber C,
Blankenstein T and Seliger B: Increased tumorigenicity, but
unchanged immunogenicity of transporter for antigen presentation
1-deficient tumors. Cancer Res. 62:2856–2860. 2002.PubMed/NCBI
|
|
49
|
Blum JS, Wearsch PA and Cresswell P:
Pathways of antigen processing. Annu Rev Immunol. 31:443–473.
2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Leonhardt RM, Keusekotten K, Bekpen C and
Knittler MR: Critical role for the tapasin-docking site of TAP2 in
the functional integrity of the MHC class I-peptide-loading
complex. J Immunol. 175:5104–5114. 2005.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Panter MS, Jain A, Leonhardt RM, Ha T and
Cresswell P: Dynamics of major histocompatibility complex class I
association with the human peptide-loading complex. J Biol Chem.
287:31172–31184. 2012.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sadasivan B, Lehner PJ, Ortmann B, Spies T
and Cresswell P: Roles for calreticulin and a novel glycoprotein,
tapasin, in the interaction of MHC class I molecules with TAP.
Immunity. 5:103–114. 1996.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhang X, Cui X, Li P, Zhao Y, Ren Y, Zhang
H, Zhang S, Li C, Wang X, Shi L, et al: EGC enhances tumor antigen
presentation and CD8+ T cell-mediated antitumor immunity
via targeting oncoprotein SND1. Cancer Lett.
592(216934)2024.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Diao C, Guo P, Yang W, Sun Y, Liao Y, Yan
Y, Zhao A, Cai X, Hao J, Hu S, et al: SPT6 recruits SND1 to
co-activate human telomerase reverse transcriptase to promote colon
cancer progression. Mol Oncol. 15:1180–1202. 2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Rajasekaran D, Jariwala N, Mendoza RG,
Robertson CL, Akiel MA, Dozmorov M, Fisher PB and Sarkar D:
Staphylococcal nuclease and tudor domain containing 1 (SND1
Protein) promotes hepatocarcinogenesis by inhibiting monoglyceride
lipase (MGLL). J Biol Chem. 291:10736–10746. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Santhekadur PK, Akiel M, Emdad L, Gredler
R, Srivastava J, Rajasekaran D, Robertson CL, Mukhopadhyay ND,
Fisher PB and Sarkar D: Staphylococcal nuclease domain containing-1
(SND1) promotes migration and invasion via angiotensin II type 1
receptor (AT1R) and TGFβ signaling. FEBS Open Bio. 4:353–361.
2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Santhekadur PK, Das SK, Gredler R, Chen D,
Srivastava J, Robertson C, Baldwin AS, Fisher PB and Sarkar D:
Multifunction protein staphylococcal nuclease domain containing 1
(SND1) promotes tumor angiogenesis in human hepatocellular
carcinoma through novel pathway that involves nuclear factor κB and
miR-221. J Biol Chem. 287:13952–13958. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Liao SY, Rudoy D, Frank SB, Phan LT,
Klezovitch O, Kwan J, Coleman I, Haffner MC, Li D, Nelson PS, et
al: SND1 binds to ERG and promotes tumor growth in genetic mouse
models of prostate cancer. Nat Commun. 14(7435)2023.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zheng HC: The molecular mechanisms of
chemoresistance in cancers. Oncotarget. 8:59950–59964.
2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Brasseur K, Gévry N and Asselin E:
Chemoresistance and targeted therapies in ovarian and endometrial
cancers. Oncotarget. 8:4008–4042. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Lu C and Shervington A: Chemoresistance in
gliomas. Mol Cell Biochem. 312:71–80. 2008.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Zhao Y, Ren P, Yang Z, Wang L and Hu C:
Inhibition of SND1 overcomes chemoresistance in bladder cancer
cells by promoting ferroptosis. Oncol Rep. 49(16)2023.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Jariwala N, Rajasekaran D, Mendoza RG,
Shen XN, Siddiq A, Akiel MA, Robertson CL, Subler MA, Windle JJ,
Fisher PB, et al: Oncogenic role of SND1 in development and
progression of hepatocellular carcinoma. Cancer Res. 77:3306–3316.
2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Wang Y, Wang Y, Fang Y, Jiang H, Yu L, Hu
H and Zeng S: SND1 regulates organic anion transporter 2 protein
expression and sensitivity of hepatocellular carcinoma cells to
5-fluorouracil. Drug Metab Dispos. 52:997–1008. 2024.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Fu X, Duan Z, Lu X, Zhu Y, Ren Y, Zhang W,
Sun X, Ge L and Yang J: SND1 promotes radioresistance in cervical
cancer cells by targeting the DNA damage response. Cancer Biother
Radiopharm. 39:425–434. 2024.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Carruthers R, Ahmed SU, Strathdee K,
Gomez-Roman N, Amoah-Buahin E, Watts C and Chalmers A: Abrogation
of radioresistance in glioblastoma stem-like cells by inhibition of
ATM kinase. Mol Oncol. 9:192–203. 2015.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y,
Zhang J, Yuan J, Wang M, Chen D, Sun Y, et al: ATM-mediated
stabilization of ZEB1 promotes DNA damage response and
radioresistance through CHK1. Nat Cell Biol. 16:864–875.
2024.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Ammazzalorso F, Pirzio LM, Bignami M,
Franchitto A and Pichierri P: ATR and ATM differently regulate WRN
to prevent DSBs at stalled replication forks and promote
replication fork recovery. EMBO J. 29:3156–3169. 2010.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Zhao Y, Dhani S, Gogvadze V and
Zhivotovsky B: The crosstalk between SND1 and PDCD4 is associated
with chemoresistance of non-small cell lung carcinoma cells. Cell
Death Discov. 11(34)2025.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Yin L, Duan JJ, Bian XW and Yu SC:
Triple-negative breast cancer molecular subtyping and treatment
progress. Breast Cancer Res. 22(61)2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Prat A, Pineda E, Adamo B, Galván P,
Fernández A, Gaba L, Díez M, Viladot M, Arance A and Muñoz M:
Clinical implications of the intrinsic molecular subtypes of breast
cancer. Breast. 24 (Suppl 2):S26–S35. 2015.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Morris GJ, Naidu S, Topham AK, Guiles F,
Xu Y, McCue P, Schwartz GF, Park PK, Rosenberg AL, Brill K and
Mitchell EP: Differences in breast carcinoma characteristics in
newly diagnosed African-American and Caucasian patients: A
single-institution compilation compared with the National Cancer
Institute's Surveillance, epidemiology, and end results database.
Cancer. 110:876–884. 2007.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod S:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Lin NU, Claus E, Sohl J, Razzak AR,
Arnaout A and Winer EP: Sites of distant recurrence and clinical
outcomes in patients with metastatic triple-negative breast cancer:
High incidence of central nervous system metastases. Cancer.
113:2638–2645. 2008.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Gu X, Xue J, Ai L, Sun L, Zhu X, Wang Y
and Liu C: SND1 expression in breast cancer tumors is associated
with poor prognosis. Ann N Y Acad Sci. 1433:53–60. 2018.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Cappellari M, Bielli P, Paronetto MP,
Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O and Sette C:
The transcriptional co-activator SND1 is a novel regulator of
alternative splicing in prostate cancer cells. Oncogene.
33:3794–3802. 2014.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Wang N, Du X, Zang L, Song N, Yang T, Dong
R, Wu T, He X and Lu J: Prognostic impact of Metadherin-SND1
interaction in colon cancer. Mol Biol Rep. 39:10497–10504.
2012.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Hossain MJ, Korde R, Singh S, Mohmmed A,
Dasaradhi PV, Chauhan VS and Malhotra P: Tudor domain proteins in
protozoan parasites and characterization of Plasmodium falciparum
tudor staphylococcal nuclease. Int J Parasitol. 38:513–526.
2008.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Yoo BK, Santhekadur PK, Gredler R, Chen D,
Emdad L, Bhutia S, Pannell L, Fisher PB and Sarkar D: Increased
RNA-induced silencing complex (RISC) activity contributes to
hepatocellular carcinoma. Hepatol. 53:1538–1548. 2011.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Blanco MA, Alečković M, Hua Y, Li T, Wei
Y, Xu Z, Cristea IM and Kang Y: Identification of staphylococcal
nuclease domain-containing 1 (SND1) as a Metadherin-interacting
protein with metastasis-promoting functions. J Biol Chem.
286:19982–19992. 2011.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Pang P, Liu S, Hao X, Tian Y, Gong S, Miao
D and Zhang Y: Exploring binding modes of the selected inhibitors
to SND1 by all-atom molecular dynamics simulations. J Biomol Struct
Dyn. 42:5536–5550. 2024.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Shen H, Ding J, Ji J, Hu L, Min W, Hou Y,
Wang D, Chen Y, Wang L, Zhu Y, et al: Discovery of novel
small-molecule inhibitors disrupting the MTDH-SND1 protein-protein
interaction. J Med Chem. 68:1844–1862. 2025.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Li P, He Y, Chen T, Choy KY, Chow TS, Wong
ILK, Yang X, Sun W, Su X, Chan TH and Chow LMC: Disruption of
SND1-MTDH interaction by a high affinity peptide results in SND1
degradation and cytotoxicity to breast cancer cells in vitro and in
vivo. Mol Cancer Ther. 20:76–84. 2021.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Chen H, Zhan M, Zhang Y, Liu J, Wang R, An
Y, Gao Z, Jiang L, Xing Y, Kang Y, et al: Intracellular delivery of
stabilized peptide blocking MTDH-SND1 interaction for breast cancer
suppression. JACS Au. 4:139–149. 2023.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Navarro-Imaz H, Ochoa B, García-Arcos I,
Martínez MJ, Chico Y, Fresnedo O and Rueda Y: Molecular and
cellular insights into the role of SND1 in lipid metabolism.
Biochim Biophys Acta Mol Cell Biol Lipids.
1865(158589)2020.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Shen H, Ding J, Ji J, Jiang B, Wang X and
Yang P: Overcoming MTDH and MTDH-SND1 complex: Driver and potential
therapeutic target of cancer. Cancer Insight. 3:55–82. 2023.
|
|
87
|
Duo L, Liu Y, Ren J, Tang B and Hirst JD:
Artificial intelligence for small molecule anticancer drug
discovery. Expert Opin Drug Discov. 19:933–948. 2024.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Adon T, Shanmugarajan D, Ather H, Ansari
SMA, Hani U, Madhunapantula SV and Honnavalli YK: Virtual screening
for identification of dual inhibitors against CDK4/6 and aromatase
enzyme. Molecules. 28(2490)2023.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Zhu J, Li K, Xu L, Cai Y, Chen Y, Zhao X,
Li H, Huang G and Jin J: Discovery of novel selective PI3Kγ
inhibitors through combining machine learning-based virtual
screening with multiple protein structures and bio-evaluation. J
Adv Res. 36:1–13. 2021.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Wang Y and Zhang P: Prediction of histone
deacetylase inhibition by triazole compounds based on artificial
intelligence. Front Pharmacol. 14(1260349)2023.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Nayarisseri A, Abdalla M, Joshi I, Yadav
M, Bhrdwaj A, Chopra I, Khan A, Saxena A, Sharma K, Panicker A, et
al: Potential inhibitors of VEGFR1, VEGFR2, and VEGFR3 developed
through deep learning for the treatment of cervical cancer. Sci
Rep. 14(13251)2024.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Di Stefano M, Galati S, Ortore G,
Caligiuri I, Rizzolio F, Ceni C, Bertini S, Bononi G, Granchi C,
Macchia M, et al: Machine learning-based virtual screening for the
identification of Cdk5 inhibitors. Int J Mol Sci.
23(10653)2022.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Liu Z, Hu M, Yang Y, Du C, Zhou H, Liu C,
Chen Y, Fan L, Ma H, Gong Y and Xie Y: An overview of PROTACs: A
promising drug discovery paradigm. Mol Biomed. 3(46)2022.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Han X and Sun Y: Strategies for the
discovery of oral PROTAC degraders aimed at cancer therapy. Cell
Rep Phys Sci. 3(101062)2022.
|