Introduction
Colon cancer remains a leading cause of
cancer-related mortality worldwide, accounting for nearly 10% of
all cancer deaths (1). While
localized tumors exhibit favorable outcomes with >90% 5-year
survival rates, metastatic disease continues to portend a dismal
prognosis, with median survival under 30 months despite advances in
systemic therapy (2). This stark
contrast underscores the critical need for robust prognostic
biomarkers and novel therapeutic strategies to improve patient
stratification and treatment outcomes.
Recent breakthroughs in tumor immunology have
reshaped our understanding of colon cancer progression. The tumor
immune microenvironment, specifically the infiltration density and
spatial organization of CD8+ cytotoxic T lymphocytes and
CD45RO+ memory T cells, represents an independent
prognostic factor beyond conventional TNM staging (3-5).
Conversely, FOXP3+ regulatory T cells (Tregs) contribute
to immunosuppression and correlate with tumor recurrence (6,7).
However, despite these advances, current immune score systems
remain incomplete, failing to incorporate emerging players such as
CEACAM6, a glycoprotein increasingly implicated in immune evasion
and metastasis (8-10).
Simultaneously, CEACAM6 (carcinoembryonic
antigen-related cell adhesion molecule 6) has emerged as a key
player in colon cancer progression (11,12).
Originally identified as a marker of poor differentiation, CEACAM6
is now recognized to promote metastasis through multiple mechanisms
including resistance to anoikis, enhancement of
epithelial-mesenchymal transition and modulation of tumor-stroma
interactions (13,14). Most recently, CEACAM6 has been
implicated in immune evasion by upregulating PD-L1 expression and
recruiting myeloid-derived suppressor cells (MDSCs) (15). These findings suggest that CEACAM6
may serve as both a prognostic marker and a potential therapeutic
target in colon cancer.
A multicenter, dual-platform investigation was
conducted, across three tertiary hospitals in China, analyzing 301
patients with colon cancer. Notably, the present study represents a
significant expansion beyond our previous single-center pilot
investigation (16), which
analyzed 120 patients from one institution. The current multicenter
design, larger sample size (301 vs. 120), incorporation of
long-term overall survival (OS) data (follow-up until 2025), and
multivariate Cox regression analysis which established CEACAM6 and
FOXP3 as independent prognostic factors, collectively provide a
higher level of evidence and move beyond initial correlation to
establish prognostic value. Furthermore, the samples used herein
are entirely new and non-overlapping with the previous publication,
having been collected between 2015 and 2020 from three distinct
medical centers. The present study investigated the associations of
tumor-infiltrating lymphocytes (TILs), FOXP3+ regulatory
T cells, and CEACAM6 expression with clinicopathological
characteristics and prognosis in colon cancer, while exploring
their roles in tumor progression and providing a theoretical
foundation for immunotherapy development.
Materials and methods
Patient specimens
A total of 301 paraffin-embedded colon cancer tissue
samples were collected from July 2015 to June 2020. The sample
inclusion period was limited to June 2020 to ensure a minimum of 5
years of follow-up data for OS analysis, with the final follow-up
date being June 30, 2025. This approach guaranteed the availability
of complete and robust survival data for meaningful Kaplan-Meier
and Cox regression analyses. The specimens for the present study
were obtained through a collaborative effort among three
independent tertiary hospitals. A total of 161 cases were collected
from the Affiliated Hospital of Jiangnan University (Wuxi, China),
while Jiangsu Provincial Veterans Hospital (Wuxi, China) and the
First Affiliated Hospital of Soochow University (Suzhou, China)
contributed 30 and 110 cases, respectively. It should be noted that
these institutions share no direct administrative affiliation;
their collaboration was solely facilitated by academic connections
within our research team. To ensure consistency across all samples,
each hospital collected specimens from their own patient archives
using identical predefined inclusion and exclusion criteria. Prior
to specimen acquisition, written informed consent was obtained from
all participants for the use of their tissues in scientific
research. Patients with prior exposure to radiotherapy,
chemotherapy, or immunotherapeutic interventions were excluded from
the study cohort. Histopathological examination confirmed the
diagnosis of colon adenocarcinoma in all cases, with tumor staging
(TNM classification) and histological grading performed in strict
accordance with the Union for International Cancer Control
guidelines (8th edition) (17,18).
The present retrospective multicenter study was conducted with
approval from the research Ethics Committee of all participating
hospitals in accordance with the Declaration of Helsinki.
IHC staining for CD3, CD4, CD8,
CD45RO, CEACAM6 and FOXP3
Sections (4-µΜ) were prepared from formalin-fixed,
paraffin-embedded (FFPE) tissue blocks and subjected to standard
deparaffinization through three xylene washes (5 min each) followed
by graded ethanol rehydration (100, 95, 75 and 50%; 5 min each).
Antigen retrieval was performed by heat-induced epitope retrieval
in 10 mmol/l sodium citrate buffer (pH 6.0) at 100˚C for 15 min,
after which endogenous peroxidase activity was blocked with 3%
H2O2 in methanol (10 min, 25˚C). Non-specific
binding was minimized by 10 min blocking with 10% normal horse
serum (Wuhan Boster Biological Technology) at room temperature,
followed by 2 h incubation in a humidified dark chamber with the
following antihuman antibodies: Monoclonal mouse IgG against CD3
(cat. no. sc-20047), polyclonal rabbit IgG against CD4 (cat. no.
sc-7219) and CD8 (cat. no. sc7188), (all from Santa Cruz
Biotechnology, Inc.; 1:100); monoclonal mouse IgG2a against CD45RO
(cat. no. ab86080; Abcam; 1:10), monoclonal rabbit IgG against
CEACAM6 (cat. no. ab134074; Abcam; 1:400) and monoclonal mouse IgG3
against FOXP3 (cat. no. ab450; Abcam; 1:50). After PBS washing,
sections were incubated with biotinylated polyclonal goat
antimouse/rabbit IgG secondary antibodies (cat. no. K5007; Dako;
Agilent Technologies, Inc.; 1:2,000) for 1 h under identical
conditions, then developed with 3,3'-diaminobenzidine
tetrahydrochloride hydrate and counterstained with hematoxylin (5
min) at room temperature. Five random high-power fields per section
were analyzed by light microscopy (BX53; Olympus Corporation),
excluding necrotic areas, with human tonsillar FFPE sections as
positive controls (obtained with donor consent) and PBS
substitution as negative controls, all procedures being performed
in accordance with institutional biosafety protocols and
manufacturer specifications.
Scoring system for IHC
IHC expression of CD3, CD4, CD8, CD45RO, CEACAM6 and
FOXP3 was evaluated using a validated two-tiered scoring system
incorporating both staining intensity and cellular distribution
(19). All slides underwent
blinded evaluation by two independent pathologists. The staining
intensity was scored as 0 (achromatic), 1 (light yellow), 2
(brownish yellow) or 3 (brown). In addition, the percentage of
positive cells was scored as 0 (<5%), 1 (5-25%), 2 (26-50%), 3
(51-75%), or 4 (>75%). The two scores were added together, and
the samples were assigned to one of four levels as follows: (-),
score 0-1; (+), score 2; (++), score 3-4; or (+++), score ≥5. ()
and (+) were defined as negative expression, (++) as weak
expression and (+++) as strong expression. Discrepant scores
between the two pathologists were resolved through joint
re-evaluation until a consensus was reached.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from FFPE tissue sections
using the RNeasy FFPE Kit (cat. no. 73504; Qiagen, Inc.). Briefly,
freshly cut sections were deparaffinized using the manufacturer's
proprietary solution, followed by tissue lysis in an optimized
buffer to release nucleic acids. To reverse formalin-induced
cross-linking, samples were heated at 80˚C, then treated with DNase
to eliminate genomic DNA contamination. Lysates were mixed with
Buffer RBC, and ethanol was added to facilitate RNA binding to
RNeasy MinElute spin columns. Purified RNA was eluted in a minimum
of 14 µl of RNase-free water. First-strand cDNA was synthesized
from total RNA using the RevertAid™ First Strand cDNA Synthesis kit
(cat. no. K1622; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. qPCR was performed with FastStart
Universal SYBR Green Master (Rox) kit (cat. no. 4913914001;
Sigma-Aldrich; Merck KGaA) under the following conditions: Initial
denaturation at 95˚C for 1 min; 45 cycles of 95˚C for 20 sec, 58˚C
for 30 sec, and 68˚C for 45 sec. β-actin served as the endogenous
control. Relative gene expression was calculated via the
2-ΔΔCq method using Cq values normalized to β-actin
(20). All primers were
synthesized by Sangon Biotech Co., Ltd., as shown in Table I.
 | Table IPrimer pairs used for reverse
transcription-quantitative PCR. |
Table I
Primer pairs used for reverse
transcription-quantitative PCR.
| Gene name (GenBank
no.) | Sequence | Product size,
bp |
|---|
| CD3
(NM_000732.4) | F:
5'-GGGAGTCTTCTGCTTTGCTG-3' | 153 |
| | R:
5'-TTGTTCCGAGCCCAGTTTC-3' | |
| CD4
(NM_000616.4) | F:
5'-GTGAACCTGGTGGTGATG-3' | 122 |
| | R:
5'-GAGACCTTTGCCTCCTTG-3' | |
| CD8
(NM_001768.7) | F:
5'-ATGGCCTTACCAGTGACCG-3' | 104 |
| | R:
5'-AGGTTCCAGGTCCGATCCAG-3' | |
| CD45RO
(NM_002838) | F:
5'-TCTGCTGGAACTGACACG-3' | 168 |
| | R:
5'-CTCATTAACATTTAGCTTTG-3' | |
| CEACAM6
(BC005008.1) | F:
5'-TCCAGCAATCCACACAAGAG-3' | 144 |
| | R:
5'-GGACAGGAGCACTTCCAGAG-3' | |
| FOXP3
(NM_014009.3) | F:
5'-TCCCAGAGTTCCTCCACAAC-3' | 122 |
| | R:
5'-ATTGAGTGTCCGCTGCTTCT-3' | |
| β-actin
(NM_001101.3) | F:
5'-CACTGTGCCCATCTACGAGG-3' | 154 |
| | R:
5'-AATGTCACGCACGATTTCC-3' | |
Statistical analysis
All statistical analyses were carried out using SPSS
(v26; IBM Corporation). Normality of continuous variables was
assessed using Shapiro-Wilk tests. Normally distributed continuous
variables are presented as the mean ± standard deviation. For
comparisons of mRNA expression levels between groups, the
independent samples t-test was used after confirming normality
(Shapiro-Wilk test, P>0.05) and homogeneity of variances
(Levene's test, P>0.05). The χ2 test was employed to
assess the association between expression levels and patient
characteristics. Survival was analyzed by Kaplan-Meier curves, and
differences were assessed using the logrank test. Variables were
selected by univariate and multivariate Cox regression analyses,
with forest plots visualized using R (version 4.5.1). P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
From July 2015 to June 2020, a total of 301 patients
with colon cancer were enrolled. The study population comprised 141
patients (46.8%) with early-stage disease (stage I-II) and 160
patients (53.2%) with advanced-stage disease (stage III-IV). Sex
distribution showed a male predominance (n=174, 57.8%) compared
with female patients (n=127, 42.2%). Tumor laterality was
distributed with 159 cases (52.8%) presenting as left-sided tumors
and 142 cases (47.2%) as right-sided tumors. Histopathological
examination revealed poorly differentiated tumors in 96 cases
(31.9%), while the majority exhibited well-to-moderately
differentiated features (n=205, 68.1%). The mean age at the time of
surgery was 64.94±10.63 years (range: 32-93 years).
Association between CD3, CD4, CD8,
CD45RO, CEACAM6 and FOXP3 expression and clinicopathological
data
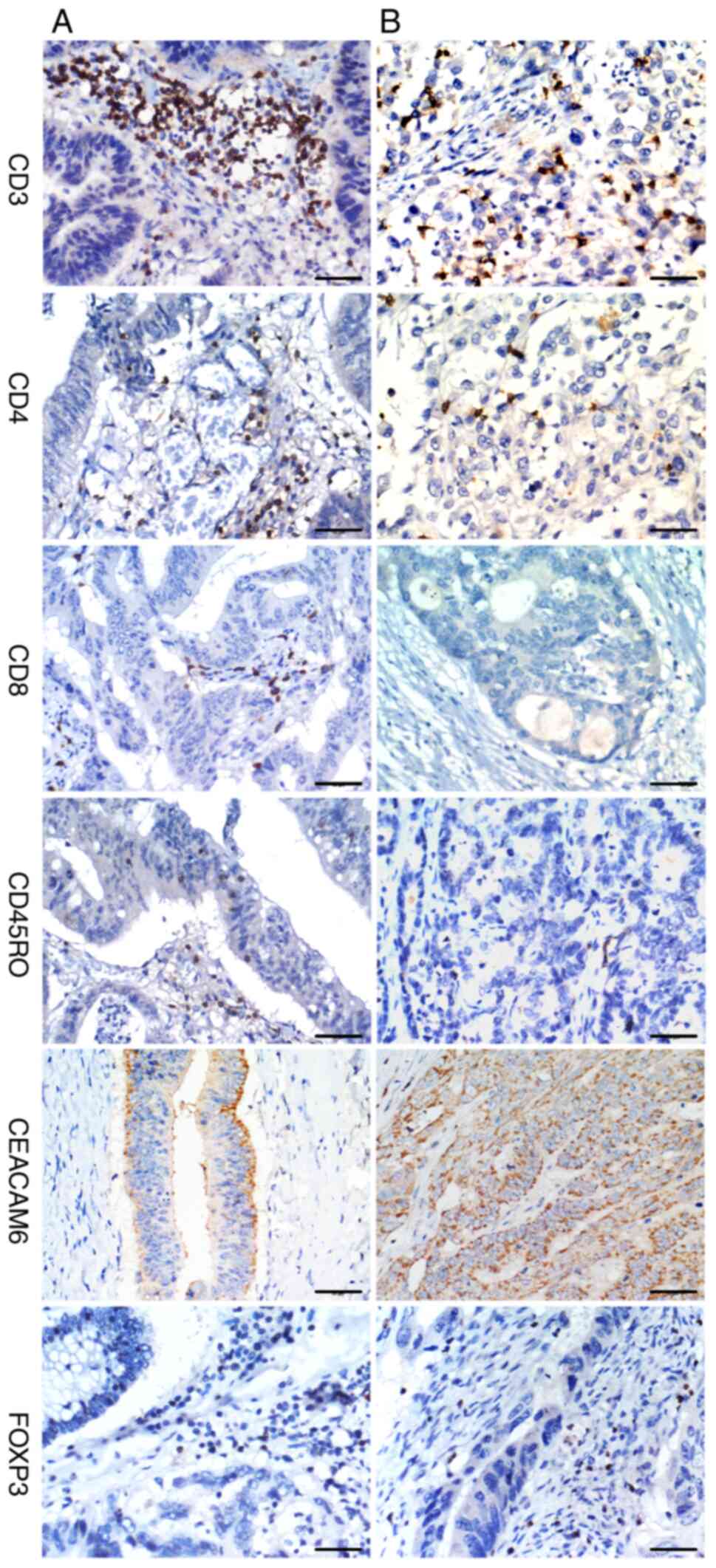
IHC analysis of colon cancer specimens revealed
distinct spatial distribution patterns of TILs, with infiltration
densities significantly associated with tumor stage and
differentiation (Table II).
Early-stage tumors (I-II) exhibited significantly higher
infiltration of CD3+ (P<0.001), CD8+
(P=0.001), and CD45RO+ T-cells (P<0.001) compared
with advanced-stage (III-IV) tumors, whereas CD4+ T-cell
infiltration showed an inverse correlation with tumor progression
(P=0.014). Similarly, well-to-moderately differentiated tumors
displayed significantly greater infiltration of CD3+
(P<0.001) and CD45RO+ T-cells (P<0.001) than
poorly differentiated tumors (Fig.
1), while CD4+ T-cell density was reduced in
higher-grade tumors (P=0.020). By contrast, no significant
associations were found between TIL subsets and patient age (all
P>0.05) or tumor location (P>0.05). Notably, sex differences
were observed only for CD45RO+ T-cells (P=0.017), with
female patients exhibiting higher infiltration levels than
males.
 | Table IIRelationship between
clinicopathological parameters and CD3+,
CD4+, CD8+ and CD45RO+ T-cell
infiltrations in patients with colon cancer. |
Table II
Relationship between
clinicopathological parameters and CD3+,
CD4+, CD8+ and CD45RO+ T-cell
infiltrations in patients with colon cancer.
| | CD3+ T
cell | | CD4+ T
cell | | CD8+ T
cell | | CD45RO+
T cell | |
|---|
| Characteristic | n | Strong | Negative/weak | χ2 | P-value | Strong | Negative/weak | χ2 | P-value | Strong | Negative/weak | χ2 | P-value | Strong | Negative/weak | χ2 | P-value |
|---|
| Age, years | | | | 0.440 | 0.507 | | | 0.17 | 0.681 | | | 2.025 | 0.155 | | | 0.031 | 0.860 |
|
≤60 | 93 | 57 | 36 | | | 29 | 64 | | | 22 | 71 | | | 29 | 64 | | |
|
>60 | 208 | 119 | 89 | | | 60 | 148 | | | 66 | 142 | | | 67 | 141 | | |
| Sex | | | | 2.549 | 0.110 | | | 0.01 | 0.909 | | | 2.269 | 0.132 | | | 5.653 | 0.017 |
|
Male | 174 | 95 | 79 | | | 51 | 123 | | | 45 | 129 | | | 46 | 128 | | |
|
Female | 127 | 81 | 46 | | | 38 | 89 | | | 43 | 84 | | | 50 | 77 | | |
| Tumor site | | | | 1.957 | 0.162 | | | 1.03 | 0.310 | | | 0.017 | 0.896 | | | 2.427 | 0.119 |
|
Left
colon | 159 | 87 | 72 | | | 43 | 116 | | | 47 | 112 | | | 57 | 102 | | |
|
Right
colon | 142 | 89 | 53 | | | 46 | 96 | | | 41 | 101 | | | 39 | 103 | | |
|
Differentiation | | | | 14.42 | <0.001 | | | 5.45 | 0.020 | | | 3.692 | 0.055 | | | 27.099 | <0.001 |
|
Well/moderately | 205 | 135 | 70 | | | 52 | 153 | | | 67 | 138 | | | 85 | 120 | | |
|
Poorly | 96 | 41 | 55 | | | 37 | 59 | | | 21 | 75 | | | 11 | 85 | | |
| Stage | | | | 16.93 | <0.001 | | | 6.02 | 0.014 | | | 10.529 | 0.001 | | | 27.165 | <0.001 |
|
I and
II | 141 | 100 | 41 | | | 32 | 109 | | | 54 | 87 | | | 66 | 75 | | |
|
III and
IV | 160 | 76 | 84 | | | 57 | 103 | | | 34 | 126 | | | 30 | 130 | | |
IHC staining demonstrated strong FOXP3 and CEACAM6
expression in 65.4% (119/182) and 61.8% (115/186) of cases,
respectively (Table III). Both
FOXP3 and CEACAM6 showed no significant associations with patient
age, sex, or tumor location (P>0.05). Notably, elevated
expression of both FOXP3 and CEACAM6 was observed in advanced-stage
(III-IV) tumors (P<0.001) and poorly differentiated colon cancer
(P<0.001) (Fig. 1). These
results suggested that FOXP3-positive regulatory T cells and
CEACAM6 may contribute to tumor progression and aggressive
biological behavior in colon cancer.
 | Table IIIRelationship between
clinicopathological parameters and CEACAM6 and FOXP3 expression
levels in patients with colon cancer. |
Table III
Relationship between
clinicopathological parameters and CEACAM6 and FOXP3 expression
levels in patients with colon cancer.
| | CEACAM6 | | FOXP3 | |
|---|
| Characteristic | n | Strong | Negative/weak | χ2 | P-value | Strong | Negative/weak | χ2 | P-value |
|---|
| Age, years | | | | 0.019 | 0.891 | | | 1.479 | 0.224 |
|
≤60 | 93 | 35 | 58 | | | 32 | 61 | | |
|
>60 | 208 | 80 | 128 | | | 87 | 121 | | |
| Sex | | | | 0.716 | 0.398 | | | 0.819 | 0.366 |
|
Male | 174 | 70 | 104 | | | 65 | 109 | | |
|
Female | 127 | 45 | 82 | | | 54 | 73 | | |
| Tumor site | | | | 0.172 | 0.678 | | | 0.831 | 0.362 |
|
Left
colon | 159 | 59 | 100 | | | 59 | 100 | | |
|
Right
colon | 142 | 56 | 86 | | | 60 | 82 | | |
|
Differentiation | | | | 41.540 | <0.001 | | | 40.138 | <0.001 |
|
Well/moderately | 205 | 53 | 152 | | | 56 | 149 | | |
|
Poorly | 96 | 62 | 34 | | | 63 | 33 | | |
| Stage | | | | 81.049 | <0.001 | | | 71.310 | <0.001 |
|
I and
II | 141 | 16 | 125 | | | 20 | 121 | | |
|
III and
IV | 160 | 99 | 61 | | | 99 | 61 | | |
Relationship between CEACAM6
expression and T cell infiltration markers
Significant associations were observed between
CEACAM6 expression and T cell infiltration markers (CD3, CD4, CD8,
CD45RO and FOXP3) in patients with colon cancer (P<0.05)
(Table IV). These findings
suggested that CEACAM6 expression is significantly correlated with
altered T cell infiltration patterns in colon cancer, particularly
showing a strong inverse association with CD3, CD8 and CD45RO but a
positive association with FOXP3.
 | Table IVRelationship between CEACAM6 and CD3,
CD4, CD8, CD45RO and FOXP3 expression levels in patients with colon
cancer. |
Table IV
Relationship between CEACAM6 and CD3,
CD4, CD8, CD45RO and FOXP3 expression levels in patients with colon
cancer.
| | CEACAM6 | |
|---|
| T cell
infiltration | n | Strong | Negative/weak | χ2 | P-value |
|---|
| CD3 | | | | 28.670 | <0.001 |
|
Strong | 176 | 45 | 131 | | |
|
Negative/weak | 125 | 70 | 55 | | |
| CD4 | | | | 11.414 | 0.001 |
|
Strong | 89 | 47 | 42 | | |
|
Negative/weak | 212 | 68 | 144 | | |
| CD8 | | | | 16.598 | <0.001 |
|
Strong | 88 | 18 | 70 | | |
|
Negative/weak | 213 | 97 | 116 | | |
| CD45RO | | | | 53.279 | <0.001 |
|
Strong | 96 | 8 | 88 | | |
|
Negative/weak | 205 | 107 | 98 | | |
| FOXP3 | | | | 168.711 | <0.001 |
|
Strong | 119 | 99 | 20 | | |
|
Negative/weak | 182 | 16 | 166 | | |
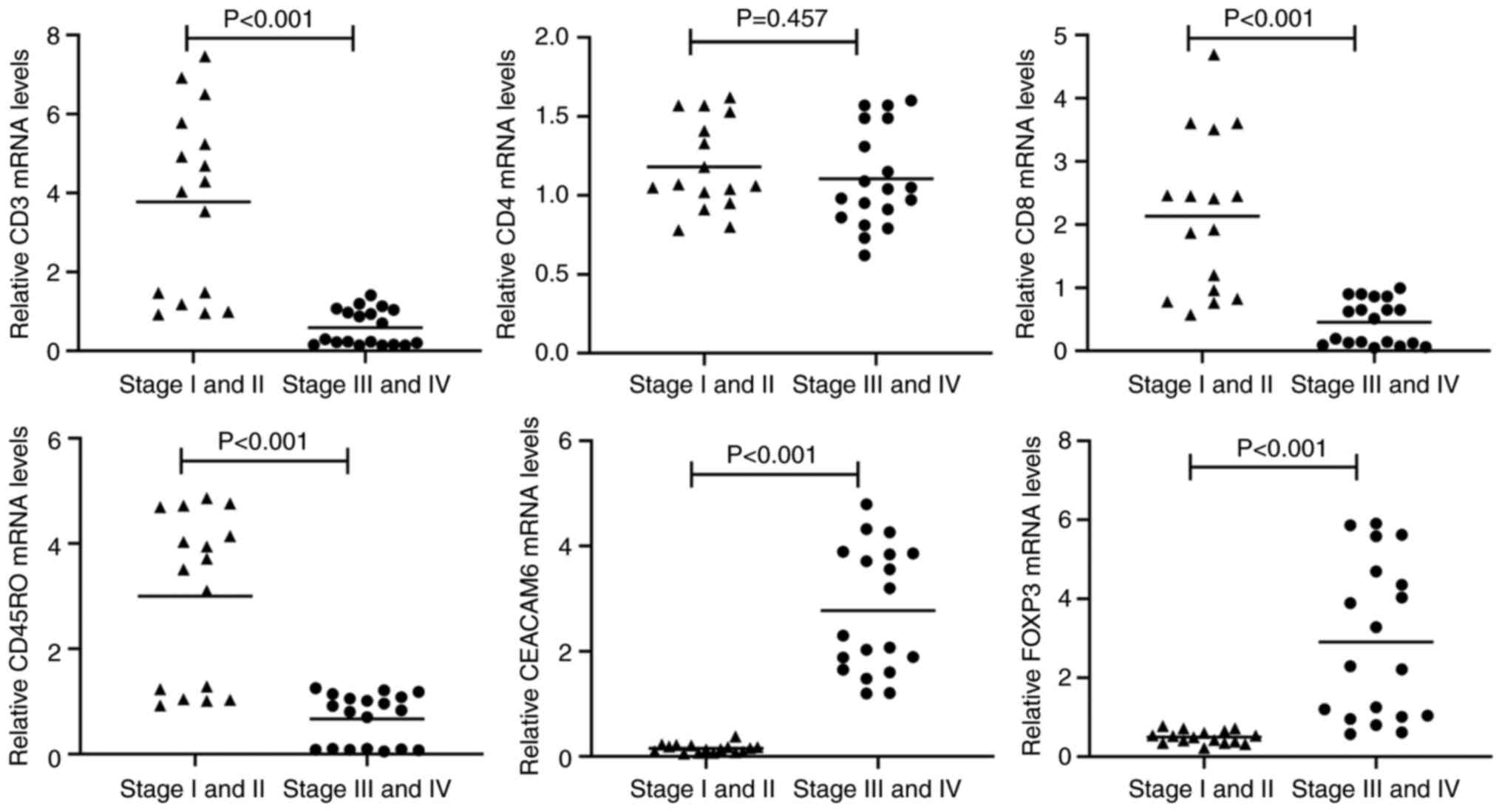
mRNA expression levels of CD3, CD4,
CD8, CD45RO, CEACAM6 and FOXP3 in stage I-II vs. stage III-IV colon
cancer
A total of 50 tumor blocks were randomly selected
from 301 paraffin-embedded specimens for mRNA extraction, with
successful RNA isolation achieved in 35 cases (16 stage I-II and 19
stage III-IV samples). Consistent with IHC findings, transcript
levels of CD3, CD8 and CD45RO were significantly downregulated in
advanced-stage (III-IV) tumors compared with early-stage (I-II)
lesions (P<0.001; Fig. 2). By
contrast, CD4 mRNA expression showed no significant intergroup
difference (P=0.457; Fig. 3). Both
CEACAM6 and FOXP3 mRNA levels were significantly elevated in
advanced-stage tumors (III-IV) relative to early-stage disease
(P<0.001; Fig. 2).
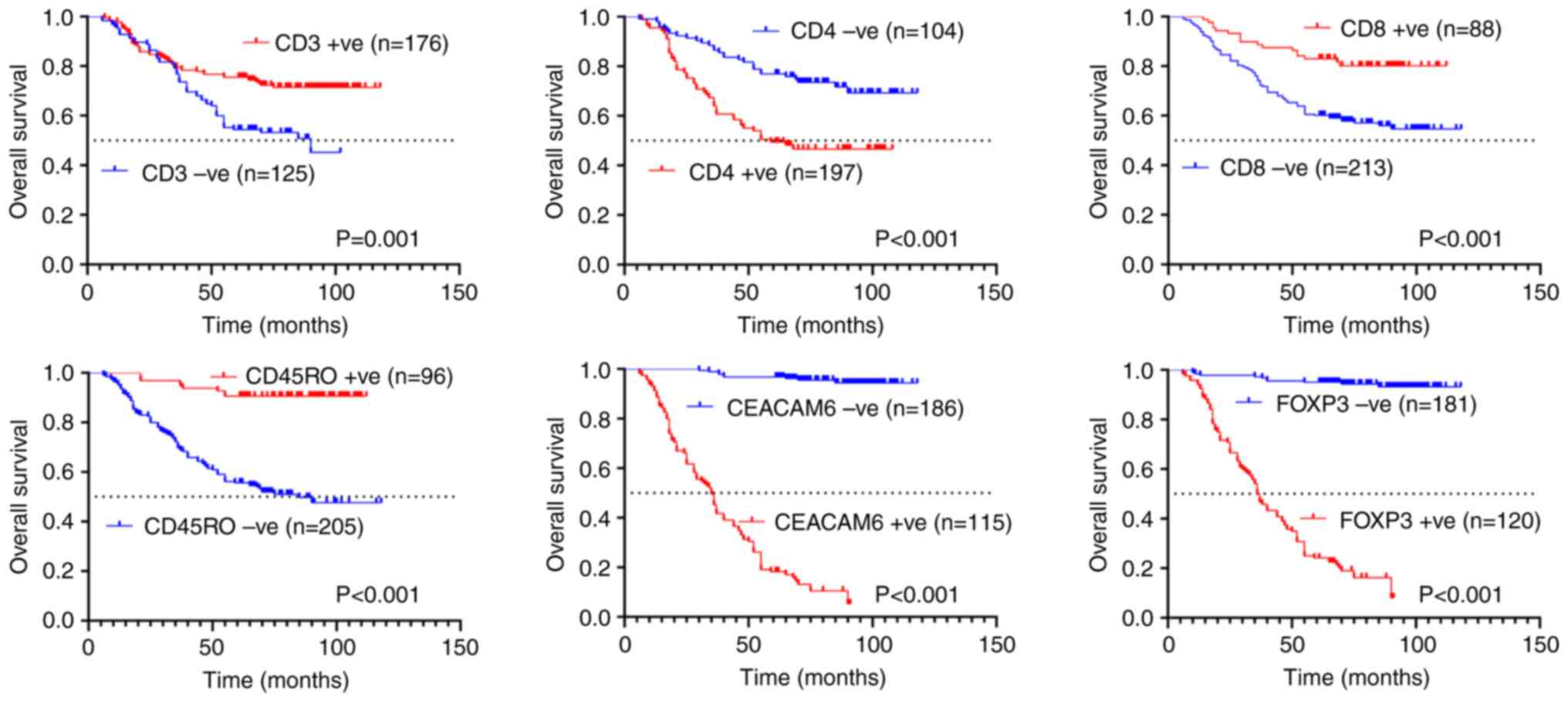
Association between CD3, CD4, CD8,
CD45RO, CEACAM6 and FOXP3 expression and the prognosis of patients
with colon cancer
In the present study, patient follow-up was
conducted until June 30, 2025, with OS as the primary endpoint
(maximum follow-up duration: 118 months). Kaplan-Meier analysis
demonstrated that patients with weakly positive or negative
expression of CD3 (median OS: 90 months) and CD45RO (median OS: 85
months) exhibited significantly shorter survival compared with
those with strongly positive expression (P=0.001 and P<0.001;
Fig. 3). By contrast, strong
positivity for CD4 (median OS: 59 months), CEACAM6 (median OS: 35
months), and FOXP3 (median OS: 36.5 months) was associated with
poorer outcomes relative to weak/negative expression (P<0.001;
Fig. 3).
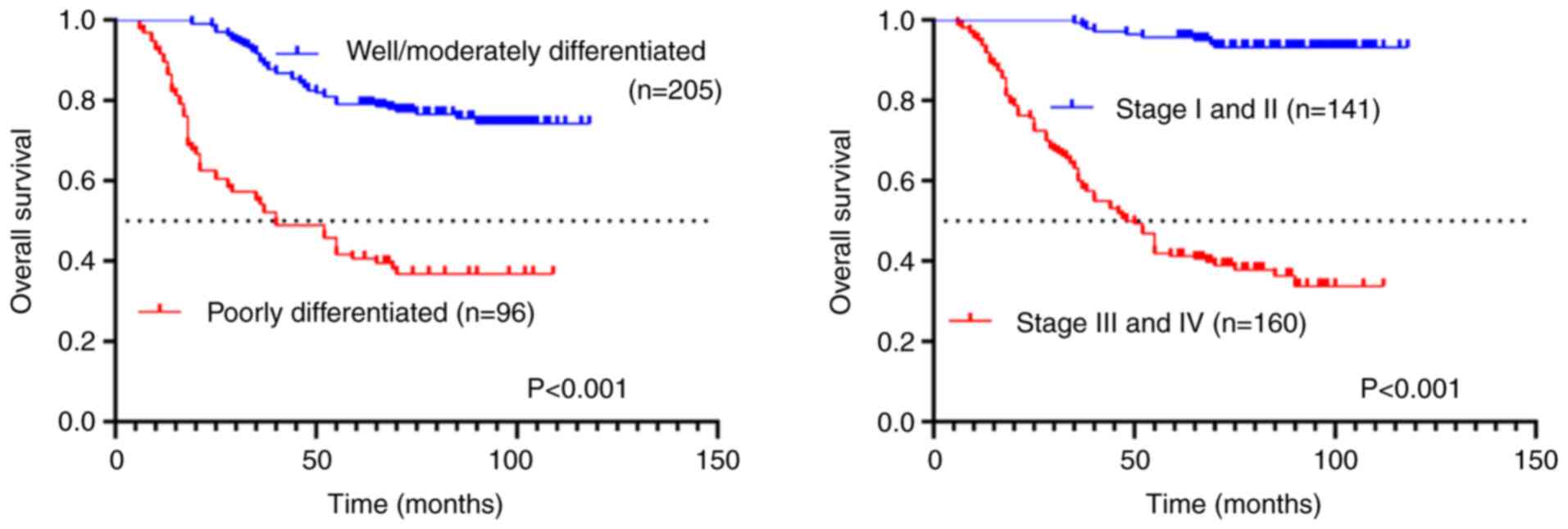
Association of survival outcomes with
tumor histological grade and TNM staging classification
Kaplan-Meier survival analysis revealed significant
prognostic differences based on tumor differentiation and TNM
staging. Patients with poorly differentiated tumors demonstrated
significantly worse outcomes, with a median OS of 40 months
compared with those with well-to-moderately differentiated tumors
(P<0.001; Fig. 4). Similarly,
advanced-stage (III-IV) cases showed significantly reduced survival
(median OS: 49 months) relative to early-stage (I-II) patients
(P<0.001; Fig. 4). These
findings underscore the strong association between pathological
grade, clinical stage and survival outcomes in this cohort.
Multivariable Cox regression analysis
of prognostic factors in patients with colon cancer
Univariate Cox regression analysis identified
multiple significant prognostic factors for colon cancer outcomes
(Table V). The statistically
significant variables from univariate Cox analysis were then
included in multivariate Cox regression modeling. This analysis
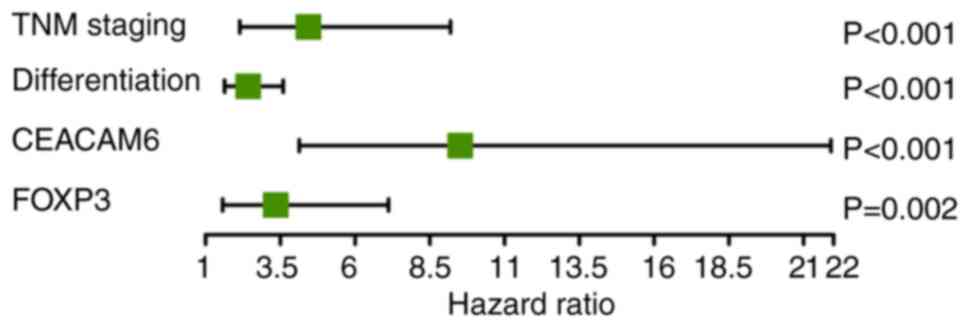
identified four independent prognostic factors (Fig. 5): TNM staging [hazard ratio
(HR)=4.437; 95% confidence interval (CI): 2.142-9.189; P<0.001),
tumor differentiation (HR=2.425; 95% CI: 1.635-3.600; P<0.001),
CEACAM6 expression (HR=9.516; 95% CI: 4.133-21.914; P<0.001) and
FOXP3 levels (HR=3.345; 95% CI: 1.572-7.118; P=0.002).
 | Table VUnivariate analysis of prognostic
factors in patients with colon cancer. |
Table V
Univariate analysis of prognostic
factors in patients with colon cancer.
|
Characteristics | HR (95% CI) | P-value |
|---|
| TNM Staging | 15.530
(7.833-30.790) | P<0.001 |
|
Differentiation | 4.092
(2.800-5.981) | P<0.001 |
| CD3 | 0.521
(0.356-0.761) | P=0.001 |
| CD4 | 2.236
(1.529-3.271) | P<0.001 |
| CD8 | 0.377
(0.225-0.633) | P<0.001 |
| CD45RO | 0.143
(0.072-0.282) | P<0.001 |
| CEACAM6 | 45.560
(22.580-91.930) | P<0.001 |
| FOXP3 | 30.030
(15.750-57.280) | P<0.001 |
Discussion
The antitumor immune response plays a pivotal role
in colon cancer progression, with T cell infiltration serving as a
key determinant of clinical outcomes (21). Colon tumors are generally
immunogenic and frequently exhibit infiltration by T lymphocytes
(22). Multiple studies have shown
that reduced T cell infiltration in colon tumors is inversely
associated with disease stage and predicts improved OS (23,24).
Our findings in poorly differentiated tumors and advanced
pathological stages further support this observation. Specifically,
it was found that increased infiltration of CD3+,
CD8+ and CD45RO+ T cells was associated with
improved prognosis, whereas CD4+ T cells showed
paradoxical associations. However, the present data highlight
limitations of current immune score systems by demonstrating that
CEACAM6 and FOXP3 refine prognostic stratification beyond TIL
density. For example, patients with tumors exhibiting high CEACAM6
or FOXP3 expression exhibited worse outcomes regardless of
CD8+ T-cell levels, suggesting that biomarker panels
integrating immune checkpoints and stromal factors may improve risk
prediction (16).
FOXP3 is a highly specific marker for Tregs
(25). Tumors frequently exhibit
increased Treg infiltration, which contributes to immunosuppression
by dampening anti-tumor immune responses (26). Multiple studies have demonstrated
that elevated CD4+CD25+FOXP3+ Treg
infiltration correlates with poor prognosis in various malignancies
(27,28). In the present study,
transcriptional analysis revealed significant downregulation of
CD3, CD8 and CD45RO mRNA levels in advanced-stage tumors,
contrasting with the upregulation of CEACAM6 and FOXP3 expression.
This differential expression pattern further supports their
respective roles in tumor suppression and disease progression.
Importantly, multivariate analysis identified CEACAM6 and FOXP3 as
independent prognostic factors, underscoring their potential for
refining risk stratification in clinical practice. These findings
align with recent studies demonstrating that FOXP3+ Treg
enrichment in advanced-stage tumors highlights their therapeutic
potential as targets for novel immunotherapies designed to disrupt
immunosuppressive networks (29,30).
The strong association between CEACAM6
overexpression and advanced tumor stage, poor differentiation, and
reduced survival aligns with its documented roles in metastasis and
therapy resistance (12). Our
findings build upon previous observations by revealing an inverse
correlation between CEACAM6 expression and tumor-infiltrating
CD3+, CD8+ and CD45RO+
lymphocytes, suggesting its potential role in mediating immune
exclusion - a key immunological feature associated with tumor
aggressiveness. Mechanistically, CEACAM6 has been shown to
contribute to an immunosuppressive microenvironment through
multiple pathways. Experimental evidence indicates that CEACAM6 can
facilitate the recruitment of MDSCs through interactions with
CEACAM1 on immune cells (8).
Furthermore, CEACAM6 signaling through Src family kinases and
PI3K/AKT pathways can lead to the upregulation of PD-L1 expression
on tumor cells (12,15). These mechanisms were demonstrated
in murine models where CEACAM6 overexpression resulted in increased
PD-L1 levels and MDSC infiltration, while a CEACAM6-targeted
vaccine combined with anti-PD-1 antibody synergistically enhanced
antitumor immunity (15). These
findings position CEACAM6 as a dual biomarker and therapeutic
target, particularly for immune checkpoint inhibitor-resistant
cases.
The independent prognostic significance of CEACAM6
and FOXP3 supports their integration into clinical decision-making.
Patients exhibiting overexpression of either CEACAM6 or FOXP3 may
benefit from intensified adjuvant therapy such as anti-CEACAM6
CAR-T cells or antibody-drug conjugates (31,32),
or agents modulating Treg function. Targeting CEACAM6 holds promise
but requires caution due to its expression on normal granulocytes
and epithelial cells, potentially leading to on-target/off-tumor
toxicities such as neutropenia or dermatitis (12). Targeting FOXP3+ Tregs
systemically carries the risk of inducing autoimmune adverse
events. Strategies focusing on specific Treg depletion within the
tumor via CTLA-4 inhibition or targeting activation markers might
offer improved windows of safety (27,28).
Combining CEACAM6 or FOXP3/Treg-targeted strategies with PD-1/PD-L1
blockade represents a rational approach to overcome immune
resistance (31). Conversely,
tumors lacking both biomarkers but showing high CD8+
T-cell infiltration may be suitable for de-escalation strategies.
Future studies should investigate dynamic changes in these
biomarkers during treatment to guide adaptive therapeutic
approaches.
Our findings on the role of CEACAM6 in promoting
immune suppression are mechanistically supported by the study of
Pinkert et al (8), which
showed that inhibiting the CEACAM6-CEACAM1 interaction potentiates
T cell-mediated cytotoxicity. Similarly, Li et al (9) identified CEACAM6 as a critical driver
of lung cancer metastasis, underscoring its importance in
advanced-stage disease across cancer types. Our observations
regarding FOXP3+ Tregs align with clinical evidence from
Martinez-Rios et al (26),
who demonstrated Treg-associated immunosuppression in colorectal
cancer, further supporting the conserved nature of this mechanism.
It should be noted, however, that the exclusive use of a patient
cohort from Chinese tertiary hospitals represents a limitation of
the present study, potentially restricting the generalizability of
our conclusions to other ethnic and geographic populations.
Variations in genetic background, environmental exposures and
regional clinical practices may influence tumor immune biology and
biomarker performance. Therefore, large-scale, prospective
multinational studies incorporating diverse ethnicities and
geographic regions will be essential to validate the universal
prognostic value of CEACAM6 and FOXP3 and to establish globally
applicable clinical thresholds.
Our multicenter study establishes CEACAM6 and FOXP3
as robust, independent prognostic biomarkers in colon cancer,
validated through integrated IHC and transcriptomic analyses. The
findings not only corroborate emerging evidence on their roles in
tumor progression and immune evasion but also reveal novel
mechanistic insights with direct clinical implications. CEACAM6
potentially contributes to an immunosuppressive microenvironment by
recruiting MDSCs and upregulating PD-L1(15), providing a mechanistic rationale
for the observed immune exclusion and suggesting combination
immunotherapy strategies targeting these pathways alongside CEACAM6
itself (32).
While our multicenter design enhances
generalizability, limitations include the retrospective nature
which limited the availability of consistent data on adjuvant
therapies, comorbidities and molecular subtypes such as
microsatellite instability status, factors that should be
incorporated into future prospective analyses and RNA extraction
challenges from FFPE samples (35/50 success rate). The mRNA subset
was randomly selected, mitigating selection bias, but future
studies should utilize more robust platforms like NanoString or
digital spatial profiling for transcriptomic analysis, the latter
also enabling spatial resolution of interactions within the tumor
microenvironment. Prospective validation using liquid biopsies or
fresh-frozen specimens is warranted. Additionally, mechanistic
studies, potentially utilizing single-cell RNA sequencing or
spatial transcriptomics to precisely map cellular interactions, are
needed to clarify whether CEACAM6 directly modulates
FOXP3+ Treg recruitment or vice versa.
In conclusion, CEACAM6 and FOXP3 were established as
critical biomarkers for colon cancer prognostication, with dual
roles in immune evasion and tumor progression. Their incorporation
into clinical practice could optimize risk stratification and
therapeutic targeting, particularly in the era of precision
immunotherapy.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81172166).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YYL and XXP conceived and designed the study,
contributed to data collection and analysis, participated in the
interpretation of results, and critically revised the manuscript.
YYL prepared the initial draft of the manuscript. JXY contributed
to the development of the research methodology and provided
supervision throughout the study. JZM carried out a part of the
data analysis. YYL, JXY, JZM and XXP confirm the authenticity of
all the raw data. All authors read and approved the final version
of the manuscript.
Ethics approval and consent to
participate
The present retrospective study analyzed anonymized
data and was approved by the research ethics committees of the
Affiliated Hospital of Jiangnan University (approval no. 2025-154;
Wuxi, China), the First Affiliated Hospital of Soochow University
(approval no. 2025-140; Suzhou, China) and Jiangsu Provincial
Veterans Hospital (approval no. 2025-001; Wuxi, China). Written
informed consent was obtained from all participants for the use of
their tissues in scientific research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Biller LH and Schrag D: Diagnosis and
treatment of metastatic colorectal cancer: A review. JAMA.
325:669–685. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Moreno V, Salazar R and Gruber SB: The
prognostic value of TILs in stage III colon cancer must consider
sidedness. Ann Oncol. 33:1094–1096. 2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Soeratram TTD, Beentjes I, Egthuijsen JMP,
Mookhoek A, Lange MM, Meershoek-Klein Kranenbarg E, Hartgrink HH,
van de Velde CJH, Ylstra B, van Laarhoven HWM and van Grieken NCT:
A biopsy-based immunoscore in patients with treatment-naive
resectable gastric cancer. Ther Adv Med Oncol.
16(17588359241287747)2024.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Doi S, Yasuda S, Miyashita M, Nagai M,
Nakamura K, Matsuo Y, Terai T, Kohara Y, Sakata T and Sho M:
Prognostic relevance of sarcopenia and tumor-infiltrating CD8(+) T
cells in patients with hepatocellular carcinoma. Ann Gastroenterol
Surg. 9:359–368. 2025.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kong D, Gao C, Yu Y, Yang L, Ma J, Tang S,
Mao Y, Li Y and Li N: The distribution characteristics of PD-1
pathway-related immune cells in esophageal cancer tissue and their
prognostic significance. PLoS One. 20(e0325349)2025.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang C, Chen F, Li J, He Y, Sun J, Zheng
Z, Liu G, Wang Y, Kang W and Ye X: Comprehensive analysis of
single-cell and bulk RNA sequencing data unveils antigen-presenting
and processing fibroblasts and establishes a predictive model in
gastric cancer. Cancer Cell Int. 25(225)2025.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Pinkert J, Boehm HH, Trautwein M, Doecke
WD, Wessel F, Ge Y, Gutierrez EM, Carretero R, Freiberg C, Gritzan
U, et al: T cell-mediated elimination of cancer cells by blocking
CEACAM6-CEACAM1 interaction. Oncoimmunology.
11(2008110)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li Y, Polyak D, Lamsam L, Connolly ID,
Johnson E, Khoeur LK, Andersen S, Granucci M, Stanley G, Liu B, et
al: Comprehensive RNA analysis of CSF reveals a role for CEACAM6 in
lung cancer leptomeningeal metastases. NPJ Precis Oncol.
5(90)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Huskey ALW, McNeely I and Merner ND:
CEACAM gene family mutations associated with inherited breast
cancer risk-A comparative oncology approach to discovery. Front
Genet. 12(702889)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ilantzis C, DeMarte L, Screaton RA and
Stanners CP: Deregulated expression of the human tumor marker CEA
and CEA family member CEACAM6 disrupts tissue architecture and
blocks colonocyte differentiation. Neoplasia. 4:151–163.
2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wu G, Wang D, Xiong F, Wang Q, Liu W, Chen
J and Chen Y: The emerging roles of CEACAM6 in human cancer
(Review). Int J Oncol. 64(27)2024.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang X, Yang C, Wang X and Duan P:
Pan-cancer analysis reveals a regulatory pattern of anoikis in
human cancers. Cell Mol Biol (Noisy-le-grand). 70:51–61.
2024.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zang M, Zhang B, Zhang Y, Li J, Su L, Zhu
Z, Gu Q, Liu B and Yan M: CEACAM6 promotes gastric cancer invasion
and metastasis by inducing epithelial-mesenchymal transition via
PI3K/AKT signaling pathway. PLoS One. 9(e112908)2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li Y, Zhu X, You J, Zhang B, Huang X and
Jin C: Efficacy of bivalent CEACAM6/4-1BBL genetic vaccine combined
with anti-PD1 antibody in MC38 tumor model of mice. Heliyon.
8(e10775)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu Y, Xia T, Jin C, Gu D, Yu J, Shi W,
Zhang KE, Zhang L, Ye J and Li L: FOXP3 and CEACAM6 expression and
T cell infiltration in the occurrence and development of colon
cancer. Oncol Lett. 11:3693–3701. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Karamchandani DM, Gonzalez RS, Lee H,
Westerhoff M, Cox B and Pai RK: Interobserver agreement and
practice patterns for grading of colorectal carcinoma: World health
organization (WHO) classification of tumours 5th edition vs.
American joint committee on cancer (AJCC) 8th edition staging
manual. Histopathology. 86:1101–1111. 2025.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA:
The 2019 WHO classification of tumours of the digestive system.
Histopathology. 76:182–188. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Condurache Hritcu OM, Ciobanu Apostol DG,
Toader SV, Solcan C, Brănișteanu DE, Toader MP and Costan VV:
Immunohistochemical assessment of maspin, β-catenin, and MMP-14 in
oral potentially malignant lesions and oral squamous cell
carcinoma: A retrospective observational study. Medicina (Kaunas).
61(1037)2025.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Menard LC, Fischer P, Kakrecha B, Linsley
PS, Wambre E, Liu MC, Rust BJ, Lee D, Penhallow B, Manjarrez Orduno
N and Nadler SG: Renal cell carcinoma (RCC) tumors display large
expansion of double positive (DP) CD4+CD8+ T cells with expression
of exhaustion markers. Front Immunol. 9(2728)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Huang CY, Chiang SF, Ke TW, Chen TW, You
YS, Chen WT and Chao KSC: Clinical significance of programmed death
1 ligand-1 (CD274/PD-L1) and intra-tumoral CD8+ T-cell infiltration
in stage II-III colorectal cancer. Sci Rep. 8(15658)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Thomas CE, Takashima Y, Buchanan DD,
Wesselink E, Qu C, Hsu L, Dias Costa A, Gallinger S, Grant RC,
Huyghe JR, et al: Density of T-cell subsets in colorectal cancer in
relation to disease-specific survival. Cancer Epidemiol Biomarkers
Prev. 34:1122–1133. 2025.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Küçükköse E, Baars MJD, Amini M, Schraa
SJ, Floor E, Bol GM, Borel Rinkes IHM, Roodhart JML, Koopman M,
Laoukili J, et al: Stromal localization of inactive CD8(+) T cells
in metastatic mismatch repair deficient colorectal cancer. Br J
Cancer. 130:213–223. 2024.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Prasongtanakij S, Soontrapa K and Thumkeo
D: The role of prostanoids in regulatory T cells and their
implications in inflammatory diseases and cancers. Eur J Cell Biol.
104(151482)2025.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Martinez-Rios J, Lopez-Pacheco CP,
Garcia-Zepeda EA and Soldevila G: CCR9 shapes the immune
microenvironment of colorectal cancer modulating the balance
between intratumoral CD8+ T cell and FoxP3+ Helios+ Treg
subpopulations. PLoS One. 20(e0321930)2025.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tanaka A and Sakaguchi S: Targeting treg
cells in cancer immunotherapy. Eur J Immunol. 49:1140–1146.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tanaka A and Sakaguchi S: Regulatory T
cells in cancer immunotherapy. Cell Res. 27:109–118.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Shah F, Giri PS, Bharti AH and Dwivedi M:
Compromised melanocyte survival due to decreased suppression of
CD4(+) & CD8(+) resident memory T cells by impaired
TRM-regulatory T cells in generalized vitiligo patients. Exp
Dermatol. 33(e14982)2024.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Panek WK, Toedebusch RG, McLaughlin BE,
Dickinson PJ, Van Dyke JE, Woolard KD, Berens ME, Lesniak MS,
Sturges BK, Vernau KM, et al: The CCL2-CCR4 axis promotes
regulatory T cell trafficking to canine glioma tissues. J
Neurooncol. 169:647–658. 2024.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Redin E, Garmendia I, Lozano T, Serrano D,
Senent Y, Redrado M, Villalba M, De Andrea CE, Exposito F, Ajona D,
et al: SRC family kinase (SFK) inhibitor dasatinib improves the
antitumor activity of anti-PD-1 in NSCLC models by inhibiting treg
cell conversion and proliferation. J Immunother Cancer.
9(e001496)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Nakazawa Y, Miyano M, Tsukamoto S, Kogai
H, Yamamoto A, Iso K, Inoue S, Yamane Y, Yabe Y, Umihara H, et al:
Delivery of a BET protein degrader via a CEACAM6-targeted
antibody-drug conjugate inhibits tumour growth in pancreatic cancer
models. Nat Commun. 15(2192)2024.PubMed/NCBI View Article : Google Scholar
|