1. Introduction
Breast cancer is a notable global health issue, with
2.3 million new cases and 670,000 mortalities worldwide in 2022 and
by 2050, new cases are expected to rise by 38%, with mortalities
increasing by 68% (1). Human
epidermal growth factor receptor (HER)2-low breast cancer refers to
tumors that have HER2 expression levels with an
immunohistochemistry (IHC) score of 1+ or 2+ without HER2 gene
amplification, which is determined using single or dual probe in
situ hybridization (ISH) tests. Clinically, these tumors are
categorized as HER2-negative and are further classified as Luminal
A/B or triple-negative breast cancer (TNBC) subtypes based on the
expression of hormone receptor (HR) (2,3). IHC
and ISH are the main biological techniques used to assess HER2
status in patients with breast cancer. Therefore, these techniques
guide therapeutic decision-making. However, different preanalytical
and analytical variables, such as the use of fixative agents or the
antigen retrieval methodology, can compromise the accuracy of the
tests, leading to discrepancies between laboratories (4). To enhance the sensitivity and
precision during the detection of HER2 expression levels, adherence
to standardized testing guidelines along with the development of
novel quantitative assays, such as automated fluorescence-based
technologies and reverse transcription-quantitative PCR, are
necessary (5-7).
Patients with HER2-positive breast cancer were
treated with a combination of chemotherapy and monoclonal
antibodies. However, advances in HER2-targeted therapies,
particularly antibody-drug conjugates (ADCs) such as trastuzumab
emtansine and trastuzumab deruxtecan (T-DXd), has enhanced the
precision of targeting cancer cells. Patients with HER2-low breast
cancer may benefit from these novel therapeutics, as the cytotoxic
agents are delivered directly to the cancer cells while the
toxicity to the healthy tissues is minimized (8,9).
Clinical studies demonstrate positive responses and clinical
benefits in patients with HER2-low breast cancer receiving ADCs
based treatments (10,11). This highlights the need to update
current guidelines to recognize HER2-low as a distinct pathological
entity. Such a shift may improve treatment selection, clinical
outcomes and the integration of innovative therapies.
The present study investigated the current pathology
and molecular biology guidelines and techniques used to determine
HER2 status in breast cancer. Additionally, the present study
reviewed the molecular biology of the HER2 receptor including its
role in breast cancer pathogenesis and progression, and highlighted
its involvement in tumor growth, survival and metastasis.
Additionally, the concept of HER2-low breast cancer was
discussed.
2. Molecular classification of breast
cancer
Breast cancer molecular subtyping was first
introduced in 2000, in a study highlighting the differences between
gene expression profiles in tumors (12). Initially, breast cancer was
classified into three groups according to the differences in gene
expression profiles, namely, Luminal, HER2-positive and basal. This
work was fundamental for identifying subgroups of patients with
breast cancer and different prognostic outcomes, which influenced
individualized clinical management (12,13).
The current guidelines divide breast cancer into
five distinct categories depending on the immunohistochemical
expression of estrogen receptor (ER), progesterone receptor (PR)
and HER2(14). Additionally, the
Ki-67 antigen, a marker of cellular proliferation, serves a crucial
role in classification, as it notably impacts disease prognosis and
survival rates (15,16). The recognized subtypes include
Luminal A and B, HER2-enriched (HER2-positive), and basal and
normal-like (16).
Luminal A
As per the 2013 St. Gallen consensus, the Luminal A
subtype of breast cancer is characterized as ER and/or PR-positive
(levels of ≥20%) and HER2-negative with low levels (<14%) of the
Ki-67 antigen, which indicates a slow tumor growth (15). Luminal A is the most common subtype
and represents 50-60% of all types of breast cancer (17). This subtype is associated with a
favorable prognosis, low risk of recurrence and high overall
survival rates (13,15). A study by Prat et al
(13) demonstrates that patients
with IHC-defined luminal A breast tumors have an increased
disease-free survival (DFS) when PR levels exceed 20% (13). Patients with Luminal A tumors
benefit from endocrine therapy but have a poor response to
chemotherapy (18).
Luminal B
Luminal B breast cancer is characterized as ER
and/or PR-positive with high levels (≥14%) of the Ki-67 antigen,
which explains the fast growth and worse prognosis compared with
Luminal A (15). Expression of
HER2 is observed in Luminal B tumors, with ~30% of cases with
HER2-positive breast cancer, as determined using IHC, being the
Luminal B subtype. Overall, Luminal B constitutes 10-20% of Luminal
breast cancer. Compared with the Luminal A subtype, Luminal B shows
increased expression of proliferation-related genes including
avian myeloblastosis viral oncogene homolog, γ-glutamyl
hydrolase, lysosomal-associated protein transmembrane-4β, nuclease
sensitive element binding protein 1 and cyclin (CCN)E1,
as well as increased expression of growth receptor signaling genes
such as the insulin-like growth factor 1 receptor and the
fibroblast growth factor receptors (17,19,20).
Compared with Luminal A tumors, Luminal B tumors have an improved
response to chemotherapy but demonstrate a higher rate of visceral
recurrence as well as lower survival rate between diagnosis and
relapse (12,15-18,21).
HER2-positive
HER2-positive tumors account for 10-15% of all types
of breast cancer and are defined by high expression levels of the
HER2 gene, as well as other genes associated with the HER2
pathway and/or HER2 amplicon located in the 17q12 chromosome
[such as erythroblastic oncogene B (ERBB)2 and growth factor
receptor-bound protein 7] (15,22).
The HER2-positive group is subcategorized according to the HR
expression levels, namely luminal HER2 (ER, PR and HER2-positive
with Ki-67 levels of 15-30%) and HER2-enriched (HER2-positive, ER
and PR-negative with Ki-67 levels >30%) (16). HER2-positive tumors are sensitive
to chemotherapy; however, they are associated with a worse
prognosis compared with Luminal tumors (17,23).
The poor outcome is mainly due to the higher risk of early relapse,
and thus requires the use of targeted agents such as the anti-HER2
monoclonal antibody (15,17,23).
Among the HER2-positive group, the HER2-enriched subtype is the
most common (accounting for 31-76%) and has higher levels of ERBB2
mRNA and protein as well as a higher activation of the EGFR-HER2
signaling pathway. The HER2-enriched tumors have an improved
response to anti-HER2 treatments in both early (adjuvant) and
pre-surgical (neoadjuvant) settings, regardless of whether HER2 is
clinically positive. Only approximately half of clinically
HER2-positive tumors (based on IHC 3+ scores) are HER2-enriched
(16). Τhis HER2-enriched subtype
can also occur in tumors that are clinically HER2-negative, which
currently are not treated with HER2-targeted therapies (16).
Basal-type
Basal-type breast cancer represents 15-25% of all
types of breast cancer and are the most homogeneous of all
subtypes. The current and commonly accepted IHC definition includes
tumors that are negative for ER, PR and HER2, but positive for
CK5/6 and/or EGFR (24). Due to
being negative for ER, PR and HER2 RNA, the term triple-negative is
widely used instead of basal, although there is only partial
overlap between the two subtypes (24). The precise classification of
basal-type breast cancer remains a notable challenge. A previous
study reveals up to a 30% mismatch between transcriptomic-based and
IHC-based identification methods (25).
Triple-negative tumors are a heterogenous group,
including both basal and non-basal subtypes, which differ in
clinical features and molecular profiles, especially in terms of
therapeutic targets. The low expression of genes that regulate
tight junctions and cell-cell adhesion, such as claudins 3, 4 and
7, occludin and E-cadherin, is a feature of one of the numerous
triple negative subtypes (26,27).
The term ‘basal’ refers to tumors defined by specific gene
expression profiles, which are identified using microarrays.
Microarray analysis distinguishes basal-type from non-basal-type
breast cancer by its unique gene expression profile, which is
characterized by the high expression of basal cytokeratin genes
(such as CK5, 14 and 17) and the notable lack of expression of
genes for the hormone receptors [such as estrogen receptor 1
(ESR1) and PR gene] and HER2. The term ‘triple-negative’
indicates the absence of ER, PR and HER2, and the term ‘basal-like’
describes tumors that are identified using IHC based on four or
five protein markers (such as high molecular weight keratins and
EGFR) (25,28). A study by Burstein et al
(29) further investigates the
different TNBC subtypes using RNA and DNA profiling. It
distinguishes four clinically relevant subtypes of TNBC, namely
Luminal-AR (LAR), mesenchymal (MES), basal-like immune-suppressed
(BLIS) and basal-like immune-activated (BLIA), which are
characterized by distinct molecular signatures (29). Epidemiologically, TNBC most
commonly occurs in premenopausal women and is strongly associated
with the presence of BRCA1 mutations. Basal types of breast cancer
are characterized by the notable poor prognosis and reduced
metastasis-free survival, despite being chemo-sensitive (15,25).
Normal-like type
Although originally described as part of the
classification, normal-like breast cancer is not included in the
prediction analysis of microarray 50 (PAM50) molecular
classification at present. Due to its low invasive tumor cell
content, it is hypothesized that this group often results from a
high proportion of normal (non-tumor) breast tissue in the sample,
which leads to a misleading expression profile (17,28).
There is evidence supporting little to no notable survival rate
differences between normal-like and Luminal A subtypes, thus
excluding the need to recognize the normal-like subtype as a
distinct subtype of breast cancer (23).
3. Molecular biology and molecular pathology
of HER2-positive breast cancer
Among the aforementioned molecular subtypes of
breast cancer, HER2-positive tumors represent a biologically
distinct group that are characterized by amplification or
overexpression of the ERBB2 gene, which encodes HER2. This
group not only has notable implications for prognosis but also
guides targeted therapeutic strategies, such as the use of
monoclonal antibodies (such as trastuzumab) and small molecule
tyrosine kinase inhibitors (such as Lapatinib) (30). Therefore, understanding the
molecular classification framework is essential for contextualizing
the role of HER2 in breast cancer pathogenesis. The following
section investigated the molecular pathology of HER2 and
highlighted its biological functions and mechanisms of
dysregulation, as well as its relevance to its clinical
characteristics.
HER2 is a member of the ERBB-EGFR family of surface
tyrosine kinase receptors. As a transmembrane receptor, it consists
of an extracellular ligand-binding domain, a single-pass
transmembrane helix and an intracellular domain with intrinsic
tyrosine kinase activity. Unlike other ERBB family members, HER2
lacks a known ligand and is primarily activated through homo- or
heterodimerization with other ERBB receptors, such as EGFR (ERBB1),
HER3 (ERBB3) and HER4 (ERBB4) (31,32).
While dimerization in the ERBB family is typically
ligand-dependent, the overexpression or high local concentration of
HER2 can promote spontaneous homo- or heterodimerization,
particularly with HER3. Upon activation, HER2 undergoes
autophosphorylation at key tyrosine residues within its
intracellular domain, initiating downstream signaling cascades,
including the PI3K/AKT and RAS/MAPK pathways, which regulate
pivotal cellular functions including proliferation, survival and
differentiation. The persistent signaling that occurs due to
HER2 gene amplification and overexpression enhances cellular
proliferation, survival and metastatic potential while inhibiting
apoptosis, which contributes to tumor aggressiveness (33). In addition, HER2 overexpression
alters the tumor microenvironment by modulating angiogenesis,
immune evasion and the epithelial-mesenchymal transition, further
facilitating disease progression (32,34,35).
HER2-positive breast cancers have been reported to occur in up to
15-30% of all cases of breast cancer (36) and is associated with an aggressive
clinical phenotype, which is characterized by increased disease
aggressiveness, higher recurrence rates and a poorer prognosis
(37).
4. HER2-low breast cancer
Previously, HER2-low types of breast cancer were
classified and treated as HER2-negative tumors and therapeutic
interventions were determined by the presence or absence of
specific biomarkers (for example HRs), expression of genetic
mutational signatures, as well as other factors such as the stage
of the disease and the overall condition of the patient (10). However, following the results of
the DESTINY-Breast04 clinical trial (10), the therapeutic management of
HER2-low breast cancer underwent a notable change. In this phase 3
trial, the efficacy of T-DXd is assessed in patients with HER2-low
metastatic breast cancer and previous chemotherapy treatment.
Compared with the control group of patients that are treated with
chemotherapy as chosen by the physician, the T-DXd-treated group
have markedly longer progression-free and overall survival. These
findings highlight the clinical relevance of the HER2-low patient
population and highlights the need to redefine the classification
of HER2 status (10).
After the aforementioned trial, the American Society
of Clinical Oncology (ASCO)-College of American Pathologists (CAP)
reevaluated and updated the guidelines regarding testing,
evaluation and interpretation of HER2 status in breast cancer
specimens (3). The updated HER2
testing guidelines from ASCO-CAP are a nuanced approach to the
classification of breast cancer and aim to improve the diagnostic
accuracy and optimize the selection of treatments. The key changes
suggest the mandatory HER2 testing for all newly diagnosed patients
with invasive breast cancer and the retesting of metastatic cases
when clinically required. While the largest invasive tumor
component remains the primary target for testing, smaller lesions
should also be assessed and documented if they exhibit a difference
in HER2 expression levels.
IHC scoring follows a 3-tier system as follows: A
score of 0 for no or faint staining in ≤10% of cells; a score of 1+
for faint and incomplete staining in >10% of the cells; a score
of 2+ for weak to moderate complete staining in >10%; and a
score of 3+ for intense complete staining in >10% of the cells.
This system classifies HER2 status as negative (a score of 0),
equivocal (a score of 2+; requiring ISH confirmation) or positive
(a score of 3+), reducing ambiguity. According to the current
guidelines, HER2-low (an IHC score of 1+ or 2+ with negative ISH
testing) is a distinct subgroup that has notable therapeutic
implications. The CAP guidelines highlight the importance of
standardized pre-analytical variables (such as optimal initial test
validation and optimal internal quality assurance), accurate
documentation of tissue handling, assay validation and the use of
quality controls in order to enhance laboratory proficiency and
ensure precise HER2 classification for optimal patient treatment
(3). Fig. 1 presents the CAP guidelines for the
laboratory workflow of HER2 assessment in breast cancer tissue, as
aforementioned.
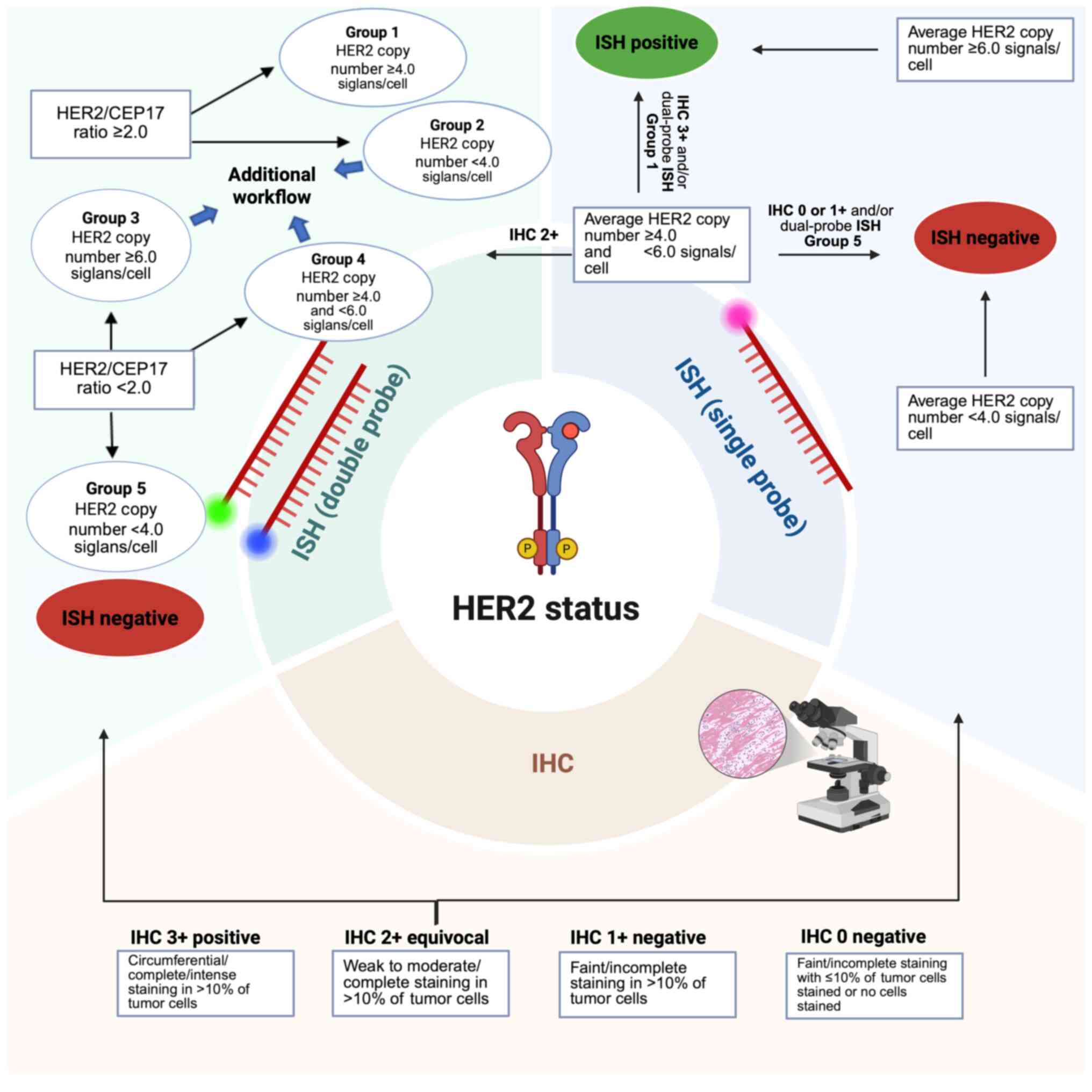 | Figure 1Laboratory workflow for HER2
assessment in tissue samples of breast cancer, based on guidelines
from the College of American Pathologists (https://www.cap.org/protocols-and-guidelines/cap-guidelines/current-cap-guidelines/recommendations-for-human-epidermal-growth-factor-2-testing-in-breast-cancer).
HER2 assessment usually begins with IHC (scores of 0-1+ are
negative, 2+ are equivocal and 3+ are positive). Molecular
investigation is required for equivocal cases. Single-probe FISH
assesses ERBB2 copy number, while dual-probe FISH compares
ERBB2 to CEP17, which improves the accuracy. IHC,
immunohistochemistry; ISH, in situ hybridization; FISH,
fluorescence ISH; ERBB, erythroblastic oncogene B;
CEP17, chromosome enumeration probe 17. Created in BioRender
(publication license, Georgiou, A. (2025); https://BioRender.com/3140k7k). |
5. Molecular and clinicopathological
features of HER2-low breast cancer
Due to advancements in diagnostic technologies and
targeted therapies, interest in elucidating the molecular and
distinct clinicopathological characteristics of HER2-low tumors has
increased. Understanding these features may refine the
classification, prediction of therapeutic responses and
optimization of the treatment strategies for patients with HER2-low
breast cancer.
A study by Schettini et al (38) investigates the molecular pathology
of HER2-low breast cancer by analyzing clinicopathological and
PAM50 gene expression level data. The findings reveal that HER2-low
tumors are notably more prevalent in patients with HR-positive
breast cancer (65.4%) compared with patients with TNBC (36.5%).
Furthermore, compared with patients with HER2 negative tumors,
HER2-low tumors are associated with more advanced
clinicopathological stages of the disease, including larger primary
tumor sizes and increased nodal involvement, as well as an older
median age at diagnosis (59 vs. 55 years). The distribution of
PAM50 intrinsic subtypes also varies between HER2-low tumors.
Tumors were predominantly classified as Luminal A (50.8%) and
Luminal B (28.8%), with smaller fractions of basal-like (13.3%),
HER2-enriched (3.5%) and Normal-like (3.5%) subtypes. In addition,
subtype distribution varies at the transcriptional level, with
Luminal A and B showing a higher ERBB2 mRNA expression level
compared with basal-like tumors that demonstrate lower levels.
Furthermore, the aforementioned study reveals distinct gene
expression profiles between HER2-low and HER2 negative tumors.
Compared with HER2 negative tumors, proliferation-associated genes
(such as CCNB1 and E1), basal-like-associated genes
(such as Keratin 14 and 5) and tyrosine-kinase receptors
(such as EGFR and fibroblast growth factor receptor 4) are
downregulated in HER2-low tumors, while Luminal-associated genes
(such as BCL-2, BCL-2-associated athanogene-1, forkhead box α 1,
ESR1, PR, G protein-coupled receptor 160 and androgen
receptor) are overexpressed. Taken together, these findings
highlight the potential implications of HR status in HER2-low
breast cancer and indicates that HR-positive/HER2-low tumors
probably constitute a distinct biological entity compared with
TNBC/HER2-low tumors (38).
In another study by Agostinetto et al
(39), a retrospective analysis of
patients with primary breast cancer from The Cancer Genome Atlas
(n=804) further elucidates the molecular characteristics of
HER2-low tumors. The aforementioned study reveals HER2-low breast
cancer accounts for 51% of the cohort, with the majority being
HR-positive (82%). Additionally, analysis of PAM50 intrinsic
subtypes reveals that HER2-low/HR-positive tumors are predominantly
classified as Luminal A (54.4%) and B (54.5%). This suggests that
in this molecular subgroup, HR signaling and expression of Luminal
genes are the primary oncogenic promoters, instead of HER2 itself.
Furthermore, HER2-low tumors demonstrate notably higher ERBB2 mRNA
levels compared with HER2-negative tumors (39).
Additionally, a study by Zhang et al
(40) investigates the molecular
profile of HER2-low breast cancer using a cohort of 523 female
patients stratified into three groups based on HER2 status. The
findings confirm that the HR-positive status is predominant in
HER2-low tumors (87.4%) and reveals that HER2-low tumors have
notably lower Ki-67 expression levels compared with both
HER2-negative and HER2-positive tumors. Furthermore, the IHC-based
molecular subtype distribution demonstrates that the majority of
patients with HER2-low breast cancer are classified as Luminal B
(58.9%) or A (28.6%) tumors, with only a small proportion
classified as TNBC (12.5%). In addition, targeted sequencing
identifies distinct mutational patterns among the three HER2
subgroups. HER2-low tumors demonstrate notably higher mutation
frequencies in PTEN, GATA binding protein 3,
core-binding factor subunit β (CBFB) and AKT1
genes compared with HER2-positive tumors, as well as increased
mutation rates in CBFB,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
α (PIK3CA), mitogen-activated protein kinase kinase
kinase 1 and AT-rich interaction domain 1A compared with
HER2-negative tumors (40).
Another similar study supports the aforementioned
findings, highlighting the predominance of Luminal B transcriptomic
features in HER2-low tumors (41).
Moreover, HER2-low tumors exhibit mutation rates for TP53
and PIK3CA that are closer to those in HER2-positive types
of cancer compared with HER2-negative types of cancer (41).
6. HR-HER2 crosstalk
The findings from the aforementioned studies
highlight the interplay between HER2 and HR and their influence on
the molecular pathobiology of HER2-low breast cancer. HR and HER2
status are critical biomarkers in the diagnosis, prognosis and
therapeutic management of breast cancer, with ~10% of patients with
breast cancer exhibiting concurrent expression of both (42). A deeper understanding of how HR
signaling influences the molecular landscape of HER2-low tumors is
essential, as it may provide insights into the mechanisms
underlying tumorigenesis and holds important implications for
optimizing therapeutic strategies for this specific subtype.
At the molecular level, an ER binding site is
present within an intronic region of the HER2 gene, where it
serves a regulatory role in modulating the expression of HER2.
Dysregulation of this pathway is particularly relevant in the
context of endocrine resistance in breast cancer (43). A study by Hurtado et al
(44) demonstrates that the paired
box 2 (PAX2) transcription factor functions as a cis-regulatory
repressor of HER2 transcription in the presence of tamoxifen. Their
findings reveal that PAX2 binding suppresses the expression of
HER2, which contributes to the therapeutic efficacy of tamoxifen
(44). Conversely, overexpression
of HER2 is a well-established feature of tamoxifen-resistance in
breast cancer, highlighting its role in mediating endocrine therapy
resistance (44). Amplified in
breast cancer-1 (AIB1), is a transcriptional co-activator of ER
that is frequently overexpressed in tamoxifen-resistant tumors.
AIB1 promotes both ER signaling and HER2-induced oncogenic
pathways, facilitating tumor progression and therapeutic resistance
in experimental models (45-47).
Additionally, the study by Hurtado et al (44) identifies a competitive interaction
between PAX2 and AIB1 at the HER2 cis-regulatory site. This
competition determines the transcriptional output of ERBB2 in
response to tamoxifen. Silencing of PAX2 using siRNA restores AIB1
occupancy at the regulatory element, which increases HER2
transcription and sustains tumor cell proliferation despite
tamoxifen treatment. These findings suggest that the balance
between PAX2 and AIB1 at ER-bound enhancer elements is a critical
determinant of tamoxifen response and resistance in ER-positive
breast cancer (44).
Emerging evidence indicates that the interplay
between HR and HER2 serves a role in shaping the molecular biology
and clinical behavior of HER2-low breast cancer. In a pooled
analysis of individual patient data from four prospective
neoadjuvant clinical trials, 2,310 patients with HER2-non-amplified
primary breast cancer treated with a neoadjuvant combination of
chemotherapy are evaluated. Among these, 1,098 tumors (47.5%)
identify as HER2-low, with the majority (64.0%) being HR-positive.
Patients with HER2-low tumors exhibit a notably lower pathological
complete response (pCR) rate compared with those with HER2-negative
tumors (29.2 vs. 39.0%). This difference is particularly evident in
the HR-positive subgroup. Patients with HR-positive and HER2-low
breast cancer demonstrate lower pCR (17.5%) compared with those
with HR-positive and HER2-negative breast cancer (23.6%), while no
notable difference in the pCR is observed in the HR-negative
subgroup (48).
7. Conclusions
Breast cancer, one of the most prevalent types of
cancer worldwide with 2.3 million new cases in 2022(1), has been a focal point of scientific
research for decades. Breakthroughs in molecular biology
revolutionize the current understanding of this disease and
redefine the therapeutic management of patients, leading to
improvements in patient survival and quality of life.
HER2-low breast cancer represents a distinct and
emerging subclass, with growing evidence suggesting its unique
molecular, pathological and clinical features. This highlights the
need to refine diagnostic criteria, incorporate advanced molecular
assays and update pathology guidelines to ensure accurate
identification and stratification of patients with HER2-low breast
cancer. The integration of the HER2-low classification into routine
clinical practice presents several challenges, including
standardized and reproductible methodologies, clear cut-off points
and updated evidence-based guidelines. Future research should focus
on refining diagnostic thresholds, validating predictive biomarkers
and developing quantitative assays in order to improve the
diagnostic accuracy. As precision oncology continues to advance,
recognizing the distinct biology of HER2-low breast cancer will not
only improve patient diagnosis, but will also enhance treatment
outcomes through the integration of novel, targeted therapeutic
strategies, under the scope of precision medicine.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
AG and ES equally contributed to the preparation of
the manuscript, including the conceptualization, review of the
literature, writing of the manuscript and preparation of the image.
VZ supervised the project, provided scientific insight and assisted
during the writing and correct use of the scientific language in
the manuscript. DS, SA and VZ revised and edited the manuscript.
Data authentication is not applicable. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
AG, ES, SA and VZ declare that they have no
competing interests. DS is the Editor-in-Chief for the journal, but
had no personal involvement in the reviewing process, or any
influence in terms of adjudicating on the final decision, for this
article.
Use of artificial intelligence tools
During the preparation of this work, artificial
intelligence tools were used to improve the readability and
language of the manuscript, and subsequently, the authors revised
and edited the content produced by the artificial intelligence
tools as necessary, taking full responsibility for the ultimate
content of the present manuscript.
References
|
1
|
Kim J, Harper A, McCormack V, Sung H,
Houssami N, Morgan E, Mutebi M, Garvey G, Soerjomataram I and
Fidler-Benaoudia MM: Global patterns and trends in breast cancer
incidence and mortality across 185 countries. Nat Med.
31:1154–1162. 2025.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Shirman Y, Lubovsky S and Shai A: HER2-low
breast cancer: Current landscape and future prospects. Breast
Cancer (Dove Med Press). 15:605–616. 2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wolff AC, Somerfield MR, Dowsett M,
Hammond MEH, Hayes DF, McShane LM, Saphner TJ, Spears PA and
Allison KH: Human epidermal growth factor receptor 2 testing in
breast cancer. Arch Pathol Lab Med. 147:993–1000. 2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Tozbikian G, Bui MM, Hicks DG, Jaffer S,
Khoury T, Wen HY, Krishnamurthy S and Wei S: Best practices for
achieving consensus in HER2-low expression in breast cancer:
Current perspectives from practising pathologists. Histopathology.
85:489–502. 2024.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gustavson MD, Bourke-Martin B, Reilly D,
Cregger M, Williams C, Mayotte J, Zerkowski M, Tedeschi G, Pinard R
and Christiansen J: Standardization of HER2 immunohistochemistry in
breast cancer by automated quantitative analysis. Arch Pathol Lab
Med. 133:1413–1419. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Larson JS, Goodman LJ, Tan Y, Defazio-Eli
L, Paquet AC, Cook JW, Rivera A, Frankson K, Bose J, Chen L, et al:
Analytical Validation of a highly quantitative, sensitive,
accurate, and reproducible assay (HERmark) for the measurement of
HER2 Total Protein and HER2 Homodimers in FFPE breast cancer tumor
specimens. Patholog Res Int. 2010(814176)2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Baehner FL, Achacoso N, Maddala T, Shak S,
Quesenberry CP Jr, Goldstein LC, Gown AM and Habel LA: Human
epidermal growth factor receptor 2 assessment in a case-control
study: Comparison of fluorescence in situ hybridization and
quantitative reverse transcription polymerase chain reaction
performed by central laboratories. J Clin Oncol. 28:4300–4306.
2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zimmerman BS and Esteva FJ:
Next-generation HER2-targeted antibody-drug conjugates in breast
cancer. Cancers (Basel). 16(800)2024.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nicolò E, Boscolo Bielo L, Curigliano G
and Tarantino P: The HER2-low revolution in breast oncology: Steps
forward and emerging challenges. Ther Adv Med Oncol.
15(17588359231152842)2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Modi S, Jacot W, Yamashita T, Sohn J,
Vidal M, Tokunaga E, Tsurutani J, Ueno NT, Prat A, Chae YS, et al:
Trastuzumab deruxtecan in previously treated HER2-low advanced
breast cancer. N Engl J Med. 387:9–20. 2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Modi S, Park H, Murthy RK, Iwata H, Tamura
K, Tsurutani J, Moreno-Aspitia A, Doi T, Sagara Y, Redfern C, et
al: Antitumor activity and safety of trastuzumab deruxtecan in
patients with HER2-low-expressing advanced breast cancer: Results
from a phase Ib study. J Clin Oncol. 38:1887–1896. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Prat A, Cheang MCU, Martín M, Parker JS,
Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen
TO and Perou CM: Prognostic significance of progesterone
receptor-positive tumor cells within immunohistochemically defined
luminal a breast cancer. J Clin Oncol. 31:203–209. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cho SY, Park SY, Bae YK, Kim JY, Kim EK,
Kim WG, Kwon Y, Lee A, Lee HJ, Lee JS, et al: Standardized
Pathology report for breast cancer. J Breast Cancer. 24:1–21.
2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Orrantia-Borunda E, Anchondo-Nuñez P,
Acuña-Aguilar LE, Gómez-Valles FO and Ramírez-Valdespino CA:
Chapter 3. Subtypes of Breast. Cancer. In: Breast Cancer
(Internet). Mayrovitz HN (ed). Exon Publications, Brisbane, AU,
2022.
|
|
16
|
Turova P, Kushnarev V, Baranov O, Butusova
A, Menshikova S, Yong ST, Nadiryan A, Antysheva Z, Khorkova S,
Guryleva MV, et al: The breast cancer classifier refines molecular
breast cancer classification to delineate the HER2-low subtype. NPJ
Breast Cancer. 11(19)2025.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yersal O and Barutca S: Biological
subtypes of breast cancer: Prognostic and therapeutic implications.
World J Clin Oncol. 5:412–424. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gao JJ and Swain SM: Luminal A breast
cancer and molecular assays: A review. Oncologist. 23:556–565.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lee JS, Tocheny CE and Shaw LM: The
insulin-like growth factor signaling pathway in breast cancer: An
elusive therapeutic target. Life (Basel). 12(1992)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Marin A, Morales F and Walbaum B:
Fibroblast growth factor receptor signaling in estrogen
receptor-positive breast cancer: Mechanisms and role in endocrine
resistance. Front Oncol. 14(1406951)2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Eliyatkın N, Yalçın E, Zengel B, Aktaş S
and Vardar E: Molecular classification of breast carcinoma: From
traditional, old-fashioned way to a new age, and a new way. J
Breast Health. 11:59–66. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Godoy-Ortiz A, Sanchez-Muñoz A, Chica
Parrado MR, Álvarez M, Ribelles N, Rueda Dominguez A and Alba E:
Deciphering HER2 breast cancer disease: Biological and clinical
implications. Front Oncol. 9(1124)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J
and Shi B: Breast cancer intrinsic subtype classification, clinical
use and future trends. Am J Cancer Res. 5:2929–2943.
2015.PubMed/NCBI
|
|
24
|
Badve S, Dabbs DJ, Schnitt SJ, Baehner FL,
Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR,
et al: Basal-like and triple-negative breast cancers: A critical
review with an emphasis on the implications for pathologists and
oncologists. Mod Pathol. 24:157–167. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bertucci F, Finetti P and Birnbaum D:
Basal breast cancer: A complex and deadly molecular subtype. Curr
Mol Med. 12:96–110. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Herschkowitz JI, Zhao W, Zhang M, Usary J,
Murrow G, Edwards D, Knezevic J, Greene SB, Darr D, Troester MA, et
al: Comparative oncogenomics identifies breast tumors enriched in
functional tumor-initiating cells. Proc Natl Acad Sci USA.
109:2778–2783. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Voutsadakis IA: Comparison of clinical
subtypes of breast cancer within the claudin-low molecular cluster
reveals distinct phenotypes. Cancers (Basel).
15(2689)2023.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Parker JS, Mullins M, Cheang MCU, Leung S,
Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al:
Supervised risk predictor of breast cancer based on intrinsic
subtypes. J Clin Oncol. 27:1160–1167. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Burstein MD, Tsimelzon A, Poage GM,
Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK,
Hilsenbeck SG, Chang JC, et al: Comprehensive genomic analysis
identifies novel subtypes and targets of triple-negative breast
cancer. Clin Cancer Res. 21:1688–1698. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Callahan R and Hurvitz S: Human epidermal
growth factor receptor-2-positive breast cancer: Current management
of early, advanced, and recurrent disease. Curr Opin Obstet
Gynecol. 23:37–43. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Dawson JP, Berger MB, Lin CC, Schlessinger
J, Lemmon MA and Ferguson KM: Epidermal growth factor receptor
dimerization and activation require ligand-induced conformational
changes in the dimer interface. Mol Cell Biol. 25:7734–7742.
2005.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Rubin I and Yarden Y: The basic biology of
HER2. Ann Oncol. 12 (Suppl 1):S3–S8. 2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Cheng X: A comprehensive review of HER2 in
cancer biology and therapeutics. Genes (Basel).
15(903)2024.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ross JS, Slodkowska EA, Symmans WF,
Pusztai L, Ravdin PM and Hortobagyi GN: The HER-2 receptor and
breast cancer: Ten years of targeted anti-HER-2 therapy and
personalized medicine. Oncologist. 14:320–368. 2009.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hsu JL and Hung MC: The role of HER2,
EGFR, and other receptor tyrosine kinases in breast cancer. Cancer
Metastasis Rev. 35:575–588. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Müller V, Bartsch R, Lin NU, Montemurro F,
Pegram MD and Tolaney SM: Epidemiology, clinical outcomes, and
unmet needs of patients with human epidermal growth factor receptor
2-positive breast cancer and brain metastases: A systematic
literature review. Cancer Treat Rev. 115(102527)2023.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Nader-Marta G, Martins-Branco D and de
Azambuja E: How we treat patients with metastatic HER2-positive
breast cancer. ESMO Open. 7(100343)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Schettini F, Chic N, Brasó-Maristany F,
Paré L, Pascual T, Conte B, Martínez-Sáez O, Adamo B, Vidal M,
Barnadas E, et al: Clinical, pathological, and PAM50 gene
expression features of HER2-low breast cancer. NPJ Breast Cancer.
7(1)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Agostinetto E, Rediti M, Fimereli D,
Debien V, Piccart M, Aftimos P, Sotiriou C and de Azambuja E:
HER2-low breast cancer: Molecular characteristics and prognosis.
Cancers (Basel). 13(2824)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang G, Ren C, Li C, Wang Y, Chen B, Wen
L, Jia M, Li K, Mok H, Cao L, et al: Distinct clinical and somatic
mutational features of breast tumors with high-, low-, or
non-expressing human epidermal growth factor receptor 2 status. BMC
Med. 20(142)2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Marchiò C, Dell'Orto P, Annaratone L,
Geyer FC, Venesio T, Berrino E, Verdun di Cantogno L, Garofoli A,
Rangel N, Casorzo L, et al: The dilemma of HER2 double-equivocal
breast carcinomas: Genomic profiling and implications for
treatment. Am J Surg Pathol. 42:1190–1200. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Howlader N, Altekruse SF, Li CI, Chen VW,
Clarke CA, Ries LA and Cronin KA: US incidence of breast cancer
subtypes defined by joint hormone receptor and HER2 status. J Natl
Cancer Inst. 106(dju055)2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Pegram M, Jackisch C and Johnston SRD:
Estrogen/HER2 receptor crosstalk in breast cancer: Combination
therapies to improve outcomes for patients with hormone
receptor-positive/HER2-positive breast cancer. NPJ Breast Cancer.
9(45)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hurtado A, Holmes KA, Geistlinger TR,
Hutcheson IR, Nicholson RI, Brown M, Jiang J, Howat WJ, Ali S and
Carroll JS: Regulation of ERBB2 by oestrogen receptor-PAX2
determines response to tamoxifen. Nature. 456:663–666.
2008.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Anzick SL, Kononen J, Walker RL, Azorsa
DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM and
Meltzer PS: AIB1, a steroid receptor coactivator amplified in
breast and ovarian cancer. Science. 277:965–968. 1997.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Torres-Arzayus MI, Font de Mora J, Yuan J,
Vazquez F, Bronson R, Rue M, Sellers WR and Brown M: High tumor
incidence and activation of the PI3K/AKT pathway in transgenic mice
define AIB1 as an oncogene. Cancer Cell. 6:263–274. 2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Fereshteh MP, Tilli MT, Kim SE, Xu J,
O'Malley BW, Wellstein A, Furth PA and Riegel AT: The nuclear
receptor coactivator amplified in breast cancer-1 is required for
Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis
in mice. Cancer Res. 68:3697–3706. 2008.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Denkert C, Seither F, Schneeweiss A, Link
T, Blohmer JU, Just M, Wimberger P, Forberger A, Tesch H, Jackisch
C, et al: Clinical and molecular characteristics of
HER2-low-positive breast cancer: Pooled analysis of individual
patient data from four prospective, neoadjuvant clinical trials.
Lancet Oncol. 22:1151–1161. 2021.PubMed/NCBI View Article : Google Scholar
|















