1. Introduction
Cervical cancer (CC) ranks as the fourth most
prevalent malignancy among females globally, with ~569,847 new
cases and 311,365 related deaths each year, according to the latest
GLOBOCAN statistics (1,2). Although early-stage CC exhibits a
high cure rate, the disease presents significant clinical
challenges, including an increasing incidence rate, a trend towards
younger onset and poor prognosis in the advanced stages of the
disease. These factors underscore CC as a critical public health
issue, with its elimination recognized as a global health priority.
Current therapeutic strategies, such as chemotherapy combined with
targeted therapy, have improved the overall survival and
progression-free survival of patients with CC. However, its
incidence continues to increase, particularly in cases of locally
advanced disease, which is often associated with low control rates,
a high metastatic potential and adverse prognostic factors.
Consequently, the 5-year survival rate remains suboptimal at ~60%
(3). These limitations highlight
the urgent need for the identification of novel biomarkers to
enhance early diagnosis, guide targeted therapies and improve
prognostic assessment.
Exosomes are bilayer lipid membrane vesicles, 30-150
nm in diameter, that facilitate intercellular communication by
transporting bioactive molecules, including proteins, lipids, DNA
and RNA. Their lipid membranes confer stability, protecting cargo
from degradation and enabling efficient delivery to recipient
cells, thereby modulating diverse physiological and pathological
processes (4). Ubiquitously
secreted by mammalian cells, such as dendritic cells, adipocytes,
endothelial cells and epithelial cells, exosomes are abundant in
plasma and other bodily fluids (5). Notably, cancer-derived exosomes are
secreted at levels 10-fold higher than those from normal cells and
play pivotal roles in tumor progression, including invasion,
angiogenesis and chemotherapeutic resistance, via autocrine,
paracrine and endocrine mechanisms (6,7). Due
to their tumor-specific cargo and stability, exosomes have emerged
as promising biomarkers for distinguishing malignant from
non-malignant tissues (8). Among
their molecular constituents, non-coding RNAs (ncRNAs), such as
microRNAs (miRNAs/miRs), long ncRNAs (lncRNAs) and circular RNAs
(circRNAs), are increasingly recognized for their regulatory roles
in the progression of CC. These ncRNAs represent potential
diagnostic, prognostic and therapeutic targets, providing new
avenues for the management of CC.
Systematic search strategy
A thorough literature search was conducted on PubMed
(https://pubmed.ncbi.nlm.nih.gov/) using
specific MeSH, including ‘exosomes’, ‘cervical cancer’, ‘lncRNAs’,
‘miRNAs’, ‘circRNAs’, ‘ncRNAs’, ‘drug resistance’, ‘angiogenesis’,
‘EMT’ and ‘lymphangiogenesis’. The search was limited to studies
published in various countries within the past 10 years
(2015-2025). As a result, a total of 57 relevant references were
identified and reviewed.
2. Exosomal miRNAs in CC
miRNAs are single-stranded non-coding RNAs (~22 nt
in length) that post-transcriptionally regulate gene expression
through mRNA degradation or translational inhibition, profoundly
influencing cellular phenotype and function (9). A previous study demonstrated that
cancer-related fibroblast-derived exosomal miR-18a-5p can regulate
the transmembrane protein 170B signaling axis, stimulating the
proliferation and migration, and inhibiting the apoptosis of CC
cells (10). Emerging as critical
regulators of tumor microenvironment homeostasis, exosomal miRNAs
exhibit distinct expression profiles in the vaginal lavage fluid of
patients with CC, including upregulated oncogenic miRNAs
(miR-483-5p and miR-1246) and downregulated tumor suppressors
(let-7d-5p and miR-20a-5p) (11).
In addition, another study demonstrated that Th17 cells induced the
expression of miR-142-5p in CC cells, and identified the subunits C
and D of the succinate dehydrogenase complex as novel targets of
miR-142-5p responsible for enhanced migration and invasion
(12). These findings position
exosomal miRNAs as promising molecular signatures for CC detection
and monitoring.
Exosomal miRNAs promote
angiogenesis
Angiogenesis, the formation of new blood vessels
from pre-existing vasculature, serves as a critical biological
process driving tumor growth, local invasion and distant metastasis
in CC (13). This process is
fundamentally required for tumor progression beyond the 1 to 2-mm
diffusion limit imposed. Malignant cells overcome this limitation
through the robust secretion of exosomes that reprogram the tumor
microenvironment to induce pathological angiogenesis (14). Given its central role in tumor
progression and metastasis, angiogenesis has emerged as a promising
therapeutic target for the management of advanced-stage CC
(15). At the molecular level,
CC-derived exosomal miRNAs orchestrate angiogenesis through
multiple mechanisms: i) miR-221-3p mediates the inhibition of
mitogen-activated protein kinase 10 (MAPK10). The exosomal transfer
of miR-221-3p from CC cells to microvascular endothelial cells
specifically targets and modulates MAPK10 activity. This
interaction stimulates endothelial cell proliferation, invasion and
migration, culminating in the formation of an aberrant tumor
vasculature (16). ii)
miR-663b-dependent vascular remodeling: CC-secreted exosomal
miR-663b potently enhances tumor vascular density by suppressing
vinculin expression in vascular endothelial cells. The
downregulation of vinculin alters endothelial cell adhesion
dynamics, thereby promoting extensive vascular network formation
and significantly augmenting the angiogenic potential of CC tissues
(17).
These findings illustrate the sophisticated,
multi-faceted regulatory networks through which exosomal miRNAs
coordinate neovascularization in CC. The diversity of these
pathways highlights both the complexity of tumor angiogenesis and
the potential for developing targeted anti-angiogenic therapies
that disrupt specific exosomal miRNA-mediated mechanisms.
Exosomal miRNAs promote
lymphangiogenesis
Lymphangiogenesis, the formation of new lymphatic
vessels from pre-existing ones, plays a critical role in cancer
metastasis. Tumor cells often exploit lymphatic vessels to enter
the circulatory system, facilitating their spread to distant organs
and aggravating disease progression (18). In a number of types of cancer,
lymphatic metastasis occurs at an early stage and serves as a major
route for cancer cell dissemination, with lymphangiogenesis being a
key driver of this process. For instance, exosomal miR-1468-5p from
CC cells promotes lymphangiogenesis and upregulates programmed
death-ligand 1 expression in lymphatic vessels. This occurs through
the suppression of homeobox containing 1 (HMBOX1)-suppressor of
cytokine signaling 1 (SOCS1) transcription and the activation of
the Janus kinase 2 (JAK2)/signal transducer and activator of
transcription 3 (STAT3) pathway, which impairs CD8+
T-cell immunity, enabling immune evasion and metastatic spread
(19). Similarly, exosomes from
cervical squamous cell carcinoma (CSCC) enhance human lymphatic
endothelial cell (HLEC) migration and tube formation in
vitro. Among their cargo, miR-221-3p is highly enriched and
delivered to HLECs, where it downregulates vasohibin 1, further
stimulating lymphangiogenesis and lymphatic metastasis (20). Given its functional role,
miR-221-3p may serve as both a therapeutic target and a diagnostic
biomarker for metastatic CSCC.
Exosomal miRNAs regulate drug
resistance
Drug resistance represents a major therapeutic
challenge in cancer management, with complex underlying mechanisms.
Current research has identified three primary exosome-mediated drug
resistance pathways in cancers: Antibody-based drug neutralization,
drug efflux and miRNA transfer via exosomes. Notably,
miRNA-mediated drug resistance is directly or indirectly associated
with multidrug resistance protein 1 expression (21). The selective packaging of miRNAs
into exosomes from drug-resistant cells can induce P-glycoprotein
overexpression, thereby enhancing the multidrug resistance
phenotype in cancer cells (22).
Zhu et al (23)
demonstrated that miR-651 expression was significantly
downregulated in the circulation of patients with CC and in
cisplatin-resistant HeLa cells (HeLa/DDP) compared with their
cisplatin-sensitive counterparts (HeLa/S). Functional analyses
revealed that miR-651 overexpression reduced cisplatin resistance
and proliferation, while promoting the apoptosis of HeLa cells.
Mechanistically, exosomal miR-651 from CC cells has been shown to
directly target autophagy-related 3 (ATG3) to suppress cisplatin
resistance. Furthermore, miR-106a/b has been implicated in the
exosome-mediated modulation of cisplatin sensitivity in HeLa cells
(24).
In addition, the study by Li et al (25) demonstrated that HIV-positive T
cell-derived exosomal miR-155-5p promoted IL-6 and IL-8 secretion
via AT-rich interaction domain 2 (ARID2) targeting and the
subsequent activation of the ERCC excision repair 5, endonuclease
(ERCC5)-NF-κB pathway, thereby accelerating epithelial-mesenchymal
transition (EMT) and enhancing cervical cancer invasiveness. In
CSCC, tumor-derived exosomal miR-223 induced IL-6 secretion from
monocytes/macrophages in vitro, creating a positive feedback
loop through STAT3 activation in CSCC cells (26). Moreover, highly metastatic HeLa
cells transferred miR-29 via exosomes to low-metastatic C-33A
cells, thereby enhancing their metastatic potential (27).
In summary, exosomal miRNAs play pivotal roles in
the progression of CC by regulating angiogenesis,
lymphangiogenesis, drug resistance and metastasis. These findings
highlight their potential as biomarkers for early diagnosis,
targets for precision therapy and indicators for prognosis
assessment in the management of CC.
3. Exosomal lncRNAs in CC
lncRNAs are ncRNAs with a length of >200
nucleotides that participate in various physiological processes,
including chromatin remodeling, epigenetic regulation,
transcriptional and post-transcriptional regulation, as well as
cell proliferation and differentiation (28). Accumulating evidence indicates that
lncRNAs are dysregulated in multiple malignant tumors and modulate
cancer cell phenotypes via the cis- and
trans-regulation of tumor-related genes, exerting oncogenic
or tumor-suppressive effects. Thus, the dysregulated expression of
lncRNAs has been proposed as one of the hallmark features of
malignant tumors (29). Previous
studies have demonstrated that lncRNAs play critical roles in CC
tumorigenesis, progression, metastasis and drug resistance,
suggesting their potential as diagnostic and prognostic biomarkers
for CC (30,31). Given these findings, exosomal
lncRNAs, functioning as intercellular communication mediators,
likely contribute to the development and progression of CC.
Exosomal lncRNAs promote the
proliferation and differentiation of CC cells
It has been well-established that cell proliferation
and differentiation are fundamental processes in the development
and progression of CC. Recent studies have demonstrated that
exosomal lncRNAs promote the proliferation and differentiation of
CC cells. For instance, it was previously demonstrated that the
level of serum exosomal lncRNA DLX6-AS1 was significantly elevated
in patients with CC compared with those with cervical
intraepithelial neoplasia (CIN) or healthy controls (32). Functionally, DLX6-AS1 enhanced the
proliferation, migration and EMT of CC cells by modulating the
miR-16-5p/CAMP-regulated phosphoprotein 1 (ARPP19) axis or
degrading fused in sarcoma protein in a xenograft mouse model
(33), suggesting that the serum
exosomal DLX6-AS1 may serve as a diagnostic and prognostic
biomarker for CC.
Additionally, lncRNAs such as HOX transcript
antisense intergenic RNA (HOTAIR) and metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) have been found to be highly
enriched in CC-derived exosomes isolated from cervicovaginal lavage
samples, with expression levels significantly higher than those in
HPV-infected patients or healthy controls (34). Mechanistically, HOTAIR and MALAT1
function as competitive endogenous RNAs (ceRNAs): HOTAIR binds to
miR-214-3p to activate the Wnt/β-catenin signaling pathway, while
MALAT1 sequesters miR-485-5p to upregulate MAT2A expression,
thereby promoting CC cell proliferation (35,36).
In a separate study, Gao et al (37) reported that exosomes derived from
CaSki cells enhanced the self-renewal and differentiation
capacities of CC stem cells, these exosomes carry lncRNA urothelial
carcinoma-associated 1 (UCA1), which functions as a ceRNA for
miR-122-5p to upregulate SRY-Box transcription factor 2 (SOX2)
expression. Notably, silencing exosomal UCA1 or overexpressing
miR-122-5p suppressed the self-renewal and differentiation of CC
stem cells, subsequently inhibiting the invasion, migration and
proliferation of CaSki cells. These interventions also induced
cancer cell apoptosis and reduced tumor volume and weight in nude
mouse models. Furthermore, lncRNA H19 was found to be upregulated
in CC cell lines and detectable in extracellular vesicles from cell
culture supernatants. Functional assays revealed that H19 promoted
cell proliferation and multicellular tumor spheroid formation
without significantly affecting apoptosis or migration, suggesting
its potential as a diagnostic and therapeutic target in CC
(38).
Exosomal lncRNAs promote
angiogenesis
Exosomal lncRNAs have been implicated in
facilitating CC metastasis through pro-angiogenic mechanisms
analogous to those mediated by miRNAs. Emerging evidence suggests
that CC-derived exosomal lncRNAs can modulate vascular endothelial
cell behavior by either upregulating pro-angiogenic factors or
sequestering anti-angiogenic miRNAs, thereby promoting tumor
vascularization (39). The seminal
study by Lei and Mou (40)
identified the lncRNA taurine upregulated 1 (TUG1) as significantly
overexpressed in both CC cells and their secreted exosomes.
Mechanistically, CC-derived exosomal TUG1 was internalized by human
umbilical vein-derived endothelial cells (HUVECs), where it
suppressed caspase-3 activity and altered the expression of
apoptosis-related proteins, ultimately enhancing endothelial cell
proliferation and survival. These findings posit exosomal TUG1 as a
potential diagnostic biomarker for early-stage CC.
Exosomal lncRNAs regulate drug
resistance
Accumulating evidence has confirmed that
tumor-derived exosomal lncRNAs play a critical role in mediating
drug resistance in cancers. In CC, the lncRNA hepatocyte nuclear
factor 1 homeobox A antisense RNA 1 (HNF1A-AS1) is highly enriched
in exosomes secreted by cervical cancer cells. Functionally,
HNF1A-AS1 acts as a ceRNA to sponge miR-34b, thereby promoting the
expression of tuftelin 1 (TUFT1). This mechanism enhances cisplatin
(DDP) resistance in CC cells, concurrently promoting proliferation,
conferring drug resistance and inhibiting apoptosis (41). Furthermore, recent studies have
revealed that exosomal MALAT1 is markedly upregulated in
DDP-resistant CC cells. The pro-resistance effect of MALAT1 can be
attenuated by targeting miR-370-3p, suggesting that the exosomal
MALAT1/miR-370-3p/STAT3 axis modulates DDP resistance via PI3K/Akt
pathway activation (42). Thus,
exosomal MALAT1 represents a potential diagnostic biomarker for
patients with CC who are resistant to DDP. Another key finding
demonstrated that lncRNA maternally expressed gene 3 (MEG3)
expression was significantly downregulated in CC tissues compared
with adjacent normal tissues. The silencing of MEG3 not only
promoted CC cell proliferation and migration, but also suppressed
apoptosis. Mechanistically, MEG3 enhanced DDP sensitivity by acting
as a ceRNA to regulate the miR-21/PTEN axis (43). Additionally, exosomal LINC01305 was
shown to contribute to chemoresistance by activating the
Wnt/β-catenin signaling pathway, thereby maintaining cancer
stemness through the upregulation of β-catenin, transcription
factor 7 and NADH dehydrogenase subunit 2(31). In summary, given the pivotal roles
of exosomal lncRNAs in modulating the tumor microenvironment and
malignant phenotypes, they emerge as promising biomarkers for CC.
Their clinical applications span early screening, therapeutic
decision-making, and prognostic evaluation, highlighting their
translational significance.
4. Exosomal circRNAs in CC
circRNAs are a newly identified class of endogenous
ncRNAs that exhibit an abundant expression in eukaryotic
transcriptomes. They are characterized by their covalently closed
circular structure, the absence of 5' caps and 3' poly(A) tails,
and exonuclease resistance due to the lack of free termini. These
features confer greater stability compared with their linear RNA
counterparts (44). Functionally,
circRNAs often contain miRNA response elements, enabling them to
modulate gene expression at transcriptional or post-transcriptional
levels through interactions with miRNAs or other RNA-binding
proteins. They can function as miRNA sponges or ceRNAs to regulate
diverse physiological and pathological processes, such as cancer
cell proliferation, invasion and metastasis. For instance,
circ_0000069 was shown to sponge miR-873-5p to de-repress tumor
suppressor candidate 3, thereby promoting CC progression (45), and circ_0005576 was demonstrated to
upregulate kinesin family member 20A by sequestering miR-153, which
accelerated the pathogenesis of CC (46). Notably, circRNAs are enriched and
stably preserved within exosomes. For example, circSLC26A4 was
detected in biofluids (such as blood and urine), rendering them
promising liquid biopsy biomarkers (47,48).
Emerging evidence highlights dysregulated exosomal circRNA profiles
in patients with CC, suggesting their clinical potential for
diagnosis and therapeutic targeting.
Exosomal circRNAs promote the EMT of
CC cells
EMT is a fundamental biological process whereby
epithelial cells undergo phenotypic conversion into mesenchymal
cells. During this process, under stimulation by external factors,
the expression of epithelial phenotypic markers is decreased, and
intercellular junctions are disrupted. Conversely, mesenchymal
phenotypic markers are re-expressed or upregulated, accompanied by
a morphological shift from a cuboidal, polarized distribution to a
spindle-shaped, randomly oriented distribution (49). As a key mechanism in cancer
progression, EMT enables epithelial-derived malignant tumor cells
to acquire migratory and invasive properties. Wang et al
(50) found that the levels of
hsa_circ_0009143 (circPVT1) in both plasma- and urine-derived
exosomes from patients with CC were significantly elevated.
Functional assays demonstrated that the overexpression of circPVT1
promoted the migration and invasion of C33A CC cells, concomitant
with the upregulation of mesenchymal markers (vimentin, N-cadherin
and Snail), and the downregulation of the epithelial marker,
E-cadherin, all established EMT biomarkers. These findings suggest
that exosomal circPVT1 may facilitate CC metastasis by activating
EMT pathways.
Exosomal circRNAs promote
angiogenesis
Exosomal circRNAs also contribute to multiple stages
of vascular development, including intercellular crosstalk, the
secretion of pro-angiogenic factors, the degradation of the
vascular basement membrane, endothelial cell proliferation, tube
formation, and alternative microvascular patterning such as
vasculogenic mimicry (51).
Previously, an in vitro study demonstrated that circ_0087432
is significantly upregulated in serum exosomes from patients with
CC. The overexpression of exosomal circ_0087432 derived from CC
cells has been shown to enhance the proliferation and migration of
HUVECs (52), indicating its
involvement in CC metastasis via angiogenesis. Wang et al
(53) found that circ_0064516 was
highly expressed in CC cells, and exosomes derived from CC cells
carried circ_0064516 to HUVECs. circ_0064516 increased
mitogen-activated protein kinase 1 (MAPK1) expression by sponging
miR-6805-3p, thereby enhancing angiogenesis.
Exosomal circRNAs regulate drug
resistance
Drug resistance remains a major challenge in cancer
therapy. Emerging evidence suggests that the aberrant expression of
exosomal circRNAs may contribute to chemoresistance. Unlike
intracellular circRNAs, exosomal circRNAs appear to be selectively
packaged and actively shuttled between exosomes and the cytoplasm
(54). Chen et al (55) analyzed 46 patients with CC who
received radiotherapy and chemotherapy combined with surgical
resection and found that circ_0074269 expression was elevated in
exosomes from DDP-resistant CC cells. Mechanistically, it may
upregulate TUFT1 by sponging miR-485-5p, thereby promoting
resistance to DDP in patients with CC. As demonstrated in another
in vivo and in vitro study, circ_0004488, as a
miR-136 sponge, increased the expression of Mex-3 RNA-binding
family member C (MEX3C), which is a direct target gene of miR-136,
ultimately increasing the resistance of CC to paclitaxel (56). These findings highlight the
potential of exosomal circRNAs as biomarkers and therapeutic
targets for CC. Further research is required however, to elucidate
their precise mechanisms in tumor invasion and metastasis, which
may provide a foundation for novel clinical interventions.
5. Conclusion and future prospects
Exosomes in malignant tumors have emerged as a key
focus of research in recent years. These vesicles facilitate
intercellular communication by transporting functional
biomolecules, including ncRNAs, which play critical roles in cancer
progression. Exosomal ncRNAs contribute to tumor growth,
proliferation, invasion, metastasis and drug resistance by
modulating key oncogenic pathways. The present review
comprehensively summarized the involvement of exosomal ncRNAs in
the development of CC, particularly their regulatory effects on
cell proliferation, differentiation, angiogenesis,
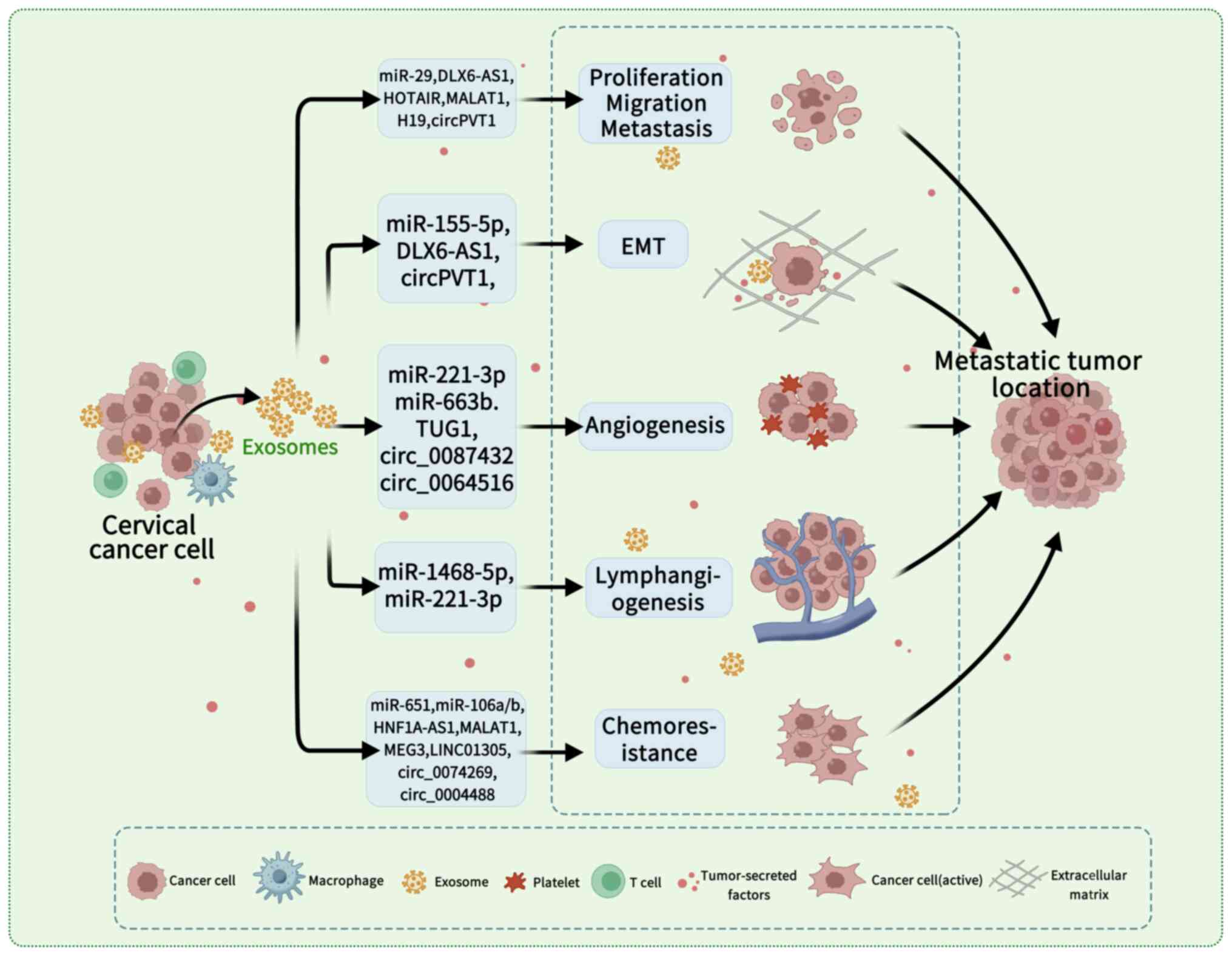
lymphangiogenesis, EMT and chemoresistance (Table I). These mechanisms collectively
promote CC initiation, progression and metastatic dissemination
(Fig. 1). However, the current
evidence base has significant limitations. A large amount of the
supporting data is derived from in vitro studies and
preclinical animal models, raising questions about the direct
translatability of these findings to the complex human tumor
microenvironment (12,23,32).
For example, numerous studies rely on HeLa cell lines, while their
well-documented genetic and phenotypic drift after decades of in
vitro culture raises questions about their representativeness
of native cancers (16,20,24).
Findings in a single cell line cannot capture the vast
heterogeneity among patients with CC. Therefore, the translational
validity of these findings in vitro is necessarily limited
at this stage. To bridge the gap between observations in
vitro and potential clinical application, a multi-tiered
validation framework is necessary. First, studies need to be
conducted in a broader range of CC cell lines to assess their
prevalence outside of a single cell line. Second, the association
between the expression levels of key molecular markers and their
clinicopathological characteristics, such as stage, grade and
prognosis need to be analyzed using patient tissue samples.
Finally, relevant animal models of xenograft tumors need to be
established to verify the effectiveness and safety of targeted
intervention in an in vivo environment. Furthermore,
attributing specific biological effects solely to exosomal ncRNAs
remains challenging due to the heterogeneity of exosome populations
and the potential co-isolation of contaminating non-exosomal
vesicles or free ncRNAs using common isolation techniques. The
functional validation of specific exosomal ncRNAs in driving CC
phenotypes often lacks rigorous genetic gain/loss-of-function
experiments within the relevant cellular context.
 | Table ISummary of exosomal non-coding RNAs
in cervical cancer. |
Table I
Summary of exosomal non-coding RNAs
in cervical cancer.
| Category | Specific
molecule | Function | Mechanism of
action | (Refs.) |
|---|
| miRNAs | miR-221-3p | Promotes
angiogenesis | Inhibits MAPK10,
stimulating endothelial cell proliferation and migration. | (16) |
| | miR-663b | Promotes
angiogenesis | Suppresses
vinculin, altering endothelial adhesion and promoting vascular
network formation. | (17) |
| | miR-1468-5p | Promotes
lymphangio-genesis and immune evasion | Suppresses
HMBOX1-SOCS1, activating JAK2/STAT3 to impair CD8+ T-cell
immunity. | (19) |
| | miR-221-3p | Promotes
lymphangiogenesis | Downregulates
VASH1, stimulating lymphatic endothelial cell migration and tube
formation. | (20) |
| | miR-651 | Suppresses
cisplatin resistance | Directly targets
ATG3 to inhibit autophagy and reduce drug resistance. | (23) |
| | miR-106a/b | Modulates cisplatin
sensitivity | Transferred via
exosomes to alter drug response in recipient cells. | (24) |
| | miR-155-5p | Promotes EMT and
invasion | Targets ARID2,
activating the ERCC5-NF-κB pathway. | (25) |
| | miR-223 | Promotes TME
inflammation | Induces IL-6
secretion from monocytes/macrophages, creating a STAT3-mediated
feedback loop. | (26) |
| | miR-29 | Promotes
metastasis | Transfer from high-
to low-metastatic cells enhances their metastatic potential. | (27) |
| lncRNAs | DLX6-AS1 | Promotes
proliferation, migration, and EMT | Modulates the
miR-16-5p/ARPP19 axis or degrades the FUS protein. | (32) |
| | HOTAIR | Promotes
proliferation | Functions as a
ceRNA for miR-214-3p, activating the Wnt/β-catenin pathway. | (35) |
| | MALAT1 | Promotes
proliferation | Functions as a
ceRNA for miR-485-5p, upregulating MAT2A expression. | (36) |
| | UCA1 | Promotes stemness
and differentiation | Functions as a
ceRNA for miR-122-5p, upregulating SOX2. | (37) |
| | H19 | Promotes
proliferation | Mechanism not fully
elucidated; enhances spheroid formation. | (38) |
| | TUG1 | Promotes
angiogenesis | Suppresses
caspase-3 activity, enhancing endothelial cell survival and
proliferation. | (40) |
| | HNF1A-AS1 | Promotes cisplatin
resistance | Functions as a
ceRNA for miR-34b, upregulating TUFT1. | (41) |
| | MALAT1
(Resistance) | Promotes cisplatin
resistance | Regulates the
miR-370-3p/STAT3 axis, activating the PI3K/Akt pathway. | (42) |
| | MEG3 | Enhances cisplatin
sensitivity | Functions as a
ceRNA to regulate the miR-21/PTEN axis, suppressing
proliferation. | (43) |
| | LINC01305 | Promotes
chemoresistance | Activates
Wnt/β-catenin signaling to maintain cancer stemness. | (31) |
| circRNAs | circPVT1 | Promotes EMT and
metastasis | Upregulates
mesenchymal markers (vimentin, N-cadherin) and downregulates
E-cadherin. | (50) |
| | circ_0087432 | Promotes
angiogenesis | Enhances the
proliferation and migration of HUVECs. | (52) |
| | circ_0064516 | Promotes
angiogenesis | Sponges
miR-6805-3p, upregulating MAPK1. | (53) |
| | circ_0074269 | Promotes cisplatin
resistance | Sponges miR-485-5p,
leading to TUFT1 upregulation. | (55) |
| | circ_0004488 | Promotes paclitaxel
resistance | Sponges miR-136,
upregulating its target MEX3C. | (56) |
Compared to synthetic nanoparticles, exosomes
exhibit superior biocompatibility, low immunogenicity, high
stability and intrinsic targeting capabilities, rendering them
promising candidates for clinical applications. Nevertheless,
translating this promise into reality faces substantial hurdles
beyond the mentioned technical challenges. First of all,
standardization is the cornerstone. The Minimal Information for
Studies of Extracellular Vesicles (MISEV) 2023 guidelines launched
by the International Society for Extracellular Vesicles (ISEV)
provide a more comprehensive framework for research; however, their
full adoption and practice still require time, which is a
prerequisite for ensuring data comparability and repeatability
(57). Secondly, the path to
clinical transformation is a complex one. The recent rejection of
Deramiocel, the first exosome mechanism therapy, by the FDA,
profoundly reveals the high standards of regulatory authorities for
substantive efficacy evidence (from rigorously designed controlled
trials) and a robust Chemistry, Manufacturing and Controls (CMC)
system, sounding the alarm for clinical development strategies
across the entire field. Furthermore, the regulatory framework is
evolving rapidly. Countries worldwide have clearly included
exosomes in the management of advanced therapeutic drugs, laying a
regulatory foundation for their development as drugs. This also
implies higher technical thresholds and quality requirements.
Finally, a critical concern is the potential for tumor-derived
exosomes themselves to exert pro-tumorigenic effects; using them as
therapeutic carriers requires meticulous engineering to eliminate
residual oncogenic cargo and ensure safety. The scalability and
cost-effectiveness of producing clinical-grade exosomes at
sufficient quantities and purity for widespread therapeutic use are
also major unresolved issues. Additionally, the immunogenicity of
exosomes, while generally low, can vary significantly, depending on
the source cell type and isolation method, necessitating careful
evaluation for each application. However, several additional
challenges need to be addressed before their translation into
therapeutic use, including the standardization of exosome isolation
and storage protocols, optimization of drug-loading efficiency and
precise control over cargo release kinetics. Furthermore, the
current understanding of exosome biogenesis, particularly the
spatiotemporal regulation of exosomal cargo sorting, molecular
transport mechanisms and definitive exosome markers, remains
limited. Crucially, current knowledge of the in vivo
biodistribution, pharmacokinetics and long-term fate of
administered therapeutic exosomes in humans remains limited, posing
significant barriers to clinical trial design and regulatory
approval.
In conclusion, future studies are required to focus
on elucidating these aspects while also prioritizing the
development of robust methods to track exosome delivery and
function in vivo, and to rigorously assess the safety
profile of engineered exosomes in relevant models, to harness the
full therapeutic potential of exosomes in oncology. Further
research is warranted to provide a more in-depth understanding of
the in vivo and molecular mechanisms of exosomes in
scientific research. Technically, it is of utmost significance to
develop efficient and controllable engineering transformation and
large-scale production processes. The highest standards of
randomized controlled trials and strict quality control norms need
to be followed in clinical translation. In terms of regulations, a
more detailed and scientific evaluation system needs to be
established. By overcoming these challenges, the marked therapeutic
potential of exosomes may prove to be beneficial to human
health.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
82002760).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XX and HG conceived the study that led to the
acquisition of data from the literature, and drafted the
manuscript. ML revised the language of the manuscript. WG and KW
designed the outline of the review and revised the manuscript. All
authors have read and approved the final manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Raghani NR, Chorawala MR, Parekh K, Sharma
A, Alsaidan OA, Alam P, Fareed M and Prajapati B: Exosomal
miRNA-based theranostics in cervical cancer: Bridging diagnostics
and therapy. Med Oncol. 42(193)2025.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Filho AM, Laversanne M, Ferlay J, Colombet
M, Piñeros M, Znaor A, Parkin DM, Soerjomataram I and Bray F: The
GLOBOCAN 2022 cancer estimates: Data sources, methods, and a
snapshot of the cancer burden worldwide. Int J Cancer.
156:1336–1346. 2025.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mileshkin LR, Moore KN, Barnes EH, Gebski
V, Narayan K, King MT, Bradshaw N, Lee YC, Diamante K, Fyles AW, et
al: Adjuvant chemotherapy following chemoradiotherapy as primary
treatment for locally advanced cervical cancer versus
chemoradiotherapy alone (OUTBACK): An international, open-label,
randomised, phase 3 trial. Lancet Oncol. 24:468–482.
2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367(eaau6977)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hao Y, Song H, Zhou Z, Chen X, Li H, Zhang
Y, Wang J, Ren X and Wang X: Promotion or inhibition of
extracellular vesicle release: Emerging therapeutic opportunities.
J Control Release. 340:136–148. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Thakur A, Ke X, Chen YW, Motallebnejad P,
Zhang K, Lian Q and Chen HJ: The mini player with diverse
functions: Extracellular vesicles in cell biology, disease, and
therapeutics. Protein Cell. 13:631–654. 2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Naderi-Meshkin H, Lai X, Amirkhah R, Vera
J, Rasko JEJ and Schmitz U: Exosomal lncRNAs and cancer: Connecting
the missing links. Bioinformatics. 35:352–360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ran Z, Wu S, Ma Z, Chen X, Liu J and Yang
J: Advances in exosome biomarkers for cervical cancer. Cancer Med.
11:4966–4978. 2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Diener C, Keller A and Meese E: Emerging
concepts of miRNA therapeutics: From cells to clinic. Trends Genet.
38:613–626. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kang C, Duo Y, Zheng L, Zhao N, Wang J,
Liu Z, Qiu L and Bi F: CAFs-derived exosomes promote the
development of cervical cancer by regulating miR-18a-5p-TMEM170B
signaling axis. Biochem Biophys Res Commun.
694(149403)2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nagamitsu Y, Nishi H, Sasaki T, Takaesu Y,
Terauchi F and Isaka K: Profiling analysis of circulating microRNA
expression in cervical cancer. Mol Clin Oncol. 5:189–194.
2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Pohlers M, Gies S, Taenzer T, Stroeder R,
Theobald L, Ludwig N, Kim YJ, Bohle RM, Solomayer EF, Meese E, et
al: Th17 cells target the metabolic miR-142-5p-succinate
dehydrogenase subunit C/D (SDHC/SDHD) axis, promoting invasiveness
and progression of cervical cancers. Mol Oncol. 18:2157–2178.
2024.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mitra T and Elangovan S: Cervical cancer
development, chemoresistance, and therapy: A snapshot of
involvement of microRNA. Mol Cell Biochem. 476:4363–4385.
2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Dittmer J and Leyh B: Paracrine effects of
stem cells in wound healing and cancer progression (Review). Int J
Oncol. 44:1789–1798. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jayson GC, Kerbel R, Ellis LM and Harris
AL: Antiangiogenic therapy in oncology: Current status and future
directions. Lancet. 388:518–529. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang L, Li H, Yuan M, Li M and Zhang S:
Cervical cancer cells-secreted exosomal microRNA-221-3p promotes
invasion, migration and angiogenesis of microvascular endothelial
cells in cervical cancer by down-regulating MAPK10 expression.
Cancer Manag Res. 11:10307–10319. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
You X, Sun W, Wang Y, Liu X, Wang A, Liu
L, Han S, Sun Y, Zhang J, Guo L and Zhang Y: Cervical
cancer-derived exosomal miR-663b promotes angiogenesis by
inhibiting vinculin expression in vascular endothelial cells.
Cancer Cell Int. 21(684)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lehuédé C, Dupuy F, Rabinovitch R, Jones
RG and Siegel PM: Metabolic plasticity as a determinant of tumor
growth and metastasis. Cancer Res. 76:5201–5208. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhou C, Wei W, Ma J, Yang Y, Liang L,
Zhang Y, Wang Z, Chen X, Huang L, Wang W and Wu S: Cancer-secreted
exosomal miR-1468-5p promotes tumor immune escape via the
immunosuppressive reprogramming of lymphatic vessels. Mol Ther.
29:1512–1528. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhou CF, Ma J, Huang L, Yi HY, Zhang YM,
Wu XG, Yan RM, Liang L, Zhong M, Yu YH, et al: Cervical squamous
cell carcinoma-secreted exosomal miR-221-3p promotes
lymphangiogenesis and lymphatic metastasis by targeting VASH1.
Oncogene. 38:1256–1268. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bhattacharjee M, Ghosh A, Das S, Sarker S,
Bhattacharya S, Das A, Ghosh S, Chattopadhyay S, Ghosh S and
Adhikary A: systemic codelivery of thymoquinone and doxorubicin by
targeted mesoporous silica nanoparticle sensitizes
doxorubicin-resistant breast cancer by interfering between the
MDR1/P-gp and miR 298 crosstalk. ACS Biomater Sci Eng.
10:6314–6331. 2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Havryliuk V, Wojtowicz K, Gagat M and
Żuryń A: Exosome-mediated mechanisms of drug resistance in lung
cancer: Molecular mechanisms and therapeutic strategies. Cell
Physiol Biochem. 59:358–374. 2025.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhu X, Long L, Xiao H and He X:
Cancer-derived exosomal miR-651 as a diagnostic marker restrains
cisplatin resistance and directly targets ATG3 for cervical cancer.
Dis Markers. 2021(1544784)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Raji GR, Sruthi TV, Edatt L, Haritha K,
Sharath Shankar S and Sameer Kumar VB: Horizontal transfer of
miR-106a/b from cisplatin resistant hepatocarcinoma cells can alter
the sensitivity of cervical cancer cells to cisplatin. Cell Signal.
38:146–158. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li H, Chi X, Li R, Ouyang J and Chen Y:
HIV-1-infected cell-derived exosomes promote the growth and
progression of cervical cancer. Int J Biol Sci. 15:2438–2447.
2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang J, Jiang M, Qian L, Lin X, Song W,
Gao Y and Zhou Y: The STAT3-miR-223-TGFBR3/HMGCS1 axis modulates
the progression of cervical carcinoma. Mol Oncol. 14:2313–2331.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cui HJ, Zhang YN and Li HT: Mechanism of
exosome miRNA29 in cervical cancer metastasis. Chin J Cancer Prev
Treat. 25:1365–1370. 2018.
|
|
28
|
Cheng T and Huang S: Roles of non-coding
RNAs in cervical cancer metastasis. Front Oncol.
11(646192)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Eptaminitaki GC, Stellas D, Bonavida B and
Baritaki S: Long non-coding RNAs (lncRNAs) signaling in cancer
chemoresistance: From prediction to druggability. Drug Resist
Updat. 65(100866)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
He J, Huang B, Zhang K, Liu M and Xu T:
Long non-coding RNA in cervical cancer: From biology to therapeutic
opportunity. Biomed Pharmacother. 127(110209)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Huang X, Liu X, Du B, Liu X, Xue M, Yan Q,
Wang X and Wang Q: LncRNA LINC01305 promotes cervical cancer
progression through KHSRP and exosome-mediated transfer. Aging
(Albany NY). 13:19230–19242. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ding XZ, Zhang SQ, Deng XL and Qiang JH:
Serum exosomal lncRNA DLX6-AS1 Is a promising biomarker for
prognosis prediction of cervical cancer. Technol Cancer Res Treat.
20(1533033821990060)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xie F, Xie G and Sun Q: Long noncoding RNA
DLX6-AS1 promotes the progression in cervical cancer by targeting
miR-16-5p/ARPP19 axis. Cancer Biother Radiopharm. 35:129–136.
2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhang J, Liu SC, Luo XH, Tao GX, Guan M,
Yuan H and Hu DK: Exosomal long noncoding RNAs are differentially
expressed in the cervicovaginal lavage samples of cervical cancer
patients. J Clin Lab Anal. 30:1116–1121. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhou Y, Wang Y, Lin M, Wu D and Zhao M:
LncRNA HOTAIR promotes proliferation and inhibits apoptosis by
sponging miR-214-3p in HPV16 positive cervical cancer cells. Cancer
Cell Int. 21(400)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tie W and Ge F: MALAT1 inhibits
proliferation of HPV16-positive cervical cancer by sponging
miR-485-5p to promote expression of MAT2A. DNA Cell Biol.
40:1407–1417. 2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Gao Z, Wang Q, Ji M, Guo X, Li L and Su X:
Exosomal lncRNA UCA1 modulates cervical cancer stem cell
self-renewal and differentiation through microRNA-122-5p/SOX2 axis.
J Transl Med. 19(229)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Iempridee T: Long non-coding RNA H19
enhances cell proliferation and anchorage-independent growth of
cervical cancer cell lines. Exp Biol Med (Maywood). 242:184–193.
2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhang W, Wang Q, Yang Y, Zhou S, Zhang P
and Feng T: The role of exosomal lncRNAs in cancer biology and
clinical management. Exp Mol Med. 53:1669–1673. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lei L and Mou Q: Exosomal taurine
up-regulated 1 promotes angiogenesis and endothelial cell
proliferation in cervical cancer. Cancer Biol Ther. 21:717–725.
2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Luo X, Wei J, Yang FL, Pang XX, Shi F, Wei
YX, Liao BY and Wang JL: Exosomal lncRNA HNF1A-AS1 affects
cisplatin resistance in cervical cancer cells through regulating
microRNA-34b/TUFT1 axis. Cancer Cell Int. 19(323)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hu Y, Li G, Ma Y, Luo G, Wang Q and Zhang
S: Effect of exosomal lncRNA MALAT1/miR-370-3p/STAT3 positive
feedback loop on PI3K/Akt pathway mediating cisplatin resistance in
cervical cancer cells. J Oncol. 2023(6341011)2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Du Y, Geng G, Zhao C, Gao T and Wei B:
LncRNA MEG3 promotes cisplatin sensitivity of cervical cancer cells
by regulating the miR-21/PTEN axis. BMC Cancer.
22(1145)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Verduci L, Strano S, Yarden Y and Blandino
G: The circRNA-microRNA code: Emerging implications for cancer
diagnosis and treatment. Mol Oncol. 13:669–680. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhang S, Chen Z, Sun J, An N and Xi Q:
CircRNA hsa_circRNA_0000069 promotes the proliferation, migration
and invasion of cervical cancer through miR-873-5p/TUSC3 axis.
Cancer Cell Int. 20(287)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Ma H, Tian T, Liu X, Xia M, Chen C, Mai L,
Xie S and Yu L: Upregulated circ_0005576 facilitates cervical
cancer progression via the miR-153/KIF20A axis. Biomed
Pharmacother. 118(109311)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Tong Y, Jia L, Li M, Li H and Wang S:
Identification of exosomal circSLC26A4 as a liquid biopsy marker
for cervical cancer. PLoS One. 19(e0305050)2024.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Preußer C, Hung LH, Schneider T, Schreiner
S, Hardt M, Moebus A, Santoso S and Bindereif A: Selective release
of circRNAs in platelet-derived extracellular vesicles. J Extracell
Vesicles. 7(1424473)2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84.
2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wang H, Wei M, Kang Y, Xing J and Zhao Y:
Circular RNA circ_PVT1 induces epithelial-mesenchymal transition to
promote metastasis of cervical cancer. Aging (Albany NY).
12:20139–20151. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhang XP, Pei JP, Zhang CD, Yusupu M, Han
MH and Dai DQ: Exosomal circRNAs: A key factor of tumor
angiogenesis and therapeutic intervention. Biomed Pharmacother.
156(113921)2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Xia H, Yang X, Chen Y, Li H and Chu GF:
Effect of cervical cancer-derived exosomal hsa_circ_0087432 on the
proliferation and migration of HUVECs. Med J Chin People's Liber
Army. 46:327–332. 2021.
|
|
53
|
Wang Y, Xie Y, Wang X, Yang N, Wu Z and
Zhang X: Tumor cells-derived extracellular vesicles carry
circ_0064516 competitively inhibit microRNA-6805-3p and promote
cervical cancer angiogenesis and tumor growth. Expert Opin Ther
Targets. 28:97–112. 2024.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18(116)2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen J, Wu S, Wang J, Sha Y and Ji Y:
Hsa_circ_0074269-mediated upregulation of TUFT1 through miR-485-5p
increases cisplatin resistance in cervical cancer. Reprod Sci.
29:2236–2250. 2022.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yi H, Han Y, Li Q, Wang X, Xiong L and Li
S: Circular RNA circ_0004488 increases cervical cancer paclitaxel
resistance via the miR-136/MEX3C signaling pathway. J Oncol.
2022(5435333)2022.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Welsh JA, Goberdhan DCI, O'Driscoll L,
Buzas EI, Blenkiron C, Bussolati B, Cai H, Di Vizio D, Driedonks
TAP, Erdbrügger U, et al: Minimal information for studies of
extracellular vesicles (MISEV2023): From basic to advanced
approaches. J Extracell Vesicles. 13(e12404)2024.PubMed/NCBI View Article : Google Scholar
|