Introduction
Oral cancer is a significant public health issue,
with approximately 300,000 new cases reported annually worldwide
(1). Approximately 90% of oral
cancer pathologies are squamous cell carcinomas (SCCs). Oral
squamous cell carcinoma (OSCC) shows high incidence and mortality
rates, particularly in developing countries. Although the
development of treatment strategies has been remarkable, the 5-year
survival rate often remains below 50% (2). Despite the good local control of
OSCC, distant metastases often result in poor outcomes. In the
existing standard treatment regimen for OSCC, surgery is the first
choice, and chemoradiotherapy is strongly recommended as
postoperative adjuvant therapy in patients with a high risk of
postoperative recurrence. High-risk factors for recurrence include
T size, resection margin positivity, extranodal extension, and
multiple lymph node metastases. However, only a few factors can be
used to evaluate future metastasis.
Liquid biopsy has attracted attention for its
usefulness in various types of cancers, including colorectal and
breast cancers. Its utility in OSCC is anticipated. In a previous
study, associations were reported between the number of circulating
tumour cells (CTC) clusters and prognosis, as well as between the
size and quantity of cfDNA and prognosis (3). In this study, next-generation
sequencing (NGS) analysis of cfDNA was performed in two patients
with inferior prognoses due to distant metastases. They were
compared with mutation data from a clinical cancer panel test and
two commonly mutated genes were detected, TP53 and
CDKN2A, in both cases. CDKN2A and TP53 are
frequently mutated genes in head and neck squamous cell carcinoma
(HNSCC) (4) and OSCC (5). CDKN2A, or p16, is an essential
tumour suppressor gene that regulates cell growth by preventing the
progression from the G1 phase to the S phase of the cell cycle
(6). TP53 is a suppressor
gene referred to as ‘the guardian of the gene’. TP53 responds to
various cellular stresses to control the expression of target
genes, which subsequently trigger cell cycle arrest, apoptosis,
senescence, or DNA repair. The IHC for CDKN2A and TP53 in patients
with pathologically positive lymph node metastases were performed,
thereby offering a new prognostic feature for metastasis.
Materials and methods
cfDNA isolation and NGS
As reported previously (3), peripheral blood (PB) was obtained
from patients 1 and 2 prior to surgery; patient 1 (48 years old,
male, Tongue cancer, Stage IVA) and patient 2 (23 years old, male,
Tongue cancer, Stage IVA) were recruited in March and January 2023,
respectively, and the blood samples were collected at Hiroshima
University Hospital. cfDNA was extracted using the MagMAX cfDNA
isolation kit (Thermo Fisher Scientific, Waltham, MA, USA)
following the manufacturer's protocol. NGS was carried out by
Macrogen Inc. (Seoul, Korea) using the Axen Cancer Panel 2
(Table SI). First, shared mutated
genes were identified between cfDNA mutation data and tissue
mutation data from a clinical cancer panel test within the same
patient. Next, the commonalities between patients 1 and 2 were
determined. The clinical relevance of the variants was identified
using the ClinVar or OncoKB database (7).
Patients and specimens
Overall, 537 patients with oral cancer visited the
Department of Oral Maxillofacial Surgery at Hiroshima University
Hospital between January 2014 and December 2023. Of them, 481
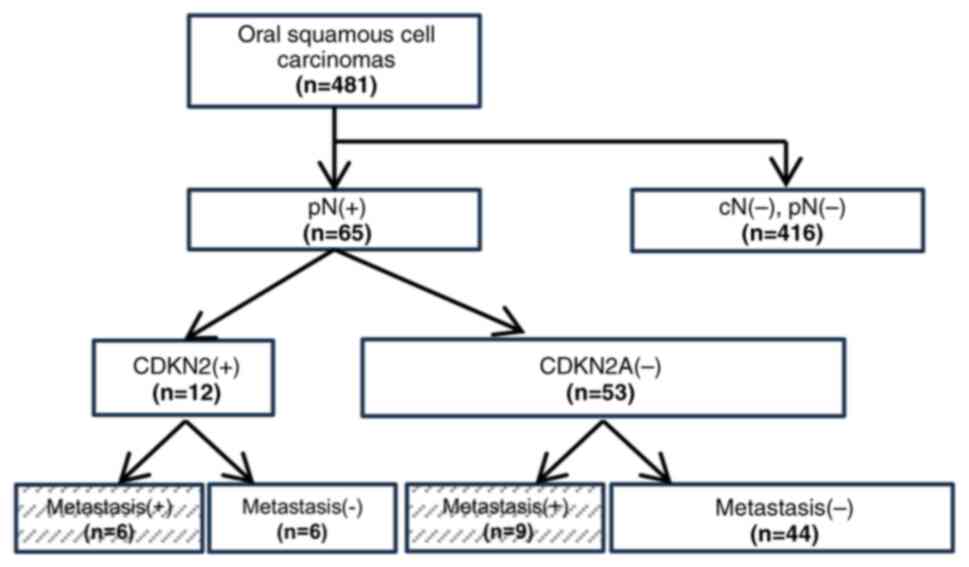
patients underwent radical surgery. Among the 481 patients, 65 with
pathologically positive lymphoid metastases (pN(+)) were selected
(Fig. 1) (Table SII). The medical records and
tissue samples of the patients included in this study were accessed
specifically for this research starting in May 2023. Inclusion
criteria included (1) absence of
distant metastases at diagnosis, (2) receipt of surgical treatment, and
(3) complete follow-up data.
Exclusion criteria included (1)
distant metastases at diagnosis, (2) non-surgical treatment, and (3) incomplete follow-up data. All medical
records, including disease characteristics, diagnostic methods, and
treatments, were retrospectively reviewed. All patients were
diagnosed according to the TNM classification of oral cancer
(8). The clinical endpoints of
this study were based on the FDA's ‘Clinical Trial Endpoints for
the Approval of Cancer Drugs and Biologics’ guidelines, which
include a 5-year evaluation period. The primary endpoint was
overall survival (OS). OS was defined as the time from the date of
the initial diagnosis to death from any cause.
IHC for CDKN2A
IHC for CDKN2A was performed as previously described
(9). Briefly, 4-µm formalin-fixed
paraffin-embedded tissue sections on amino silane-coated glass
slides (MATSUNAMI, Osaka, Japan) were deparaffinised in xylene
(FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) and
rehydrated using an alcohol gradient (FUJIFILM Wako). Antigen
retrieval was performed using 10 mM sodium citrate buffer (pH 6.0)
in an autoclave (HV-5; HIRAYAMA, Saitama, Japan). Sections were
incubated overnight at 4˚C with a 1:100 dilution of mouse
monoclonal anti-CDKN2SA/p16INK4A antibody (sc-56330, Santa Cruz
Biotechnology, TX, USA) or a 1:100 dilution of mouse monoclonal
anti-p53 antibody (DO-1, Santa Cruz Biotechnology). Detection was
performed using EnVision+ System-HRP anti-mouse (Dako, Agilent
Technologies, Santa Clara, CA, USA) for 30 min, followed by
visualisation with 3,3'-diaminobenzidine (DAB) (Dako) and
counterstaining with haematoxylin (FUJIFILM Wako). The slides were
dehydrated, mounted, and covered with coverslips (NEO cover glass;
MATSUNAMI). Images were captured using a Nikon DS-Ri2 camera (Nikon
Corporation, Tokyo, Japan) and a Nikon Eclipse E800 microscope
(Nikon Corporation).
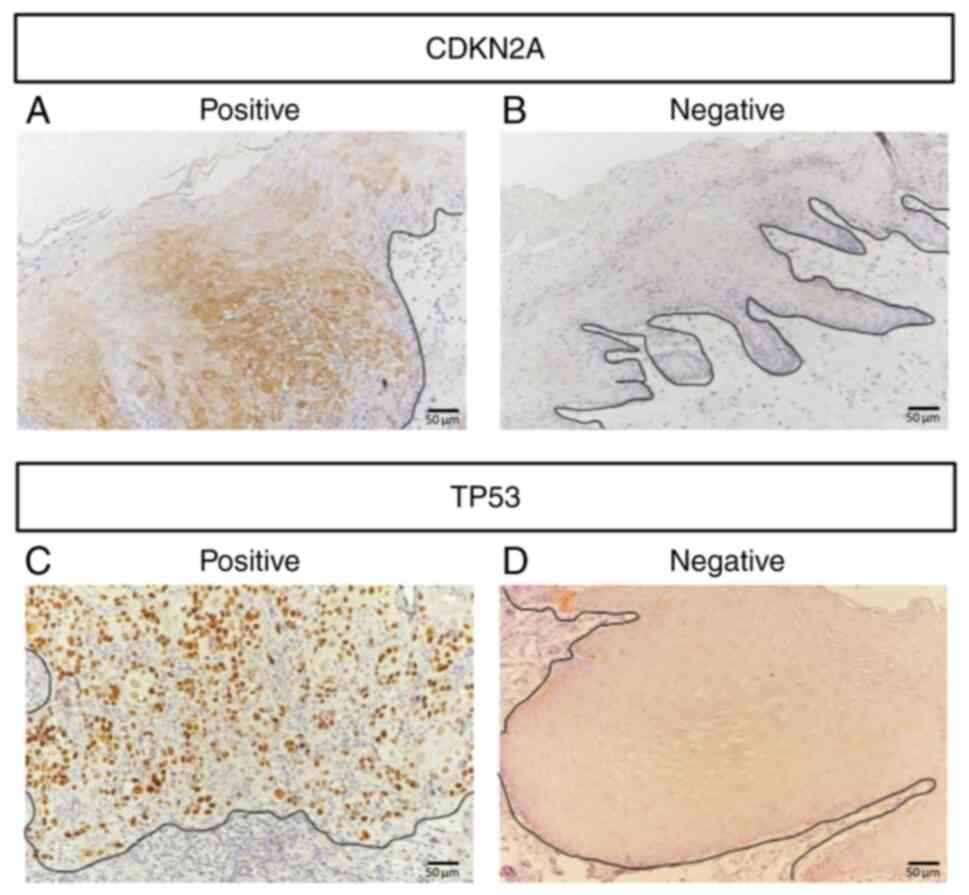
Classification of IHC positivity
Immunohistochemical staining was evaluated based on
the intensity and percentage of positively stained cells. Cases
were considered ‘positive’ when the CDKN2A-positive region was
expressed more than 70%, and the TP53-positive region was expressed
more than 60%, according to previously reported thresholds
(10-12).
Ethical consideration
This study was approved by the Ethics Committee of
Hiroshima University (approval numbers: epidemiology 2016-9191 and
epidemiology 2023-0025).
Statistical analysis
All statistical analyses were conducted using JMP
Pro (version 16.2.0, SAS Institute Inc., Cary, NC, USA). The
association between OS and factors and the IHC results of
CDKN2A/TP53 expression and OS were examined using Kaplan-Meier
survival curves and the log-rank test. To further assess the
prognostic significance of CDKN2A positivity, a multivariate Cox
proportional hazards regression analysis was performed.
Results
Result of NGS
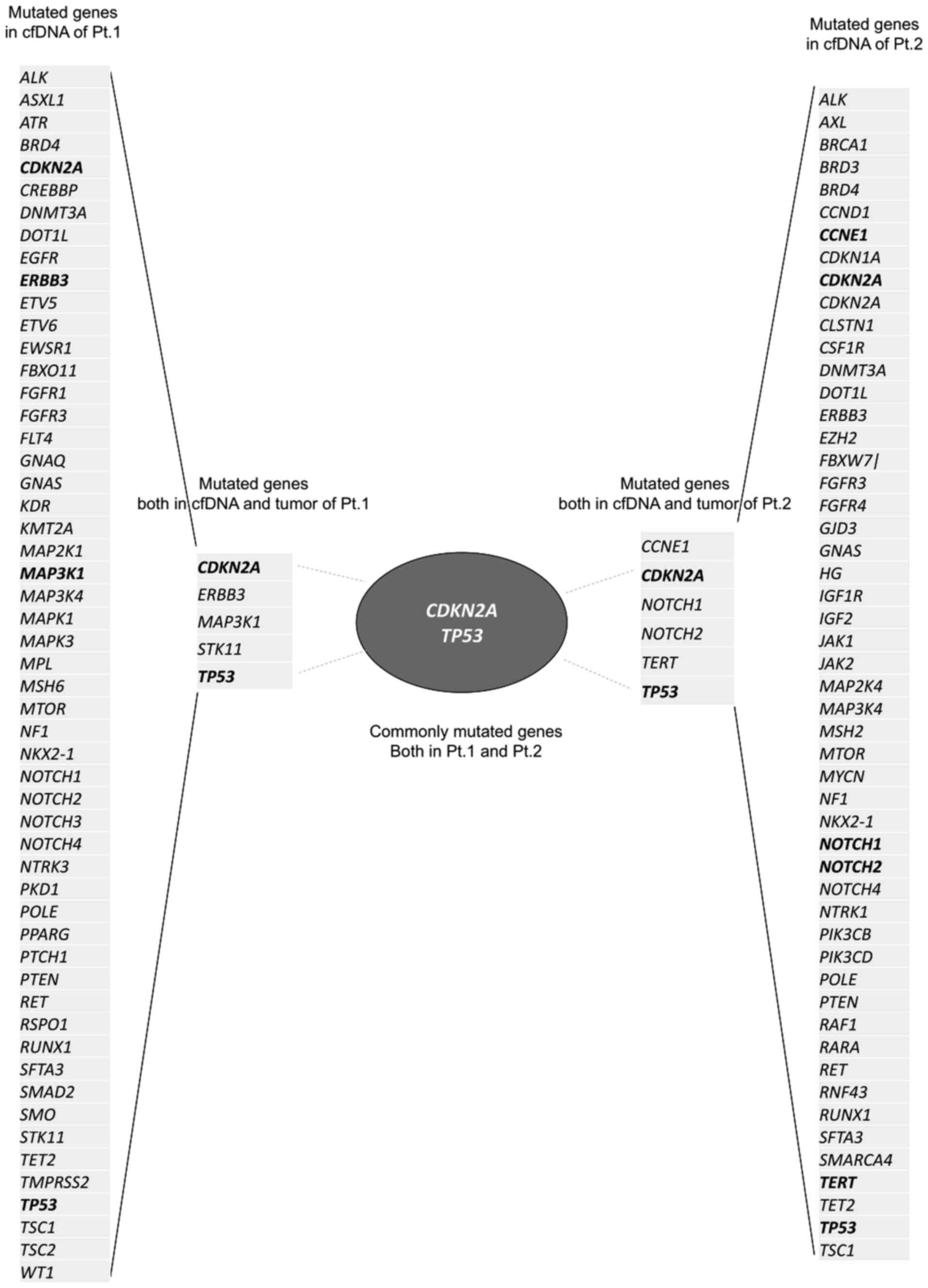
cfDNA NGS was performed for patients 1 and 2 as
preliminary exploratory observations. Forty-one and 40 genetic
mutations were detected in cfDNAs of patients 1 and 2,
respectively. Some common tumour tissue mutations were previously
detected in each patient. CDKN2A, ERBB3,
MAP3K1, STK11, and TP53 were common mutations
identified in patient 1. In patient 2, CCNE1, CDKN2A,
NOTCH1, NOTCH2, TERT, and TP53 were
common between tissues and cfDNA. Among these, two genes,
CDKN2A and TP53, were shared between patients 1 and 2
(Fig. 2). Specifically,
TP53_c.524G>A_p.R175H/CDKN2A_ c.340C>T_p. P114 and
TP53_c.569delC_p.P190fs*57, _c.733G>A
_p.G245S/CDKN2A_c.247C>A_p. H83N were shared both in tumour
tissue and cfDNA, respectively, in patients 1 and 2 (Table I). Although the positions of the
mutated base differed between patients 1 and 2, they were all
pathological mutations. Therefore, the relationship between IHC
staining for CDKN2A and TP53 and OSCC prognosis was
investigated.
 | Table ITP53 and CDKN2A mutations in cfDNA and
tumour. |
Table I
TP53 and CDKN2A mutations in cfDNA and
tumour.
| Patient no. | Origin | Gene | Protein change | Annotation | AF (%) |
|---|
| 1 | Tumour | TP53 | R175H | Pathogenic | 62.8 |
| | | CDKN2A | P114S | Pathogenic/VUS | 58.1 |
| | cfDNA | TP53 | R175H | Pathogenic | 1.6 |
| | | | G245D | Pathogenic | 0.3 |
| | | CDKN2A | P114S | Pathogenic/VUS | 1.8 |
| 2 | Tumour | TP53 | P190fs*57 | Pathogenic | 28.5 |
| | | | G245S | Pathogenic | 12.3 |
| | | CDKN2A | H83N | Likely
Pathogenic | 56 |
| | cfDNA | TP53 | P190fs*57 | Pathogenic | 14.3 |
| | | | G245S | Pathogenic | 6.7 |
| | | CDKN2A | H83N | Likely
pathogenic | 24.3 |
| | | | G89C | VUS | 23.2 |
Patient characteristics
The characteristics of the 65 patients with SCC
included in this study are summarised in Table II. Of the 65 patients, 43 (66.2%)
were males and 22 (33.8%) were females; the average age was 68
years. The primary tumour sites included the tongue (32.3%),
mandibular gingiva (27.7%), the floor of the mouth (16.9%), buccal
mucosa (10.8%), maxillary gingiva (10.8%), and others (1.5%).
Tumour size distribution showed that 17 patients (26.2%) had
Tis/T1/T2 tumours, and 48 patients (73.8%) had T3/T4 tumours.
 | Table IIPatient characteristics. |
Table II
Patient characteristics.
| Characteristic | Number | % |
|---|
| Age (median),
years | 68 (21-89) | - |
| Sex | | - |
|
Male | 43 | 66.2 |
|
Female | 22 | 33.8 |
| Site | | |
|
Tongue | 21 | 32.3 |
|
Mandibular
gingiva | 18 | 27.7 |
|
Floor of
mouth | 11 | 16.9 |
|
Buccal
mucosa | 7 | 10.8 |
|
Maxillary
gingiva | 7 | 10.8 |
|
Other | 1 | 1.5 |
| Tumour size | | |
|
Tis/T1-2 | 17 | 26.2 |
|
T3-4 | 48 | 73.8 |
| Metastasis | | |
|
No | 49 | 75.4 |
|
Yes | 16 | 24.6 |
CDKN2A-positive group shows metastasis
in 50% of OSCC cases
IHC images were assessed, as shown in Fig. 3. CDKN2A expression was positive in
12 patients (18.5%) and negative in 53 (81.5%). Of the
CDKN2A-positive and CDKN2A-negative patients, six (6/12, 50%) and
nine (9/53, 16.9%) showed metastasis (Fig. 1). In the Fisher's exact test for
distant metastasis, the IHC CDKN2A-positive group showed a
significantly higher metastasis rate (Table III).
 | Table IIIFisher's exact test for distant
metastasis and other factors. |
Table III
Fisher's exact test for distant
metastasis and other factors.
| Characteristic | No metastasis, n
(%) | Metastasis, n
(%) | Total, n (%) | P-value |
|---|
| Sex | | | | |
|
Male | 32 (49.2) | 10 (15.4) | 42 (62.6) | 1.0 |
|
Female | 18 (27.7) | 5 (7.7) | 23 (35.4) | |
| Age | | | | |
|
≤65 | 23 (35.4) | 3 (4.6) | 26 (40.0) | 0.06 |
|
>65 | 27 (41.5) | 12 (18.5) | 39 (60.0) | |
| Tumor size | | | | |
|
Tis/T1-T2 | 15 (23.1) | 2 (3.1) | 17 (26.2) | 0.2 |
|
T3-T4 | 35 (53.9) | 13 (20.0) | 48 (73.9) | |
| IHC (TP53) | | | | |
|
Negative | 29 (44.6) | 7 (10.8) | 36 (55.4) | 0.46 |
|
Positive | 21 (32.3) | 8 (12.3) | 29 (44.6) | |
| IHC (CDKN2A) | | | | |
|
Negative | 44 (67.7) | 9 (13.9) | 53 (81.5) | 0.046 |
|
Positive | 6 (9.2) | 6 (9.2) | 12 (18.5) | |
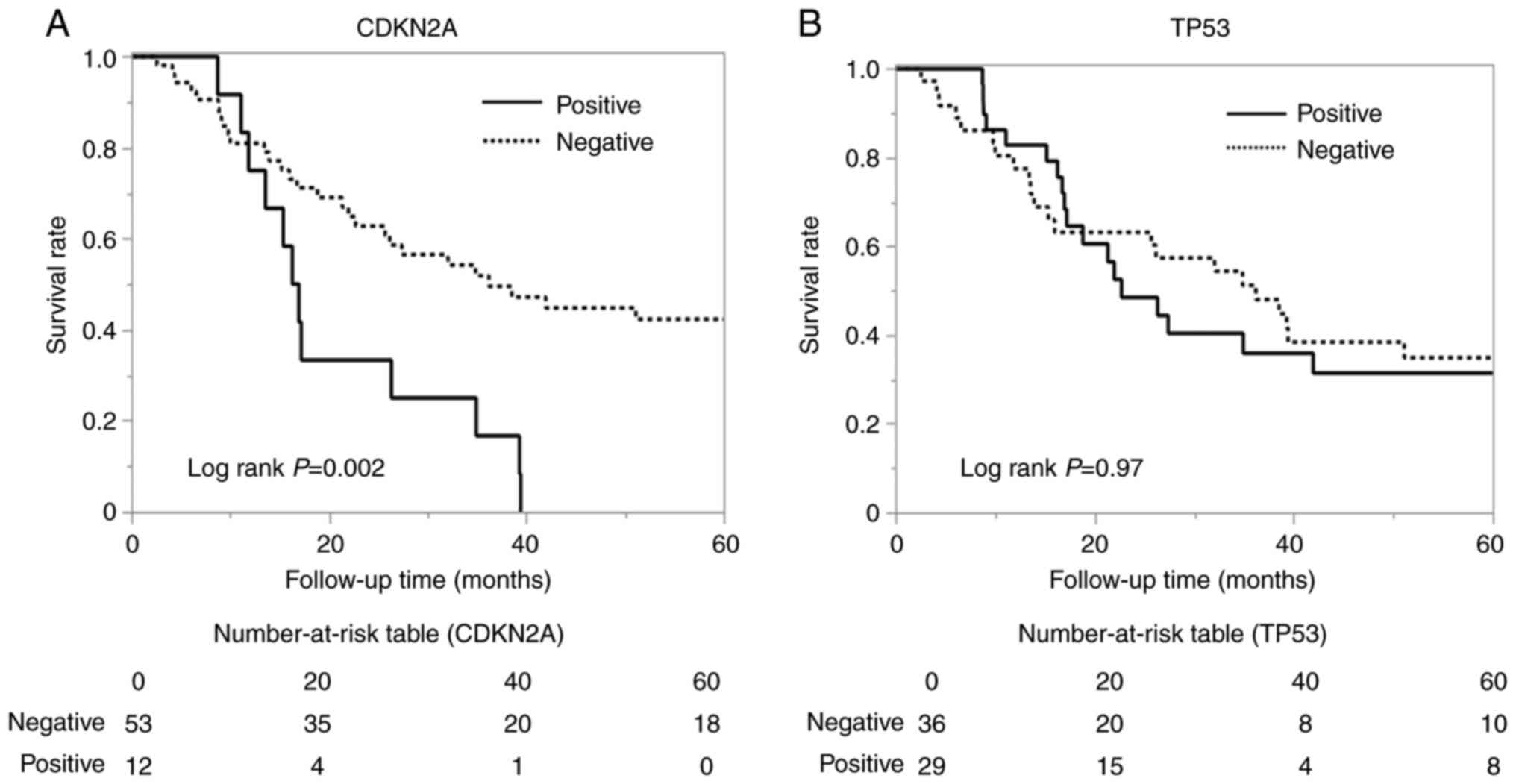
CDKN2A-positive group shows shorter OS
in OSCC
Five-year survival rates were examined based on the
positive and negative immunohistochemical results for CDKN2A and
TP53. The CDKN2A-positive group showed a significantly decreased
survival rate (hazard ratio=3.63, 95% CI: 1.36-9.68, P=0.01)
(Fig. 4A), but there were no
significant differences for TP53 (Fig.
4B). Furthermore, the results of the multivariate cox
proportional hazards model indicated that CDKN2A positivity
remained a significant independent predictor of poor prognosis
after adjusting IHC results, age, smoking history, alcohol dose,
and tumour size (hazard ratio=1.75, 95% CI: 017-1.60,
P-value=0.0179) (Table IV).
 | Table IVMultivariable cox proportional
hazards model for overall survival. |
Table IV
Multivariable cox proportional
hazards model for overall survival.
| Variable | HR | 95% CI | P-value |
|---|
| P16 positivity ≥70%
(yes vs. no) | 1.75 | 0.17-1.60 | 0.0179 |
| TP53 positivity
≥60% (yes vs. no) | 0.01 | -0.64-0.64 | 0.9787 |
| Age >60 years
(yes vs. no) | 0.95 | -0.12-1.16 | 0.1126 |
| Smoking history
(never vs. ever) | 0.27 | -0.56-1.22 | 0.5321 |
| Alcohol dose (never
vs. ever) | 0.05 | -0.95-1.01 | 0.8980 |
| T stage (T1-2 vs.
T3-4) | 0.13 | -0.72-0.55 | 0.7415 |
Discussion
This study was designed based on the commonalities
of tumour tissue and cfDNA sequencing data to identify new
prognostic features of metastasis. Most of the mutations detected
in cfDNA sequencing are not pathogenic mutations. However, a few
pathogenic mutated genes, TP53 and CDKN2A, were
common in both tumour tissues and cfDNA. TP53_p.R175H/_p.G245S
mutation, previously reported pathogenic variant, is located in the
DNA binding domain, resulting in loss of function. TP53_p.R175H is
found in acute myeloid leukaemia (13). TP53_p.G245S has been frequently
found in hepatocellular carcinoma (14). TP53_p.P190fs*57 is a truncated
mutation, resulting in the loss of function. Those truncated
mutations show a relatively strong association with poor prognosis
in HNSCC (15). CDKN2A_p.
P114S/_p. H83N mutation is located in the ankyrin repeats of the
p16/INK4A protein, resulting in loss of function. CDKN2A_p. P114S,
previously reported pathogenic variant, was identified as a
germline variant in families affected by melanoma (16). Though TP53_c.569delC_p.P190fs*57
and CDKN2A_c.247C>A_p. H83N are not reported mutations, they are
predicted as pathogenic mutation in silico analysis using mutation
taster (https://www.mutationtaster.org/index.html).
cfDNA was previously assessed at two points, pre-
and post-operation, and reported that the group with distant
metastases showed significantly higher amounts of cfDNA than those
without distant metastases both preoperatively and postoperatively
(3). In reference 3, we showed
that though only three genes (TP53, HRAS, and MLH1) were classified
as pathogenic in 13 patients, two patients, who commonly possessed
TP53_c.215C>G_p.Pro72Arg, showed short-term distant metastasis.
Despite the limited overlap in mutations among patients, these
findings underscore the clinical relevance of cfDNA mutation
profiling (3). If the same genetic
mutations in cfDNA can be detected both preoperatively and
postoperatively or the comparison of those genetic mutations can be
performed, it would further support the clinical relevance of such
findings.
Specifically, the relationship between CDKN2A/TP53
IHC expression and prognosis was investigated in 65 patients with
pN(+) OSCC. pN(+) was selected to evaluate the high-risk group for
metastasis because, as commonly reported, the pN(+) group had a
higher distant metastasis rate of 22% (15/65), than those of the
pN(-) group, 5% (2/40) (Table
SII). The following were key findings: in 65 patients with
pN(+) OSCC, CDKN2A expression was positive in 18.5% (12/65) of the
patients and negative in 81.5% (53/65), whereas no such difference
was found for TP53. The CDKN2A-positive group resulted in
metastasis in 50% of cases (6/12). This result contradicts our
expectations. Regarding the oropharynx, Fakhry et al
(17) assessed whether HPV
infection was present or absent and concluded that the HPV-positive
group (CDKN2A-positive group) in the oropharynx had a better
prognosis, showing better radiosensitivity. This present case may
have been different because it consisted of 86.2% (56/65) cases of
surgical alone and 13.8% (9/65) surgical plus adjuvant radiation
therapy with or without chemotherapy. Hong et al (18) reported that, in oropharyngeal
cancer, while surgery alone cases were deemed insufficient in
number for their multivariable analysis, HPV status predicts better
outcomes treated with surgery plus adjuvant radiotherapy as well as
with definitive radiation therapy with or without chemotherapy. In
Fakhry et al (17) and Hong
et al (18), the assessment
was not based on IHC-CHKN2A negativity or positivity but also
considered the presence of HPV16 DNA using in situ hybridisation or
PCR. Ni et al (19)
reported that CDKN2A overexpression decoupled from HPV infection
was not a prognostic marker for patients with OSCC.
CDKN2A is widely recognised as a tumour
suppressor gene that controls cell proliferation by preventing the
progression from the G1 phase to the S phase of the cell cycle
(20). In normal cells with
proliferative potential, the expression of CDKN2A is very low, and
CDKN2A has little function. However, when normal cells reach the
mitotic lifespan or undergo oncogenic stress, CDKN2A gene
expression is markedly elevated, and cellular senescence occurs
(21). Beyond phenomena related to
invasion and metastasis, Shi et al (22), reported that in the colorectal
cancer cell line, HT-29, CDKN2A could induce the
epithelial-mesenchymal transition, showing that knock downed CDKN2A
expression was followed by enhanced E-cadherin expression and
suppression of N-cadherin and vimentin expression. Cheng et
al (23), reported that CDKN2A
mediates cuproptosis, a type of cell death characterised by
excessive copper-lipid reactions in the tricarboxylic acid cycle,
resistance through regulating glycolysis and copper homeostasis,
accompanied by a malignant phenotype and pro-tumour niche, also
using colorectal cancer cell line. They concluded that radiation
and chemotherapy are expected to potentially serve as therapeutic
approaches for cuproptosis-resistant colorectal cancer with high
CDKN2A expression (23). Given the
limited literature investigating these phenomena in OSCC, future
studies should aim to 1) elucidate the molecular pathways by which
CDKN2A influences cell motility, epithelial-mesenchymal transition,
and invasion using in vitro OSCC models 2) evaluate
metastatic potential in vivo between CDKN2A-over expressing
and CDKN2A-deficient OSCC xenografts 3) use single-cell
transcriptomics or proteomics analyses to gain deeper insight into
how CDKN2A expression modulates cellular behavior at the invasive
front of OSCC lesions.
Despite these promising findings, the limitations of
this study warrant further consideration. First, the retrospective
nature of this study might have induced selection bias, as only
patients who underwent surgery and had available pathology reports
were included. Second is the lack of HPV status assessment. Since
CDKN2A (p16) overexpression is often considered a surrogate marker
for HPV-related oncogenesis, especially in head and neck cancers,
the inability to account for HPV infection status may confound the
interpretation of CDKN2A expression in this cohort. Although the
prognostic significance of CDKN2A expression has been well
established in oropharyngeal cancers, particularly in HPV-positive
cases, less attention has been given to CDKN2A's role in OSCC. Most
studies focus on the strong association between p16 expression and
HPV status in oropharyngeal cancer, where p16 overexpression is
often considered a surrogate marker for HPV infection. However,
OSCC, which is primarily HPV-negative, presents a different
biological context. Future studies should incorporate HPV testing
to better clarify this relationship. This study addresses this gap
by investigating the independent role of CDKN2A expression in OSCC,
irrespective of HPV infection. Evidence that CDKN2A dysregulation
in OSCC may be associated with tumour progression, metastasis, and
poorer prognosis was provided, independent of HPV status. This
finding is particularly important as it expands the understanding
of CDKN2A as a prognostic marker in head and neck cancers beyond
HPV-positive oropharyngeal cancers. Results suggest that CDKN2A
could serve as a valuable biomarker in OSCC, offering insights into
its potential role in cancer progression and therapeutic targeting.
Third is the sample size. Because the sample size is very small,
two samples for NGS and twelve samples for IHC CDKN2A-positive
patients, these results should be seen as preliminary and
interpreted with caution. Although further studies involving a
larger patient cohort are necessary to confirm these preliminary
observations, these findings suggest a potential prognostic role
for CDKN2A expression. Assessing CDKN2A expression could help
stratify patients based on their risks and tailor treatment
strategies accordingly. For example, the CDKN2A-positive group,
which we assumed to be a high-metastasis group, could be managed
with more aggressive treatment regimens or closer follow-up.
Further research will contribute to the development of personalised
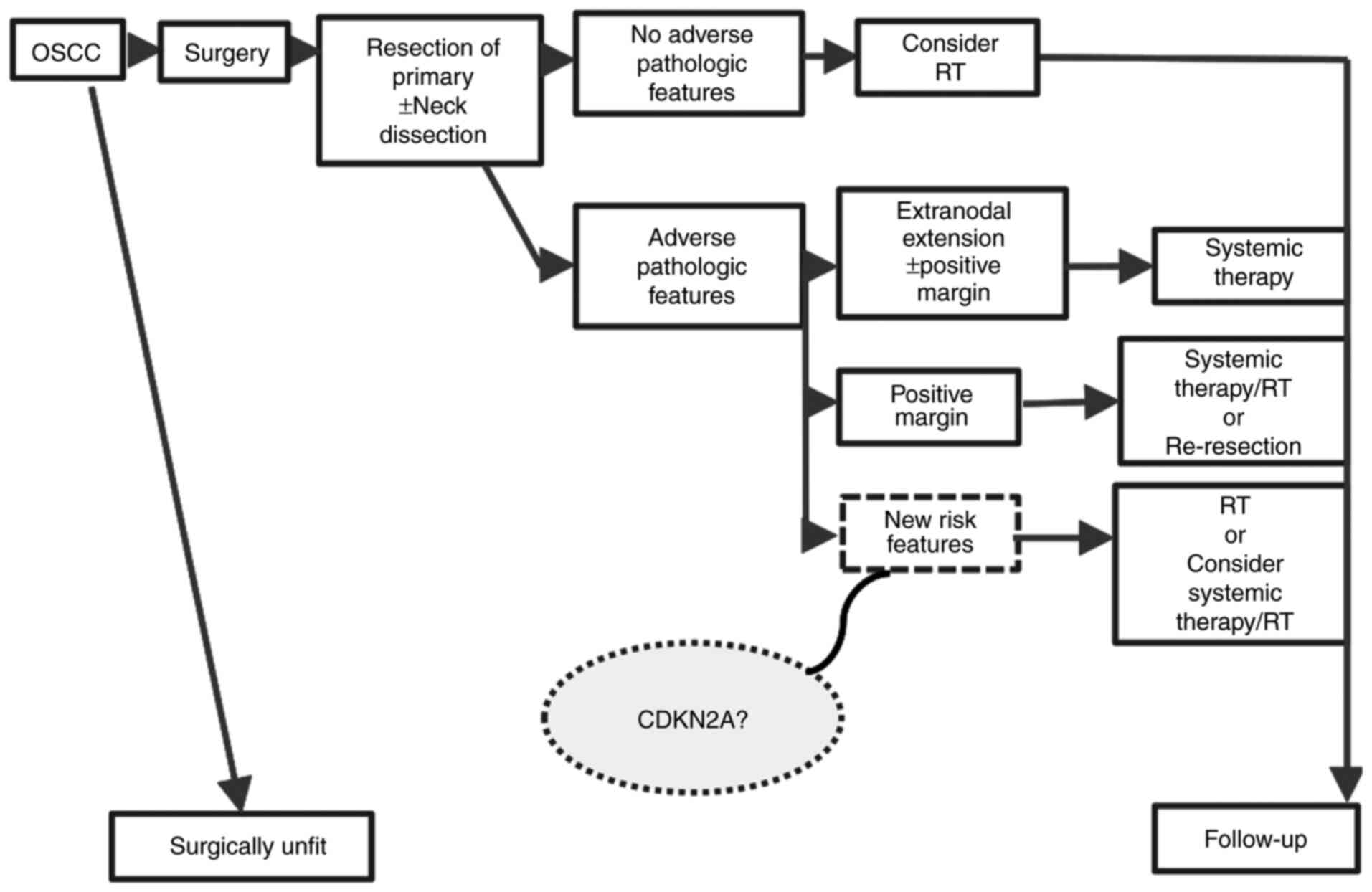
cancer therapies and improve OSCC prognosis (Fig. 5).
This study addresses this gap by investigating the
independent role of CDKN2A expression in OSCC, irrespective of HPV
infection. We provide evidence that CDKN2A dysregulation in OSCC
may be associated with tumour progression, metastasis, and poorer
prognosis, independent of HPV status. This finding is particularly
important as it expands the understanding of CDKN2A as a prognostic
marker in head and neck cancers beyond HPV-positive oropharyngeal
cancers. The results suggest that CDKN2A could serve as a valuable
biomarker in OSCC, offering insights into its potential role in
cancer progression and therapeutic targeting. In conclusion, this
study demonstrates that the CDKN2A-positive group had a high
metastasis rate, resulting in a poorer prognosis. CDKN2A expression
is a potential prognostic marker for OSCC.
Supplementary Material
Genes included in Cancer Axen Panel
2.
Patient characteristics.
Acknowledgements
We are grateful to Professor T. Hinoi, Dr H. Niitsu
and Dr H. Nakahara (Department of Clinical and Molecular Genetics,
Hiroshima University Hospital, Hiroshima, Japan) for kindly
allowing us to share the clinical cancer panel test and advising us
for the ethical application.
Funding
Funding: This research was supported in part by Grants-in-Aid
for Scientific Research (B) (grant no. 22H03292), Grant-in-Aid for
Scientific Research (C) (grant no. 22K10146), and Grant-in-Aid for
Scientific Research (C) (grant no. 22K10148) from the Japanese
Ministry of Education, Culture, Sports, Science and Technology.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The following three
mutations, NM_000546.6(TP53):c.524G>A (p.Arg175His),
NM_000546.6(TP53):c.733G>A (p.Gly245Ser) and
NM_000077.5(CDKN2A):c.340C>T (p.Pro114Ser), are reported
mutations and their URLs are as follows, https://www.ncbi.nlm.nih.gov/clinvar/RCV000013173.22/,
https://www.ncbi.nlm.nih.gov/clinvar/variation/12365/,
and https://www.ncbi.nlm.nih.gov/clinvar/RCV001915587/,
respectively. The NGS data have been deposited with links to
Biosample accession number SAMD01605716 (patient 1) and
SAMD01611905 (patient 2) in the DDBJ Biosample database, https://ddbj.nig.ac.jp/search/entry/biosample/SAMD01605716
and https://ddbj.nig.ac.jp/search/entry/biosample/SAMD01611905.
Authors' contributions
SoY and AH designed the experiments. NE, AH, AT, MH,
FO, NI, SaY, RT, TS, KK and SoY performed the experiments. NE, AH
and SoY confirmed the authenticity of all the raw data, and wrote
and reviewed the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The Research Ethics Board of Hiroshima University
approved this study. The studies were conducted in accordance with
the local legislation and institutional requirements. Patients 1
and 2 provided written informed consent to participate in the
study, which was approved by the Ethics Committee of Hiroshima
University (approval no. epidemiology 2016-9191). For IHC, written
informed consent was not obtained from patients for publication
because this was a retrospective study. This retrospective
observational study was approved by the Research Ethics Board of
Hiroshima University (approval no. epidemiology 2023-0025).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Ervik M, Lam F, Laversanne M,
Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I and Bray F:
Global cancer observatory: Lip, oral cavity. International Agency
for Research on Cancer, Lyon, 2024. https://gco.iarc.who.int/today/en/fact-sheets-cancers.
|
|
2
|
Bosetti C, Carioli G, Santucci C,
Bertuccio P, Gallus S, Garavello W, Negri E and La Vecchia C:
Global trends in oral and pharyngeal cancer incidence and
mortality. Int J Cancer. 147:1040–1049. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Eboshida N, Hamada A, Higaki M, Obayashi
F, Ito N, Yamasaki S, Tani R, Shintani T, Koizumi K and Yanamoto S:
Potential role of circulating tumor cells and cell-free DNA as
biomarkers in oral squamous cell carcinoma: A prospective
single-center study. PLoS One. 19(e0309178)2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Morris LGT, Chandramohan R, West L, Zehir
A, Chakravarty D, Pfister DG, Wong RJ, Lee NY, Sherman EJ, Baxi SS,
et al: The molecular landscape of recurrent and metastatic head and
neck cancers: Insights from a precision oncology sequencing
platform. JAMA Oncol. 3:244–255. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Saito Y, Kage H, Kobayashi K, Yoshida M,
Fukuoka O, Yamamura K, Mukai T, Oda K and Yamasoba T: TERT promoter
mutation positive oral cavity carcinomas, a clinically and
genetically distinct subgroup of head and neck squamous cell
carcinomas. Head Neck. 45:3107–3118. 2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rayess H, Wang MB and Srivatsan ES:
Cellular senescence and tumor suppressor gene p16. Int J Cancer.
130:1715–1725. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Landrum MJ, Lee JM, Benson M, Brown GR,
Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al:
ClinVar: Improving access to variant interpretations and supporting
evidence. Nucleic Acids Res. 46 (D1):D1062–D1067. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lydiatt WM, Patel SG, O'Sullivan B,
Brandwein MS, Ridge JA, Migliacci JC, Loomis AM and Shah JP: Head
and Neck cancers-major changes in the American joint committee on
cancer eighth edition cancer staging manual. CA Cancer J Clin.
67:122–137. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nguyen TQ, Hamada A, Yamada K, Higaki M,
Shintani T, Yoshioka Y, Toratani S and Okamoto T: Enhanced KRT13
gene expression bestows radiation resistance in squamous cell
carcinoma cells. In Vitro Cell Dev Biol Anim. 57:300–314.
2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shelton J, Purgina BM, Cipriani NA, Dupont
WD, Plummer D and Lewis JS Jr: p16 immunohistochemistry in
oropharyngeal squamous cell carcinoma: A comparison of antibody
clones using patient outcomes and high-risk human papillomavirus
RNA status. Mod Pathol. 30:1194–1203. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ribeiro EA and Maleki Z: p16
immunostaining in cytology specimens: Its application, expression,
interpretation, and challenges. J Am Soc Cytopathol. 10:414–422.
2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ogino S, Kawasaki T, Kirkner GJ, Yamaji T,
Loda M and Fuchs CS: Loss of nuclear p27 (CDKN1B/KIP1) in
colorectal cancer is correlated with microsatellite instability and
CIMP. Mod Pathol. 20:15–22. 2007.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Loizou E, Banito A, Livshits G, Ho YJ,
Koche RP, Sánchez-Rivera FJ, Mayle A, Chen CC, Kinalis S, Bagger
FO, et al: A gain-of-function p53-mutant oncogene promotes cell
fate plasticity and myeloid leukemia through the pluripotency
factor FOXH1. Cancer Discov. 9:962–979. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yu X, Mao SQ, Shan YY, Huang Y, Wu SD and
Lu CD: Predictive value of the TP53. p.G245S mutation frequency for
the short-term recurrence of hepatocellular carcinoma as detected
by pyrophosphate sequencing. Genet Test Mol Biomarkers. 26:476–484.
2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lindenbergh-van der Plas M, Brakenhoff RH,
Kuik DJ, Buijze M, Bloemena E, Snijders PJ, Leemans CR and
Braakhuis BJ: Prognostic significance of truncating TP53 mutations
in head and neck squamous cell carcinoma. Clin Cancer Res.
17:3733–3741. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kannengiesser C, Brookes S, del Arroyo AG,
Pham D, Bombled J, Barrois M, Mauffret O, Avril MF, Chompret A,
Lenoir GM, et al: Functional, structural, and genetic evaluation of
20 CDKN2A germ line mutations identified in melanoma-prone families
or patients. Hum Mutat. 30:564–574. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fakhry C, Westra WH, Li S, Cmelak A, Ridge
JA, Pinto H, Forastiere A and Gillison ML: Improved survival of
patients with human papillomavirus-positive head and neck squamous
cell carcinoma in a prospective clinical trial. J Natl Cancer Inst.
100:261–269. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hong AM, Dobbins TA, Lee CS, Jones D,
Harnett GB, Armstrong BK, Clark JR, Milross CG, Kim J, O'Brien CJ
and Rose BR: Human papillomavirus predicts outcome in oropharyngeal
cancer in patients treated primarily with surgery or radiation
therapy. Br J Cancer. 103:1510–1517. 2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ni Y, Zhang X, Wan Y, Dun Tang K, Xiao Y,
Jing Y, Song Y, Huang X, Punyadeera C and Hu Q: Relationship
between p16 expression and prognosis in different anatomic subsites
of OSCC. Cancer Biomark. 26:375–383. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liggett WH Jr and Sidransky D: Role of the
p16 tumor suppressor gene in cancer. J Clin Oncol. 16:1197–1206.
1998.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hara E, Smith R, Parry D, Tahara H, Stone
S and Peters G: Regulation of p16CDKN2 expression and its
implications for cell immortalization and senescence. Mol Cell
Biol. 16:859–867. 1996.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shi WK, Li YH, Bai XS and Lin GL: The cell
cycle-associated protein CDKN2A may promotes colorectal cancer cell
metastasis by inducing epithelial-mesenchymal transition. Front
Oncol. 12(834235)2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cheng X, Yang F, Li Y, Cao Y, Zhang M, Ji
J, Bai Y, Li Q, Yu Q and Gao D: The crosstalk role of CDKN2A
between tumor progression and cuproptosis resistance in colorectal
cancer. Aging (Albany NY). 16:10512–10538. 2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Joint Committee for the Revision of the
Japanese Oral Cancer Treatment Guidelines: The Clinical Practice
Guidelines for Oral Cancer. Kanehara & Co., Ltd., Tokyo,
pp19-62, 2023. https://www.sciencedirect.com/science/article/pii/S0901502724004466.
|