Introduction
Colorectal cancer (CRC) represents a pressing global
health burden. Contemporary CRC management adopts a multimodal
approach integrating surgical resection, chemotherapy,
radiotherapy, immunotherapy, and complementary modalities such as
traditional Chinese medicine (TCM) (1). Surgical intervention remains the
cornerstone for localized disease, while systemic therapy is
predominantly based on chemotherapy regimens incorporating
fluorouracil, oxaliplatin and irinotecan. Radiotherapy plays a
crucial role in the management of rectal cancer, particularly in
the neoadjuvant setting where it reduces local recurrence (3). Recent advances in immunotherapy,
especially for mismatch repair-deficient tumors, have shown
encouraging clinical outcomes (4).
Additionally, TCM and novel nanomedicine approaches are being
increasingly explored as complementary approaches to enhance
treatment efficacy and reduce side effects (5,6).
Despite these diverse treatment options, CRC remains one of the
most common malignancies globally and ranks as the third leading
cause of cancer-related mortality (7). In the United States alone, ~153,020
new CRC cases were projected in 2023, with 52,550 deaths, including
19,550 cases and 3,750 deaths among individuals under 50 years of
age (8). Although regular
screening, surveillance and high-quality treatment can prevent a
substantial proportion of CRC morbidity and mortality (9), recurrence and metastasis remain
frequent. Among patients with metastatic CRC, ~70-75% survive
beyond 1 year, 30-35% remain alive at 3 years, while the 5-year
survival rate falls below 20% (10). Therefore, there is an urgent need
to identify reliable biomarkers that can enable early and accurate
prognostic assessment.
In a variety of solid tumors, both the density and
polarization status of TAMs are strongly associated with patient
prognostic outcomes. In CRC, high infiltration of M1-polarized TAMs
is associated with a favorable prognosis across various disease
stages (11), whereas elevated
levels of M2 TAMs often portend a poorer prognosis, a phenomenon
observed in multiple cancer types, including thyroid, lung, stomach
and breast cancers (12). While
macrophage enrichment is usually linked to adverse tumor outcomes,
this correlation appears to be reversed in CRC (13). Nevertheless, the role of TAMs
within the CRC tumor microenvironment (TME) remains complex, and
some studies have shown that TAMs are associated with poor
prognosis in patients with CRC (14). Several investigations have found
that CD68+ TAMs are predominantly distributed in the CRC
tumor stroma, especially at the invasive front, and that
CD68+ TAMs infiltration at these sites correlates with
improved prognosis in patients with CRC (15). However, TAMs of distinct subtypes
and spatial distributions exert different prognostic significance
in CRC: For instance, infiltration of CD68+ TAMs and
M2-type TAMs is linked to unfavorable outcomes (16). Collectively, TAMs play diverse
roles in the prognosis of CRC, and their number, subtype, and
spatial distribution within the TME, as well as their impact on
patient outcomes, are complex. These findings emphasize the
importance of in-depth studies of TAM properties and the search for
specific TAM markers to inform therapeutic strategies and improve
prognostic assessment.
In recent years, cancer stem cells (CSCs) have been
extensively studied as key drivers underlying core hallmarks of
tumor progression, including distant metastasis, recurrence and
drug resistance (17). CSCs
exhibit long-term self-renewal capacity and metastatic potential
across a variety of malignancies, including CRC (18). In CRC,
CD133+/CD44+ CSCs populations have been shown
to correlate negatively with both disease-free survival (DFS) and
overall survival (OS) (19).
Combined detection of CD133/CD44 expression enhances the
identification of colorectal CSCs (20). The TME engages in complex crosstalk
with CSCs, which are often characterized by immune suppression and
low immunogenicity in CRCs (21).
Furthermore, Luo et al (22) reported that CSCs can recruit TAMs
into the TME and accelerate their polarization into tumor-promoting
phenotypes, whereas TAMs in turn maintain CSC stemness and
construct niches unfavorable to the survival of patients with CSC.
Although the prognostic values of TAMs or CSC markers have been
investigated individually, studies analyzing their combined effects
and synergistic interactions on patient prognosis are scarce. Most
previous studies have examined TAMs or CSC markers in isolation,
failing to capture the prognostic power embedded within their
interplay. Therefore, it was hypothesized that a combined biomarker
panel reflecting this TAM-CSC synergy may provide superior
prognostic stratification compared with individual markers
alone.
In the present study, the expression of key TAM
markers (CD86 for M1-like and CD163 for M2-like phenotypes) and CSC
markers (CD44 and CD133) was simultaneously investigated in a
cohort of patients with CRC. The primary objective of the present
study was to evaluate the prognostic impact of their combined
expression and to determine whether this synergistic biomarker
panel could serve as an independent predictor of survival,
potentially offering a more refined tool for risk assessment in
CRC.
Materials and methods
Patients and specimens
A total of 71 patients with CRC who underwent
surgical resection from April 2018 to April 2020 at Luoyang Central
Hospital Affiliated with Zhengzhou University (Luoyang, China) were
included in the present study. In addition, 20 adjacent normal
tissue samples were collected as controls. Inclusion criteria were
as follows: i) age ≥18 years; ii) availability of complete clinical
and pathological data; and iii) follow-up duration >6 months.
Exclusion criteria included: i) receipt of neoadjuvant therapy
prior to surgery; ii) presence of other synchronous malignancies;
and iii) incomplete medical records or loss to follow-up.
Clinicopathological parameters, including age, sex, tumor
differentiation, depth of invasion and TNM stage, were obtained
from pathology reports and electronic surgical records. All cases
were histologically confirmed as primary colorectal adenocarcinoma.
Patients were followed up until April 10, 2024. Tissue samples from
these patients were subsequently subjected to immunohistochemical
(IHC) analysis. The study was approved (approval no.
LWLL-2018-03-07-01) by the Institutional Review Board and Human
Ethics Committee of Luoyang Central Hospital Affiliated with
Zhengzhou University. Written informed consent was obtained from
all participants or their legal guardians prior to inclusion in the
study.
IHC
For IHC reactions, tissue specimens were fixed in
10% buffered formalin for 24-48 h, embedded in paraffin, and
sectioned at a thickness of 4 µm. Sections were dewaxed in xylene
and rehydrated through a graded series of ethanol. Endogenous
peroxidase activity was blocked by incubating sections in 3%
hydrogen peroxide (H2O2) for 10 min, followed
by microwave heating for 3 min to reduce nonspecific binding
activity. Nonspecific binding sites were further blocked with 5%
bovine serum albumin (Boster Biological Technology) at 37˚C or 30
min. Slides were then incubated overnight at 4˚C with primary
antibodies against CD86 (1:200; cat. no. 26903-1-AP), CD163 (1:200;
cat no. 16646-1-AP), CD44 (1:200; cat no. 15675-1-AP), or CD133
(1:200; cat no. 18470-1-AP; all from Proteintech Group, Inc.).
While CD86 is expressed on various antigen-presenting cells,
including dendritic cells, its expression on TAMs has been well
documented in CRC and serves as a valuable marker for assessing
antitumor immune responses within the TME (23). The current analysis specifically
focused on CD86+ cells within the tumor stroma and their
correlation with clinical outcomes. The next day, slides were
washed three times with phosphate-buffered saline and then treated
with a biotin-labeled secondary antibody (cat. no. BA1003; Boster
Biological Technology) for 30 min at room temperature, followed by
treatment with streptavidin-biotin complex. Slides were then washed
and stained with 3,3 -diaminobenzidine (cat. no. AR1022;
Bausch Biotechnology, Inc.), counterstained with hematoxylin,
dehydrated, and sealed with neutral resin. Immunostaining for CD86,
CD163, CD44 and CD133 was quantified after digital scanning under
consistent lighting conditions. IHC staining for all markers was
quantified using Image-Pro Plus 6.0 software (Media Cybernetics,
Inc.). The staining intensity was graded on a 0-3 scale by two
independent pathologists, and a final immunoreactivity score (range
0-300) was calculated for each sample by multiplying the intensity
score by the percentage of positive cells (0-100%).
Statistical analyses
All statistical analyses were conducted using SPSS
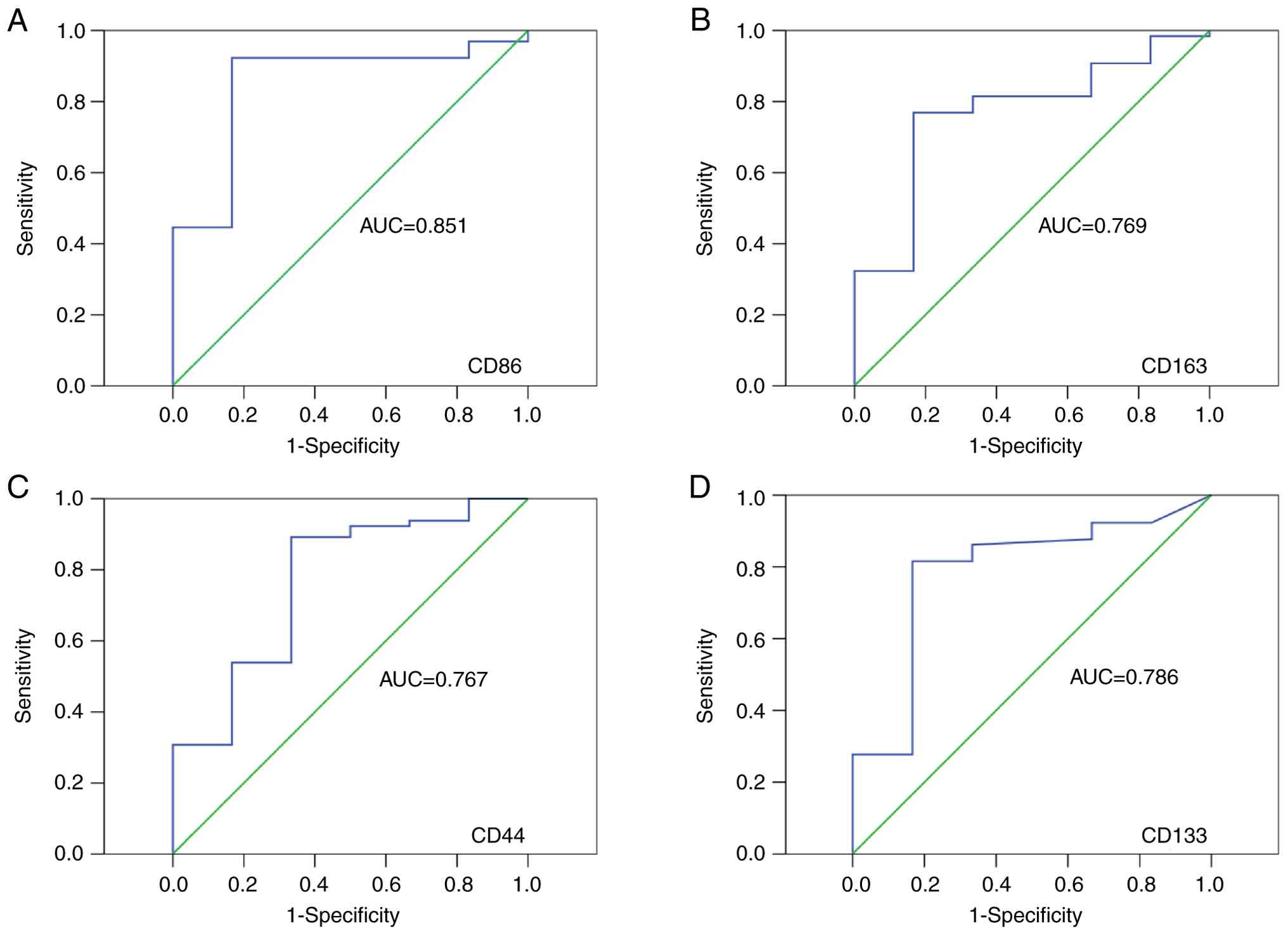
21.0 (IBM Corp.). Optimal cut-off values for CD86, CD163, CD44 and
CD133 IHC scores were determined using Receiver Operating
Characteristic (ROC) curve analysis, with OS as the endpoint.
Cut-off points were selected based on the maximum Youden Index
(sensitivity + specificity-1). The area under the curve (AUC)
values were as follows: CD86: 0.851, CD163: 0.769, CD44: 0.767,
CD133: 0.786. Correlations between CD86, CD163, CD44, CD133 and
clinicopathological parameters were analyzed using Pearson s
chi-square test. Survival differences and prognostic factors were
determined using the Kaplan-Meier method and the log-rank test.
Correlations among CD86, CD163, CD44 and CD133 were assessed using
Pearson s correlation coefficient with corresponding P-values.
Variable selection for the multivariate Cox regression model was
performed using a stepwise forward method. The Variance Inflation
Factor (VIF) was employed to assess multicollinearity among all
included variables, with all VIF values below 2, indicating no
significant multicollinearity concerns. OS was analyzed using Cox
proportional hazards regression models restricted to variables with
P<0.05 in the univariate analysis. Survival curves for the
relevant factors were plotted using the Kaplan-Meier method. In all
statistical analyses, P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient characteristics and expression
of TAM and CSC biomarkers in human CRC
A total of 71 patients with CRC were included in the
survival analysis cohort, with detailed clinicopathologic
characteristics of patients with CRC after resection listed in
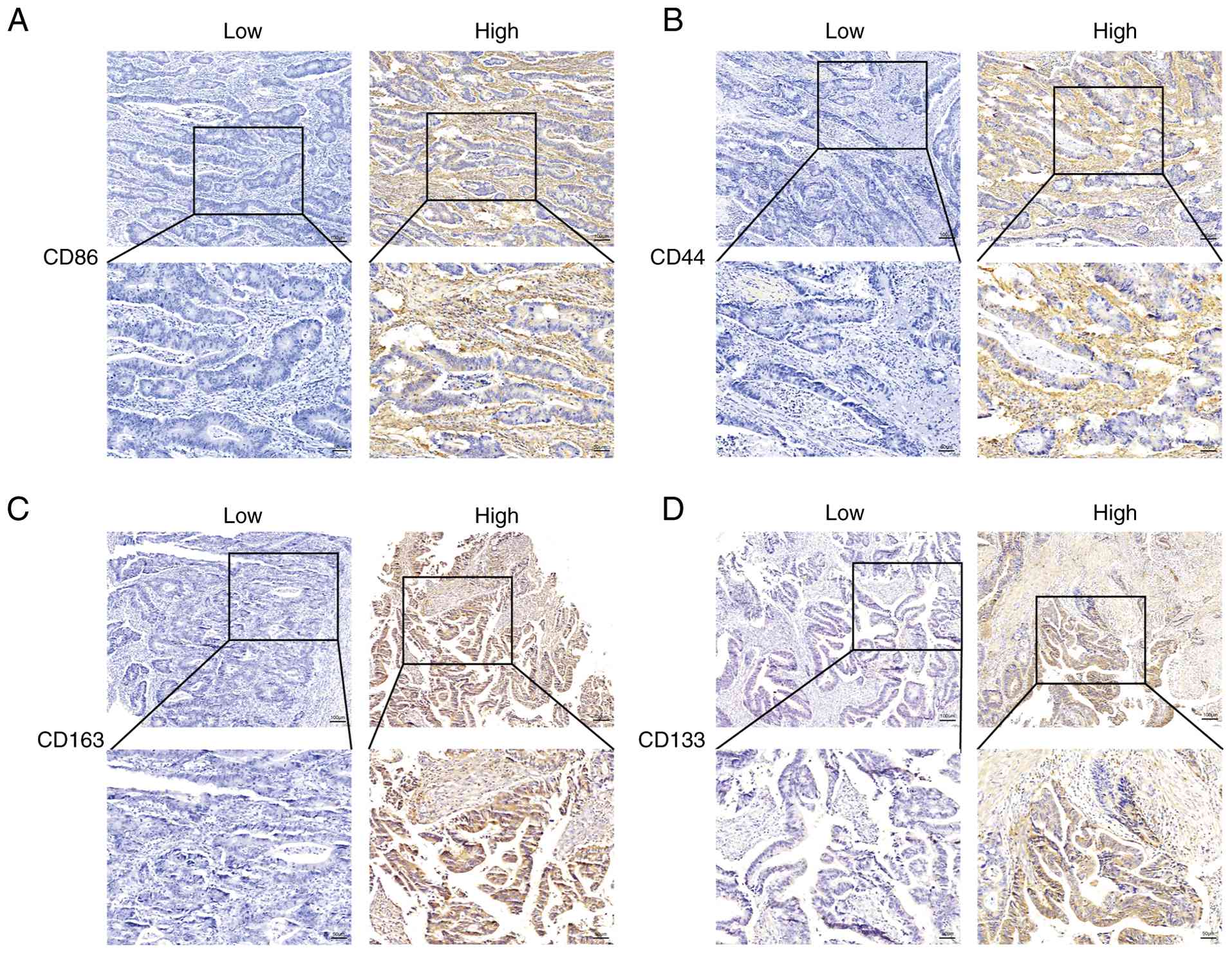
Tables I and II. CD86 was selected as a representative
marker of M1-like TAMs, and CD163 as a representative marker of
M2-like phenotype. Combined detection of multiple markers can
improve the accuracy of CSC identification. In CRC, the use of a
marker panel, including CD133, CD44, CD166, ALDH1 and CD16, has
been shown to reliably identify CSCs (24). To evaluate potential differences in
the expression levels of CD86, CD163, CD44 and CD133 in CRC
tissues, IHC staining was performed (Fig. 1). Using ROC analysis, optimal
cut-off values for distinguishing high and low levels of these
markers were determined in a cohort of 71 CRC samples (Fig. 2). The established thresholds were
as follows: for CD86, high expression was defined as an IHC score
>93.38, and low expression as <93.38; for CD163, high
expression corresponded to an IHC score >16.13, and low
expression to <16.13; for CD44, high expression was classified
as >5.37, and low as <5.37; and for CD133, high expression
was defined as >1.45, with low expression below this value.
Based on these cut-off values, 61 patients (85.9%) were categorized
as CD86-low and 10 (14.1%) as CD86-high. Among the cohort, 20
patients (28.2%) were designated as CD163-low and 51 (71.8%) as
CD163-high. Likewise, 11 patients (15.5%) were categorized as
CD44-low and 60 (84.5%) as CD44-high. 17 patients (23.9%) were
classified as CD133-low and 54 (76.1%) as CD133-high.
 | Table IUnivariate and multivariate Cox
proportional hazards analysis of disease-free survival for patients
with colorectal cancer. |
Table I
Univariate and multivariate Cox
proportional hazards analysis of disease-free survival for patients
with colorectal cancer.
| Variables | Univariate analysis
HR (95% CI) | P-value | Multivariate
analysis HR (95% CI) | P-value |
|---|
| Age, years | | | | |
|
≤60 | 1.000 | 0.688 | | |
|
>60 | 0.902
(0.546-1.492) | | | |
| Sex | | | | |
|
Male | 1.000 | 0.580 | | |
|
Female | 0.869
(0.528-1.430) | | | |
| Tumor location | | | | |
|
Colon | 1.000 | 0.071 | | |
|
Rectal | 1.578
(0.957-2.603) | | | |
| Tumor size, cm | | | | |
|
<3 | 1.000 | 0.003 | | |
|
≥3 | 0.386
(0.198-0.752) | | | |
|
Differentiation | | | | |
|
Well/moderate | 1.000 | 0.077 | | |
|
Poor/undifferentiated | 0.596
(0.334-1.065) | | | |
| T stage | | | | |
|
T1-T2 | 1.000 | <0.001 | 1.000 | 0.006 |
|
T3-T4 | 0.320
(0.182-0.564) | | 0.444
(0.250-0.789) | |
| TNM stage | | | | |
|
I | 1.000 | <0.001 | | |
|
II | 0.114
(0.038-0.338) | | | |
|
III | 0.295
(0.101-0.866) | | | |
|
IV | 0.425
(0.156-1.153) | | | |
| Lymph node
metastasis | | | | |
|
No | 1.000 | <0.001 | | |
|
Yes | 0.413
(0.250-0.685) | | | |
| Distant
metastasis | | | | |
|
No | 1.000 | 0.016 | | |
|
Yes | 0.296
(0.103-0.857) | | | |
| Preoperative CEA
level (ng/ml) | | | | |
|
≤5 | 1.000 | 0.036 | | |
|
>5 | 0.599
(0.365-0.982) | | | |
| Preoperative CA19-9
level (U/ml) | | | | |
|
<37 | 1.000 | <0.001 | 1.000 | <0.001 |
|
≥37 | 0.370
(0.0.9-0.654) | | 0.314
(0.168-0.588) | |
| CD86 protein
expression | | | | |
|
Low | 1.000 | <0.001 | | |
|
High | 4.553
(1.784-11.616) | | | |
| CD163 protein
expression | | | | |
|
Low | 1.000 | <0.001 | 1.000 | <0.001 |
|
High | 0.210
(0.110-0.402) | | 0.259
(0.128-0.522) | |
| CD44 protein
expression | | | | |
|
Low | 1.000 | <0.001 | | |
|
High | 0.228
(0.102-0.512) | | | |
| CD133 protein
expression | | | | |
|
Low | 1.000 | <0.001 | 1.000 | <0.001 |
|
High | 0.149
(0.070-0.319) | | 0.203
(0.091-0.457) | |
 | Table IIUnivariate and multivariate Cox
proportional hazards analysis of overall survival for patients with
colorectal cancer. |
Table II
Univariate and multivariate Cox
proportional hazards analysis of overall survival for patients with
colorectal cancer.
| Variables | Univariate analysis
HR (95% CI) | P-value | Multivariate
analysis HR (95% CI) | P-value |
|---|
| Age, years | | | | |
|
≤60 | 1.000 | 0.797 | | |
|
>60 | 0.936
(0.566-1.549) | | | |
| Sex | | | | |
|
Male | 1.000 | 0.525 | | |
|
Female | 0.851
(0.518-1.398) | | | |
| Tumor location | | | | |
|
Colon | 1.000 | 0.076 | | |
|
Rectal | 0.637
(0.386-1.052) | | | |
| Tumor size, cm | | | | |
|
<3 | 1.000 | 0.002 | | |
|
≥3 | 0.362
(0.184-0.710) | | | |
|
Differentiation | | | | |
|
Well/moderate | 1.000 | 0.057 | | |
|
Poor/undifferentiated | 0.573
(0.320-1.024) | | | |
| T stage | | | | |
|
T1-T2 | 1.000 | <0.001 | 1.000 | 0.003 |
|
T3-T4 | 0.305
(0.173-0.541) | | 0.420
(0.236-0.748) | |
| TNM stage | | | | |
|
I | 1.000 | <0.001 | | |
|
II | 0.115
(0.039-0.341) | | | |
|
III | 0.327
(0.112-0.954) | | | |
|
IV | 0.444
(0.164-1.203) | | | |
| Lymph node
metastasis | | | | |
|
No | 1.000 | <0.001 | | |
|
Yes | 0.407
(0.244-0.677) | | | |
| Distant
metastasis | | | | |
|
No | 1.000 | 0.023 | | |
|
Yes | 0.316
(0.110-0.911) | | | |
| Preoperative CEA
level (ng/ml) | | | | |
|
≤5 | 1.000 | 0.043 | | |
|
>5 | 0.609
(0.370-1.001) | | | |
| Preoperative CA19-9
level (U/ml) | | | | |
|
<37 | 1.000 | 0.001 | 1.000 | <0.001 |
|
≥37 | 0.379
(0.215-0.670) | | 0.333
(0.179-0.618) | |
| CD86 protein
expression | | | | |
|
Low | 1.000 | <0.001 | | |
|
High | 4.437
(1.751-11.242) | | | |
| CD163 protein
expression | | | | |
|
Low | 1.000 | <0.001 | 1.000 | <0.001 |
|
High | 0.212
(0.111-0.404) | | 0.274
(0.136-0.549) | |
| CD44 protein
expression | | | | |
|
Low | 1.000 | <0.001 | | |
|
High | 0.208
(0.092-0.471) | | | |
| CD133 protein
expression | | | | |
|
Low | 1.000 | <0.001 | 1.000 | <0.001 |
|
High | 0.126
(0.054-0.290) | | 0.177
(0.074-0.426) | |
TAM and CSC biomarkers in relation to
clinicopathological characteristics in CRC
The expression of TAM-associated biomarkers CD86 and
CD163, as well as the CSC-associated biomarkers CD44 and CD133,
showed significant associations with a variety of clinicopathologic
characteristics in CRC (Table
III). Elevated CD86 expression was significantly correlated
with smaller tumor size (P=0.025) and a lower incidence of distant
metastasis (P=0.034). By contrast, patients with high CD163
expression had significantly larger tumors (P=0.027), more advanced
T stage (P=0.006), higher TNM stage (P=0.015), and increased lymph
node metastases (P=0.033). In addition, high CD44 expression was
associated with deeper tumor invasion (T stage, P=0.032), more
advanced TNM stage (P=0.044), and a higher frequency of lymph node
metastasis (P=0.009). Similarly, high CD133 expression was
significantly associated with larger tumor volume (P=0.035),
advanced T-stage (P=0.004), later TNM stage (P=0.009), increased
lymph node metastasis (P=0.041) and higher preoperative
carcinoembryonic antigen (CEA; P=0.009) and CA19-9 levels
(P=0.045).
 | Table IIIExpression of CD86, CD163, CD44 and
CD133 protein and clinicopathological parameters in colorectal
cancer tissues. |
Table III
Expression of CD86, CD163, CD44 and
CD133 protein and clinicopathological parameters in colorectal
cancer tissues.
| Variables | CD86 High | Low | P-value | CD163 High | Low | P-value | CD44 High | Low | P-value | CD133 High | Low | P-value |
|---|
| Age | | | | | | | | | | | | |
|
≤60 | 5 | 23 | 0.461 | 22 | 6 | 0.308 | 23 | 5 | 0.657 | 19 | 9 | 0.191 |
|
>60 | 5 | 38 | | 29 | 14 | | 37 | 6 | | 35 | 8 | |
| Sex | | | | | | | | | | | | |
|
Male | 5 | 34 | 0.735 | 28 | 11 | 0.994 | 33 | 6 | 0.978 | 28 | 11 | 0.353 |
|
Female | 5 | 27 | | 23 | 9 | | 27 | 5 | | 26 | 6 | |
| Tumor location | | | | | | | | | | | | |
|
Colon | 5 | 23 | 0.461 | 23 | 5 | 0.119 | 25 | 3 | 0.369 | 23 | 5 | 0.332 |
|
Rectal | 5 | 38 | | 28 | 15 | | 35 | 8 | | 31 | 12 | |
| Tumor size, cm | | | | | | | | | | | | |
|
<3 | 5 | 11 | 0.025 | 8 | 8 | 0.027 | 12 | 4 | 0.232 | 9 | 7 | 0.035 |
|
≥3 | 5 | 50 | | 43 | 12 | | 48 | 7 | | 45 | 10 | |
|
Differentiation | | | | | | | | | | | | |
|
Well/moderate | 10 | 45 | 0.066 | 37 | 18 | 0.113 | 47 | 8 | 0.682 | 41 | 14 | 0.580 |
|
Poor | 0 | 16 | | 14 | 2 | | 13 | 3 | | 13 | 3 | |
| T stage | | | | | | | | | | | | |
|
T1-T2 | 5 | 20 | 0.291 | 13 | 12 | 0.006 | 18 | 7 | 0.032 | 14 | 11 | 0.004 |
|
T3-t4 | 5 | 41 | | 38 | 8 | | 42 | 4 | | 40 | 6 | |
| TNM stage | | | | | | | | | | | | |
|
I | 5 | 18 | 0.128 | 11 | 12 | 0.015 | 16 | 7 | 0.044 | 12 | 11 | 0.009 |
|
II | 1 | 14 | | 12 | 3 | | 12 | 3 | | 12 | 3 | |
|
III | 2 | 26 | | 23 | 5 | | 27 | 1 | | 25 | 3 | |
|
IV | 2 | 3 | | 5 | 0 | | 5 | 0 | | 5 | 0 | |
| Lymph node
metastasis | | | | | | | | | | | | |
|
No | 6 | 33 | 0.728 | 24 | 15 | 0.033 | 29 | 10 | 0.009 | 26 | 13 | 0.041 |
|
Yes | 4 | 28 | | 27 | 5 | | 31 | 1 | | 28 | 4 | |
| Distant
metastasis | | | | | | | | | | | | |
|
No | 8 | 59 | 0.034 | 47 | 20 | 0.197 | 56 | 11 | 0.378 | 50 | 17 | 0.248 |
|
Yes | 2 | 2 | | 4 | 0 | | 4 | 0 | | 4 | 0 | |
| Preoperative CEA
level (ng/ml) | | | | | | | | | | | | |
|
≤5 | 7 | 32 | 0.301 | 25 | 14 | 0.110 | 30 | 9 | 0.051 | 25 | 14 | 0.009 |
|
>5 | 3 | 29 | | 26 | 6 | | 30 | 2 | | 29 | 3 | |
| Preoperative CA19-9
level (U/ml) | | | | | | | | | | | | |
|
<37 | 8 | 46 | 0.753 | 37 | 17 | 0.269 | 45 | 9 | 0.626 | 38 | 16 | 0.045 |
|
≥37 | 2 | 15 | | 14 | 3 | | 15 | 2 | | 16 | 1 | |
Correlations between TAM and CSC
biomarker expression
Correlations between TAM- and CSC-associated
proteins are presented in Table
IV. CD86, a TAM biomarker, was negatively correlated with the
CSC biomarkers CD44 (P=0.008, r=-0.286) and CD133 (P=0.016,
r=-0.342). By contrast, CD163, a TAMs biomarker, exhibited a
positive correlation with the CSC biomarkers CD44 (P<0.001,
r=0.549) and CD133 (P<0.001, r=0.529).
 | Table IVThe relationship between the
expression of CD86, CD163, CD44 and CD133 protein. |
Table IV
The relationship between the
expression of CD86, CD163, CD44 and CD133 protein.
| | CD86 | CD163 |
|---|
| Characteristic | Low | High | P-value | r | Low | High | P-value | r |
|---|
| CD44 | | | | | | | | |
|
Low | 6 | 5 | 0.001 | -0.286 | 9 | 2 | 0.000 | 0.511 |
|
High | 55 | 5 | | | 11 | 49 | | |
| CD133 | | | | | | | | |
|
Low | 11 | 6 | 0.004 | -0.342 | 12 | 5 | 0.000 | 0.529 |
|
High | 50 | 4 | | | 8 | 46 | | |
Follow-up evaluation and prognostic
impact of TAM and CSC biomarker expression in CRC
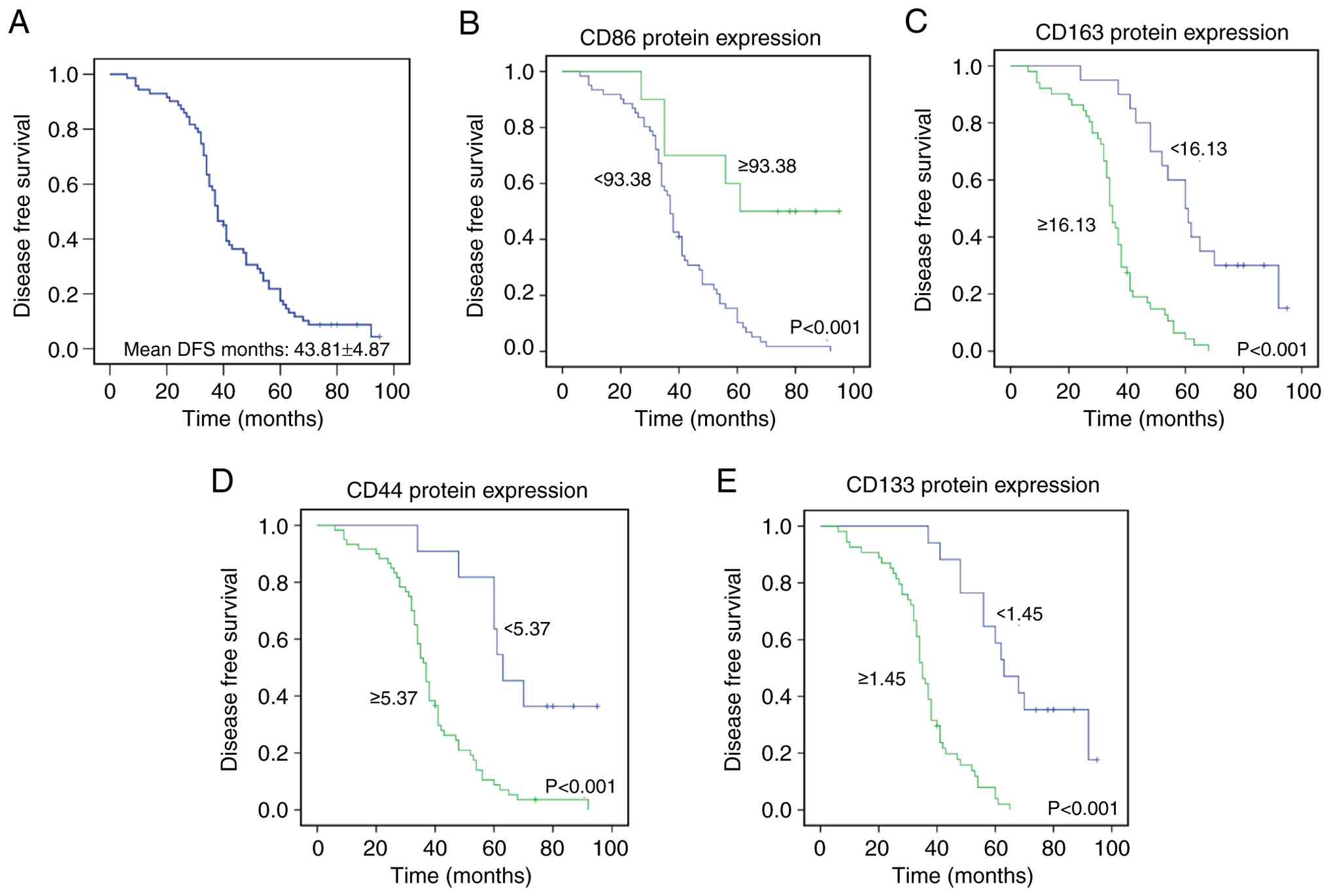
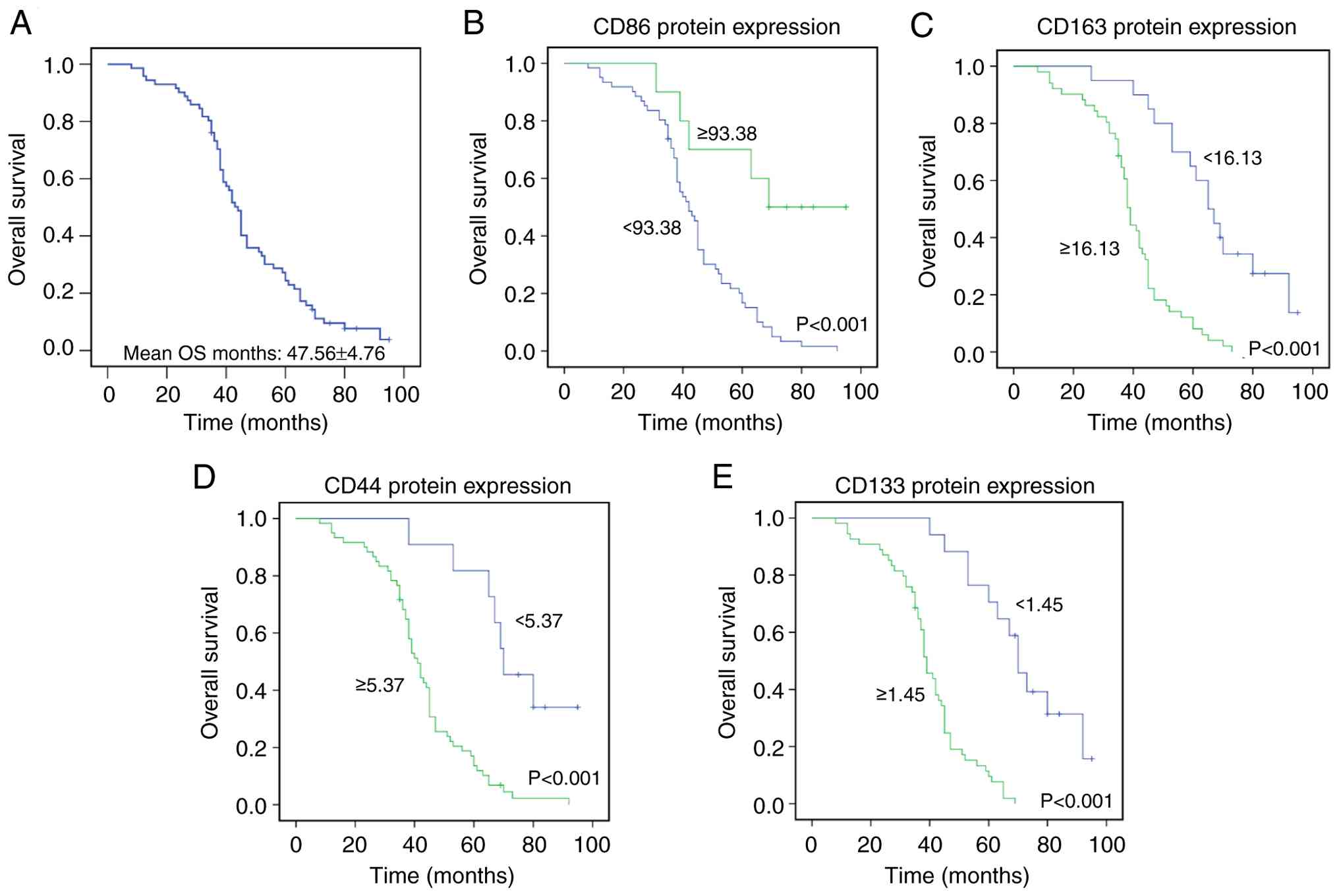
The mean and median DFS of the 71 patients were
43.81±4.87 and 38.00±3.14 months, respectively (Fig. 3A). Mean and median OS were
47.56±4.76 and 44.00±2.97 months, respectively (Fig. 4A). To investigate the prognostic
impact of TAM- and CSC-associated markers, DFS and OS were compared
among patients stratified by the expression levels of CD86, CD163,
CD44 and CD133. Kaplan-Meier survival analysis demonstrated that
high CD86 expression was significantly associated with prolonged
DFS and OS (P<0.001) (Figs. 3B
and 4B), whereas patients with
high CD163 expression had shorter DFS and OS (P<0.001) (Figs. 3C and 4C). Similarly, elevated CD44 expression
was significantly linked to decreased DFS and OS (P<0.001)
(Figs. 3D and 4D), and high CD133 expression was
significantly associated with shorter DFS and OS (P<0.001)
(Figs. 3E and 4E).
CD163 and CD133 expression, T Stage
and preoperative CA19-9 level as independent prognostic factors for
DFS and OS
Kaplan-Meier survival analyses of DFS and OS were
performed according to tumor size, T stage, TNM stage, lymph node
status, M stage, preoperative CEA and CA19-9 levels, as well as the
expression of CD86, CD163, CD44 and CD133 (Figs. S1 and S2). The results showed that DFS was
significantly associated with tumor size (P=0.003), T stage
(P<0.001), TNM stage (P<0.001), lymph node metastasis
(P<0.001), distant metastasis (P=0.016), preoperative CEA level
(P=0.036), preoperative CA19-9 level (P<0.001), and the
expression of CD86 (P<0.001), CD163 (P<0.001), CD44
(P<0.001) and CD133 (P<0.001). Multivariate Cox regression
analysis identified that CD163 expression (P<0.001), CD133
expression (P<0.001), T stage (P=0.006), and preoperative CA19-9
level (P<0.001) were independent prognostic factors
significantly associated with DFS (Table I). Similarly, OS was significantly
associated with tumor size (P=0.002), T stage (P<0.001), TNM
stage (P<0.001), lymph node metastasis (P<0.001), distant
metastasis (P=0.023), preoperative CEA level (P=0.043),
preoperative CA19-9 level (P<0.001), CD86 expression
(P<0.001), CD163 expression (P<0.001), CD44 expression
(P<0.001) and CD133 expression (P<0.001). For these factors
included in the multivariate Cox analysis, CD163 expression
(P<0.001), CD133 expression (P<0.001), T stage (P=0.003) and
preoperative CA19-9 level (P<0.001) were identified as
independent prognostic factors for OS (Table II).
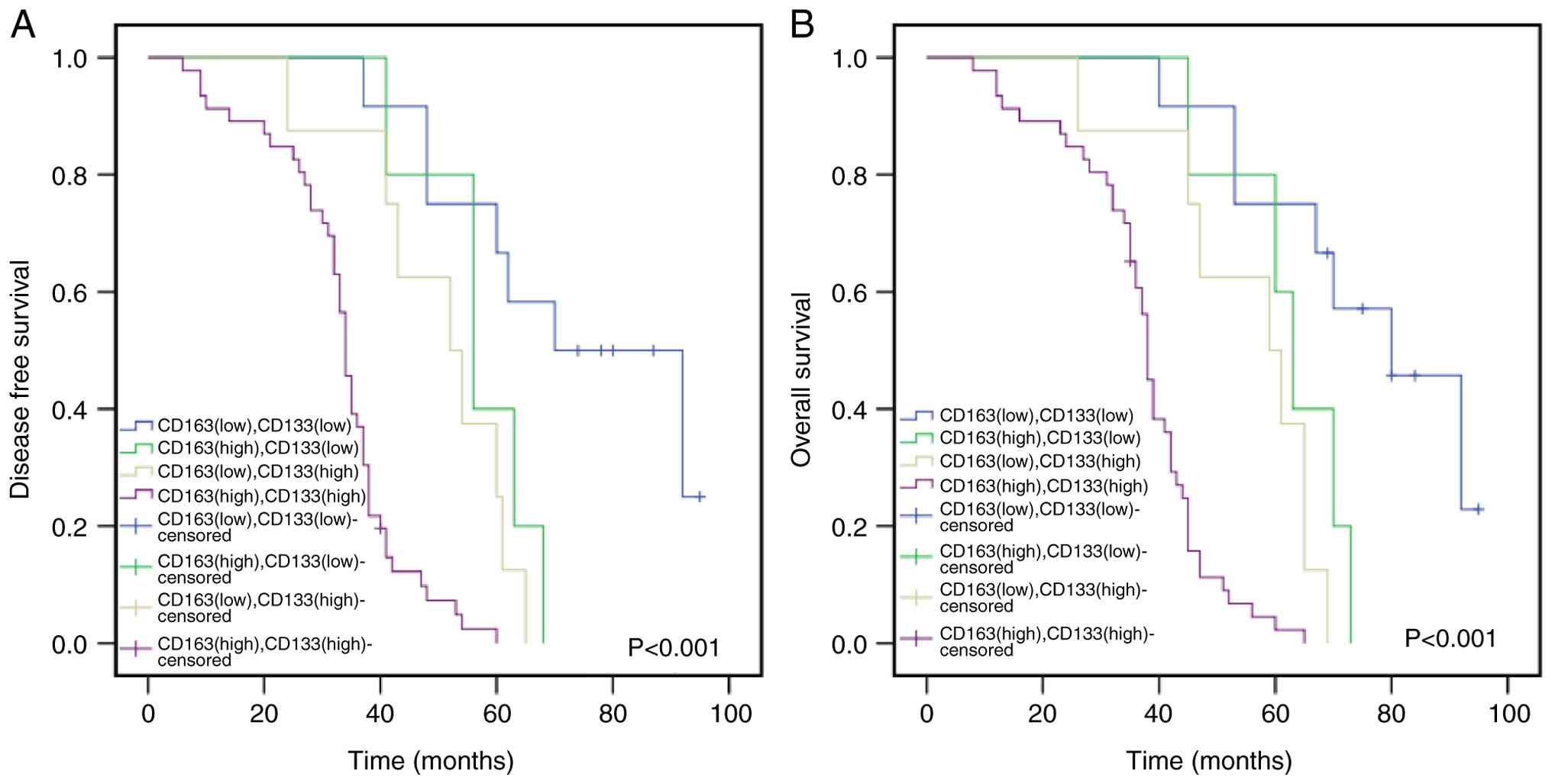
Combined expression of CD163 and CD133
as a prognostic indicator in CRC
Multivariate Cox regression analysis identified
CD163 and CD133 expression as independent prognostic factors for
DFS and OS (Tables I and II). The combined impact of CD163 and
CD133 expression on patient prognosis was therefore evaluated
(Fig. 5). Patients with concurrent
high expression of both markers exhibited significantly shorter DFS
and OS (P<0.001; Fig. 5),
indicating that this combination serves as a robust predictor of
unfavorable prognosis in CRC.
Discussion
CRC ranks as the third most commonly diagnosed
gastrointestinal malignancy worldwide (25). Current treatments for CRC include
surgical resection, chemotherapy, radiotherapy and immunomodulatory
approaches (26). Despite these
interventions, nearly 40% of patients develop disease recurrence or
distant metastasis, solidifying CRC s status as the third
leading cause of cancer-related mortality (27). TNM staging, while a major
prognostic factor, has limitations in accurately predicting CRC
patient prognosis (26).
Therefore, it was aimed to identify biomarkers with a stronger
prognostic correlation with CRC. Given that the combined detection
of multiple markers can improve the specificity of CSC and TAM
identification, the joint expression of CSC and TAM biomarkers was
evaluated to determine a panel with improved prognostic
correlation, thereby providing a potential basis for more precise
clinical diagnosis and therapeutic decision-making.
CSCs regulate the TME by recruiting immune cells via
paracrine signaling. For instance, glioma stem cells promote tumor
progression by secreting periostin, recruiting TAMs, and
constructing the TME (28). In
addition, CSCs secrete cytokines, such as TGF-β, IL-10, IL-4 and
IL-13 into the TME, which exert an inhibitory effect on multiple
immune cells (29) and thereby
promote tumor progression. Reciprocally, TAMs regulate CSC stemness
and maintain self-renewal capacity through secretion of cytokines,
chemokines, and participation in complex signaling networks. Key
pathways mediating this bidirectional crosstalk include the
IL-6/STAT3 axis, wherein TAM-derived IL-6 activates STAT3 signaling
in CSCs to promote self-renewal and stemness maintenance (30). Additionally, TGF-β signaling plays
a pivotal role in mediating the bidirectional communication between
TAMs and CSCs. TAM-secreted TGF-β not only enhances CSC stemness
but also induces epithelial-mesenchymal transition, further
contributing to tumor progression and metastasis (31). Other important pathways, such as
NF-κB, can be activated by TAM-derived IL-1 and TNF-α, creating an
inflammatory niche that further supports CSC persistence (32).
The biological significance of these
cytokine-mediated interactions lies in their ability to establish a
sustainable niche that maintains CSC populations, drives tumor
heterogeneity, and confers therapeutic resistance. For instance, in
hepatocellular carcinoma, TAMs can induce STAT3 activation via
IL-6, stimulating further cytokine release and forming a positive
feedback loop that amplifies CSC self-renewal (33). Similarly, in breast cancer, TAMs
have been reported to enhance the stemness characteristics of CSCs
through Ephrin-EphA4 interactions, which in turn stimulate CSCs to
produce inflammatory cytokines, including IL-1, IL-6 and
IL-8(34).
In summary, a bidirectional communication exists
between CSCs and TAMs. To investigate this relationship, the
protein expression of TAM and CSC biomarkers in CRC tumor tissues
was detected by IHC. It was found that high expression of CD163,
CD44 and CD133 was associated with poor prognosis, while high CD86
expression was correlated with favorable prognosis. Indeed, the
prognostic role of CD86+ TAMs in CRC is complex and
context-dependent. Certain studies suggested that CD86 is
associated with improved survival, and patients with high CD86 gene
expression have higher OS rates than those with low CD86
expression. This indicates that low expression of this marker is
linked to tumor progression and aggressiveness (35), which is in line with the present
findings. While certain studies report CD86 as a general M1 marker
associated with antitumor effects, the present data suggest its
prognostic significance may be context-dependent (36,37).
Discrepancies with previous studies could stem from heterogeneity
in TMEs, differences in antibody clones, or variations in cohort
demographics. By contrast, CD163+ TAMs are known to
drive tumor progression by suppressing antitumor immunity,
enhancing angiogenesis, and promoting metastasis. The association
between CD163+ M2-like TAMs and poor prognosis has been
more consistently reported across various cancer types, including
CRC (38,39). Therefore, Certain studies suggested
that combined analysis of CD86+ TAMs and
CD163+ TAMs appears to be more suitable for determining
relapse and mortality rates (40,41).
In addition, the correlations between TAM and CSC markers,
clinicopathologic parameters and patient prognosis were
investigated. CD86 expression was markedly correlated with tumor
size and distant metastasis, whereas CD163 expression was
significantly correlated with tumor size and T-stage. Additionally,
CD44 and CD133 expression were strongly linked to TNM staging and
lymph node metastasis, respectively. Moreover, both CD163 and CD133
were identified as independent prognostic factors in CRC. The
present analysis revealed significant prognostic value for both
CD86 (M1-like) and CD133, as well as a negative correlation between
them. While the combination of CD86 and CD133 is of interest, the
present study was strategically designed to investigate the
synergistic pro-tumorigenic interaction between TAMs and CSCs. In
this context, the concurrent high expression of CD163 (M2-like
TAMs) and CD133 represents a functionally coherent and biologically
synergistic unit that collectively fosters an immunosuppressive and
pro-stemness TME, leading to the most aggressive disease phenotype.
This is substantiated by the present multivariate Cox regression
analysis, which identified both CD163 and CD133 as the most robust
and independent prognostic factors (P<0.001 for both DFS and
OS). Therefore, prioritizing the
CD163+/CD133+ profile was a deliberate
strategy to most directly test our core hypothesis and to establish
a clinically actionable biomarker for identifying the highest-risk
patient subgroup.
The present analysis revealed significant prognostic
value for both CD86 and CD133, as well as a negative correlation
between them. While the combination of CD86 and CD133 is of
interest, the present study was strategically designed to
investigate the synergistic pro-tumorigenic interaction between
TAMs and CSCs. In this context, the concurrent high expression of
CD163 and CD133 represents a functionally coherent and biologically
synergistic unit that collectively fosters an immunosuppressive and
pro-stemness TME, leading to the most aggressive disease phenotype.
This is substantiated by our multivariate Cox regression analysis,
which identified both CD163 and CD133 as the most robust and
independent prognostic factors (P<0.001 for both DFS and OS).
Therefore, prioritizing the CD163+/CD133+
profile was a deliberate strategy to most directly test our core
hypothesis and to establish a clinically actionable biomarker for
identifying the highest-risk patient subgroup.
Current studies of TAMs and CSCs in CRC remain
relatively limited, typically focusing either on the prognostic
significance of different TAM subtypes or on the relationship
between CSC expression and CRC occurrence and progression (42,43).
By contrast, the present study combines the assessment of TAM and
CSC biomarkers, specifically CD163 and CD133, providing a more
robust prognostic tool than evaluating either marker alone in CRC.
This integrative approach distinguishes the current study from
prior works that primarily focused on single-marker systems. By
assessing TAMs in conjunction with CSCs, the roles of different
subtypes of TAMs and CSCs in CRC were more comprehensively and
accurately explored. Both univariate and multivariate analyses
identified CD163+ TAMs and CD133+ CSCs as
independent prognostic factors for DFS and OS. The prognostic
significance of their combined expression was therefore further
examined, finding that concurrent high levels of CD163 and CD133
were strongly associated with shorter DFS and OS, indicating poor
prognosis. These results suggested that the TAM-CSC functional
unit, rather than either component in isolation, represents a key
determinant of tumor aggressiveness in CRC.
In conclusion, the novel contribution of our
research lies in establishing and validating a combined CD163/CD133
biomarker profile that captures the synergistic interaction between
pro-tumorigenic TAMs and stem-like cancer cells. While TNM staging
provides an anatomical framework, the
CD163+/CD133+ profile directly reflects the
pro-tumorigenic and treatment-resistant potential of the TME. This
dual-marker panel identifies a high-risk patient subgroup with
significantly shorter DFS and OS, thereby significantly improving
the accuracy of prognostic predictions in patients with CRC. These
findings carry important clinical implications for developing more
effective prognostic assessment tools and personalized treatment
strategies. Future research should focus on validating these
biomarkers in larger, multicenter cohorts and exploring targeted
therapies that disrupt the TAM-CSC interaction axis to improve
patient outcomes. Furthermore, this approach holds promise for
predicting responses to both conventional chemotherapy and emerging
immunotherapies, potentially guiding more effective, personalized
treatment strategies.
Supplementary Material
Kaplan-Meier survival curves showing
DFS for clinicopathologic factors in CRC. (A) Tumor size. (B)
T-stage. (C) TNM stage. (D) Lymph nodes status. (E) M-stage. (F)
Preoperative CEA level. (G) Preoperative CA19-9 level. P-values
were derived by log-rank test. CEA, carcinoembryonic antigen.
Kaplan-Meier survival curves showing
the influence of clinicopathological factors on colorectal cancer.
(A) T stage. (B) Tumor size. (C) TNM stage. (D) Lymph node status.
(E) M-stage. (F) Preoperative CEA level. (G) Preoperative CA19-9
level. P-values were obtained by log-rank test. CEA,
carcinoembryonic antigen.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81803780), the Natural
Science Foundation of Jiangsu (grant no. BK20180928) and the
Science and Technology Tackling Project of Henan (grant no.
232102310131).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors contributions
YK supervised the study and wrote the original
draft. YunW proposed the research concept and designed the research
plan. BH managed the planning and execution of the research
activities. YuqW collected and interpretated data. RY developed
methodology and validated data. HT and FG conducted investigation
and data validation, and prepared figures and tables. YK and YunW
confirm the authenticity of all the raw data. All authors wrote,
reviewed and edited the manuscript. All authors read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board and Human Ethics Committee of Luoyang Central Hospital
Affiliated with Zhengzhou University (approval no.
LWLL-2018-03-07-01; Luoyang, China). All participants or their
legal guardians signed an informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang F, Chen G, Zhang Z, Yuan Y, Wang Y,
Gao YH, Sheng W, Wang Z, Li X, Yuan X, et al: The Chinese society
of clinical oncology (CSCO): Clinical guidelines for the diagnosis
and treatment of colorectal cancer, 2024 update. Cancer Commun
(Lond). 45:332–379. 2025.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hao VDT, Tri PM, My DT, Anh LT, Trung LV,
Bac NH and Vuong NL: FOLFOXIRI for first-line treatment of
unresectable colorectal cancer with liver metastases in a
resource-limited setting. J Gastrointest Cancer.
56(12)2025.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sebag-Montefiore D, Stephens RJ, Steele R,
Monson J, Grieve R, Khanna S, Quirke P, Couture J, de Metz C, Myint
AS, et al: Preoperative radiotherapy versus selective postoperative
chemoradiotherapy in patients with rectal cancer (MRC CR07 and
NCIC-CTG C016): A multicentre, randomised trial. Lancet.
373:811–820. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Xiao BY, Zhang X, Cao TY, Li DD, Jiang W,
Kong LH, Tang JH, Han K, Zhang CZ, Mei WJ, et al: Neoadjuvant
immunotherapy leads to major response and low recurrence in
localized mismatch repair-deficient colorectal cancer. J Natl Compr
Cancer Netw. 21:60–66. 2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Guo KC, Wang ZZ and Su XQ: Chinese
medicine in colorectal cancer treatment: From potential targets and
mechanisms to clinical application. Chin J Integr Med: Sep 27, 2024
(Epub ahead of print).
|
|
6
|
Jiang Y, Wang C, Zu C, Rong X, Yu Q and
Jiang J: Synergistic potential of nanomedicine in prostate cancer
immunotherapy: Breakthroughs and prospects. Int J Nanomedicine.
19:9459–9486. 2024.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33.
2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Siegel RL, Wagle NS, Cercek A, Smith RA
and Jemal A: Colorectal cancer statistics, 2023. CA Cancer J Clin.
73:233–254. 2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Winawer SJ and Zauber AG: The advanced
adenoma as the primary target of screening. Gastrointest Endosc
Clin N Am. 12:1–9. 2002.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Biller LH and Schrag D: Diagnosis and
treatment of metastatic colorectal cancer: A review. JAMA.
325:669–685. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Edin S, Wikberg ML, Dahlin AM, Rutegård J,
Öberg Å, Oldenborg PA and Palmqvist R: The distribution of
macrophages with a M1 or M2 phenotype in relation to prognosis and
the molecular characteristics of colorectal cancer. PLoS One.
7(e47045)2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Khan SU, Khan MU, Din MA, Khan IM, Khan
MI, Bungau S and Hassan SSU: Reprogramming tumor-associated
macrophages as a unique approach to target tumor immunotherapy.
Front Immunol. 14(1166487)2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bruni D, Angell HK and Galon J: The immune
contexture and Immunoscore in cancer prognosis and therapeutic
efficacy. Nat Rev Cancer. 20:662–680. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang H, Tian T and Zhang J:
Tumor-associated macrophages (TAMs) in colorectal cancer (CRC):
From mechanism to therapy and prognosis. Int J Mol Sci.
22(8470)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li J, Li L, Li Y, Long Y, Zhao Q, Ouyang
Y, Bao W and Gong K: Tumor-associated macrophage infiltration and
prognosis in colorectal cancer: Systematic review and
meta-analysis. Int J Colorectal Dis. 35:1203–1210. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wei C, Yang C, Wang S, Shi D, Zhang C, Lin
X, Liu Q, Dou R and Xiong B: Crosstalk between cancer cells and
tumor associated macrophages is required for mesenchymal
circulating tumor cell-mediated colorectal cancer metastasis. Mol
Cancer. 18(64)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Massague J and Ganesh K:
Metastasis-initiating cells and ecosystems. Cancer Discov.
11:971–994. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gao W, Chen L, Ma Z, Du Z, Zhao Z, Hu Z
and Li Q: Isolation and phenotypic characterization of colorectal
cancer stem cells with organ-specific metastatic potential.
Gastroenterology. 145:636–646. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Fekir K, Dubois-Pot-Schneider H, Désert R,
Daniel Y, Glaise D, Rauch C, Morel F, Fromenty B, Musso O, Cabillic
F and Corlu A: Retrodifferentiation of human tumor hepatocytes to
stem cells leads to metabolic reprogramming and chemoresistance.
Cancer Res. 79:1869–1883. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Nagata T, Sakakura C, Komiyama S,
Miyashita A, Nishio M, Murayama Y, Komatsu S, Shiozaki A, Kuriu Y,
Ikoma H, et al: Expression of cancer stem cell markers CD133 and
CD44 in locoregional recurrence of rectal cancer. Anticancer Res.
31:495–500. 2011.PubMed/NCBI
|
|
21
|
Miranda A, Hamilton PT, Zhang AW, Pattnaik
S, Becht E, Mezheyeuski A, Bruun J, Micke P, de Reynies A and
Nelson BH: Cancer stemness, intratumoral heterogeneity, and immune
response across cancers. Proc Natl Acad Sci USA. 116:9020–9029.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Luo S, Yang G, Ye P, Cao N, Chi X, Yang WH
and Yan X: Macrophages are a double-edged sword: Molecular
crosstalk between tumor-associated macrophages and cancer stem
cells. Biomolecules. 12(850)2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sato T, Takagi K, Higuchi M, Abe H,
Kojimahara M, Sagawa M, Tanaki M, Miki Y, Suzuki T and Hojo H:
Immunolocalization of CD80 and CD86 in non-small cell lung
carcinoma: CD80 as a potent prognostic factor. Acta Histochem
Cytochem. 55:25–35. 2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Murar M and Vaidya A: Cancer stem cell
markers: Premises and prospects. Biomark Med. 9:1331–1342.
2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Schreuders EH, Ruco A, Rabeneck L, Schoen
RE, Sung JJ, Young GP and Kuipers EJ: Colorectal cancer screening:
A global overview of existing programmes. Gut. 64:1637–1649.
2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Basak D, Uddin MN and Hancock J: The role
of oxidative stress and its counteractive utility in colorectal
cancer (CRC). Cancers (Basel). 12(3336)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
DeDecker L, Coppedge B, Avelar-Barragan J,
Karnes W and Whiteson K: Microbiome distinctions between the CRC
carcinogenic pathways. Gut Microbes. 13(1854641)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhou W, Ke SQ, Huang Z, Flavahan W, Fang
X, Paul J, Wu L, Sloan AE, McLendon RE, Li X, et al: Periostin
secreted by glioblastoma stem cells recruits M2 tumour-associated
macrophages and promotes malignant growth. Nat Cell Biol.
17:170–182. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Clara JA, Monge C, Yang Y and Takebe N:
Targeting signalling pathways and the immune microenvironment of
cancer stem cells-a clinical update. Nat Rev Clin Oncol.
17:204–232. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xu J, Lin H, Wu G, Zhu M and Li M: .:
IL-6/STAT3 is a promising therapeutic target for hepatocellular
carcinoma. Front Oncol. 11(760971)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fasano M, Pirozzi M, Miceli CC, Cocule M,
Caraglia M, Boccellino M, Vitale P, De Falco V, Farese S, Zotta A,
et al: TGF-β modulated pathways in colorectal cancer: New potential
therapeutic opportunities. Int J Mol Sci. 25(7400)2024.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cornice J, Verzella D, Arboretto P,
Vecchiotti D, Capece D, Zazzeroni F and Franzoso G: NF-κB:
Governing macrophages in cancer. Genes (Basel).
15(197)2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wan S, Zhao E, Kryczek I, Vatan L,
Sadovskaya A, Ludema G, Simeone DM, Zou W and Welling TH:
Tumor-associated macrophages produce interleukin 6 and signal via
STAT3 to promote expansion of human hepatocellular carcinoma stem
cells. Gastroenterology. 147:1393–1404. 2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lu H, Clauser KR, Tam WL, Fröse J, Ye X,
Eaton EN, Reinhardt F, Donnenberg VS, Bhargava R, Carr SA and
Weinberg RA: A breast cancer stem cell niche supported by
juxtacrine signalling from monocytes and macrophages. Nat Cell
Biol. 16:1105–1117. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
35
|
Aytekin EC, Unal B, Bassorgun CI and Ozkan
O: Clinicopathologic evaluation of CD80, CD86, and PD-L1
expressions with immunohistochemical methods in malignant melanoma
patients. Turk Patoloji Derg. 40:16–26. 2024.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kinouchi M, Miura K, Mizoi T, Ishida K,
Fujibuchi W, Ando T, Yazaki N, Saito K, Shiiba K and Sasaki I:
Infiltration of CD14-positive macrophages at the invasive front
indicates a favorable prognosis in colorectal cancer patients with
lymph node metastasis. Hepatogastroenterology. 58:352–358.
2011.PubMed/NCBI
|
|
37
|
Beider K, Bitner H, Leiba M, Gutwein O,
Koren-Michowitz M, Ostrovsky O, Abraham M, Wald H, Galun E, Peled A
and Nagler A: Multiple myeloma cells recruit tumor-supportive
macrophages through the CXCR4/CXCL12 axis and promote their
polarization toward the M2 phenotype. Oncotarget. 5:11283–11296.
2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Jääskeläinen MM, Tumelius R, Hämäläinen K,
Rilla K, Oikari S, Rönkä A, Selander T, Mannermaa A, Tiainen S and
Auvinen P: High numbers of CD163+tumor-associated macrophages
predict poor prognosis in HER2+breast cancer. Cancers (Basel).
16(634)2024.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Molina OE, LaRue H, Simonyan D, Hovington
H, Têtu B, Fradet V, Lacombe L, Toren P, Bergeron A and Fradet Y:
High infiltration of CD209+ dendritic cells and CD163+ macrophages
in the peritumor area of prostate cancer is predictive of late
adverse outcomes. Front Immunol. 14(1205266)2023.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Xu G, Jiang L, Ye C, Qin G, Luo Z, Mo Y
and Chen J: The ratio of CD86+/CD163+macrophages predicts
postoperative recurrence in stage II-III colorectal cancer. Front
Immunol. 12(724429)2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liu J, Deng Y, Liu Z, Li X, Zhang M, Yu X,
Liu T, Chen K and Li Z: Identification of genes associated with
prognosis and immunotherapy prediction in triple-negative breast
cancer via M1/M2 macrophage ratio. Medicina (Kaunas).
59(1285)2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fattahi F, Zanjani LS, Vafaei S, Shams ZH,
Kiani J, Naseri M, Gheytanchi E and Madjd Z: Expressions of TWIST1
and CD105 markers in colorectal cancer patients and their
association with metastatic potential and prognosis. Diagn Pathol.
16(26)2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Inagaki K, Kunisho S, Takigawa H, Yuge R,
Oka S, Tanaka S, Shimamoto F, Chayama K and Kitadai Y: Role of
tumor-associated macrophages at the invasive front in human
colorectal cancer progression. Cancer Sci. 112:2692–2704.
2021.PubMed/NCBI View Article : Google Scholar
|