Introduction
Colorectal cancer (CRC) is one of the most common
malignant tumors and is the second leading cause of
cancer-associated mortality worldwide (1); it is becoming increasingly prevalent in
younger individuals (2). Screening
and early detection can significantly reduce the mortality rate
associated with CRC. However, even following tumor resection and
systemic treatment, the 5-year survival rate is only 40% for
patients without tumor metastasis and ~20% for those with
metastatic CRC (3), and the quality
of life of these patients is markedly reduced. Currently, the
etiology of CRC development is not yet fully understood. Therefore,
it is critical to identify molecular targets that can lead to the
development of novel therapeutic approaches for patients with
CRC.
Ubiquitination is a post-translational modification
that involves the covalent attachment of ubiquitin to target
proteins. Ubiquitination relies on ubiquitin-activating enzymes
(E1s), ubiquitin-conjugating enzymes (E2s), ubiquitin-ligase
enzymes (E3s), the 26S proteasome, and de-ubiquitinating enzymes
(4,5). Ubiquitination mediated by the
ubiquitin-proteasome system is essential for maintaining cellular
protein homeostasis. The binding of ubiquitin to substrates
typically involves three key steps: The initiation step catalyzed
by E1, the intermediate step of covalently attaching ubiquitin to
E2, and the final step of transferring ubiquitin from E2 to the
protein substrate, usually facilitated by E3(6). Protein degradation controlled by the
ubiquitin system removes misfolded proteins and plays a critical
role in regulating cellular signal transduction (7,8).
Ubiquitin-dependent protein degradation mediated by the
ubiquitin-proteasome system regulates the cell cycle, DNA repair,
immune function and other cellular processes (7).
Members of the E2 family are key components of the
ubiquitin-proteasome system and play roles in the development of
malignant tumors, including CRC. Takahashi et al (9) found that the ubiquitin-conjugating
enzyme E2 C (Ube2C) gene was highly expressed in 50% of patients
with CRC and that Ube2C played a significant role in the liver
metastasis of advanced-stage CRC. Ubiquitin-conjugating enzyme E2
variant 1 (Ube2v1), also known as Uev1A, is a mammalian homolog of
yeast MMS2 and an auxiliary factor of the ubiquitin-conjugating
enzyme, Ube2n (10). Ube2v1 is a
unique sub-member of the E2 family as it lacks the conserved
catalytic cysteine in E2(11).
Ube2v1 complexes with Ube2n and activates the nuclear factor κB
(NF-κB) signaling pathway, which is involved in the regulation of
cancer (12). Previous studies have
demonstrated that Ube2v1 activates the NF-κB signaling pathway via
the Ube2v1-Ubc13 complex and promotes CRC metastasis by
epigenetically suppressing autophagy (6,13).
However, the clinical and pathological relevance of Ube2v1 in CRC
has not yet been elucidated, at least to the best of our knowledge.
The present study thus examined Ube2v1 expression in CRC tissues,
and identified associations between Ube2v1 expression and the
clinical and pathological characteristics of patients with CRC in
order to determine the clinical significance of Ube2v1 expression
in this type of cancer.
Patients and methods
Study population
Patients with CRC who underwent surgical treatment
at the Sunshine Union Hospital (Weifang, China) from July, 2022 to
June, 2023 were selected retrospectively and their cancer tissues
were collected. The inclusion criteria were as follows: i) Patients
who were confirmed to have CRC by a colonoscopy biopsy and
post-operative pathological analysis; ii) patients who did had not
received any pre-operative radiotherapy, chemotherapy or other
related treatments; iii) all specimens were fixed promptly,
processed correctly, and met the standards for slide preparation in
which the cancer tissues contained all the layers of the tumor; and
iv) the patient medical records were complete. The exclusion
criterion was a history of malignant tumor treatment in other parts
of the body. The inclusion of clinical data was based on written
informed consent provided by the patient and approval from the
Ethics Committee of Sunshine Union Hospital (Approval no.
2022-04-0043). A total of 37 cases were included, with 19 males and
18 females. The ages of the patients ranged from 43-80 years, with
a median age of 63 years. There were 12 patients <60 years of
age and 25 patients ≥60 years of age. There were 20 cases of CRC in
the colon and 17 cases of CRC in the rectum. There were 20 cases
with tumor diameters <5 cm and 17 cases with tumor diameters ≥5
cm. There were 20 cases with moderate to high pathological
differentiation and 17 cases with poor differentiation. There were
9 cases with invasion depths of T1-T2 and 28 cases with invasion
depths of T3-T4. There were 6 cases with vascular invasion and 31
cases with no vascular invasion. There were 7 cases with perineural
invasion and 30 cases with no perineural invasion. There were 16
cases with lymph node metastasis and 21 cases with no lymph node
metastasis. The clinicopathological characteristics of the patients
are presented in Table I.
 | Table IAssociation between Ube2v1 expression
and the clinicopathological characteristics of patients with
colorectal cancer. |
Table I
Association between Ube2v1 expression
and the clinicopathological characteristics of patients with
colorectal cancer.
| | Ube2v1
expression | |
|---|
| Characteristic | No. of patients | Positive | Negative | χ2 | P-value |
|---|
| Sex | | | | 0.755 | 0.385 |
|
Male | 19 | 9 | 10 | | |
|
Female | 18 | 6 | 12 | | |
| Age (years) | | | | - | 0.724a |
|
<60 | 12 | 4 | 8 | | |
|
≥60 | 25 | 11 | 14 | | |
| Location of
tumor | | | | 0.359 | 0.549 |
|
Colon | 20 | 9 | 11 | | |
|
Rectum | 17 | 6 | 11 | | |
| Tumor size (cm) | | | | 0.359 | 0.549 |
|
≥5 | 17 | 6 | 11 | | |
|
<5 | 20 | 9 | 11 | | |
| Tumor
differentiation | | | | 2.006 | 0.157 |
|
Well or
moderate | 20 | 6 | 14 | | |
|
Poorly | 17 | 9 | 8 | | |
| pT stage | | | | - | 0.056a |
|
pT1-2 | 9 | 1 | 8 | | |
|
pT3-4 | 28 | 14 | 14 | | |
| Vascular
invasion | | | | - | 0.670a |
|
Present | 6 | 3 | 3 | | |
|
Absent | 31 | 12 | 19 | | |
| Perineural
invasion | | | | - |
>0.999a |
|
Present | 7 | 3 | 4 | | |
|
Absent | 30 | 12 | 18 | | |
| Lymph node
metastasis | | | | - |
<0.001a |
|
Present | 16 | 12 | 4 | | |
|
Absent | 21 | 3 | 18 | | |
Bioinformatics analysis
The ‘Diff Exp’ module in the TIMER database
(https://cistrome.shinyapps.io/timer/)
(14) shows differentially expressed
genes in various cancers and normal tissues. TIMER was utilized to
predict differences in Ube2v1 expression between cancer and normal
tissues from multiple cancer patients. The association between
Ube2v1 expression and the prognosis of patients with CRC was also
investigated using the Gene Expression Profiling Interactive
Analysis (GEPIA)2 database (http://gepia2.cancer-pku.cn) and the Human Protein
Atlas database (HPA, https://www.proteinatlas.org/).
Immunohistochemical staining and
evaluation
CRC specimens were fixed in 10% neutral formalin,
embedded in paraffin, and cut into 3-µm-thick sections for
immunohistochemical staining. The sections were incubated with the
primary antibody Ube2v1 (cat. no. E-AB-18501, Elabscience
Biotechnology Co., Ltd.; diluted 1:100) overnight at 4˚C.
Subsequent procedures were performed according to the enhanced
polymer method (Data S1), and
finally, the slides were sealed with neutral balsam. The results of
immunohistochemistry were evaluated by two independent
pathologists. The staining intensity was scored on a scale of 0-3
with 0 indicating negative staining, 1 indicating weak staining, 2
indicating medium staining, and 3 indicating strong staining. The
extent of staining, which was defined as the percent of the tumor
that stained positive relative to the whole tumor, was scored on a
scale of 0-4 with 0 indicating 0%, 1 indicating 1-25%, 2 indicating
26-50%, 3 indicating 51-75%, and 4 indicating 76-100%. An overall
protein expression score ranging from 0-12 was calculated by
multiplying the staining intensity and staining extent scores
(6). Overall scores <6 indicated
a low expression, and scores ≥6 indicated a high expression.
Statistical analysis
Statistical analyses were performed using SPSS
version 26.0 software (IBM Corp.). The two-sided Chi-squared
(χ2) test was used to evaluate significance of the
associations between Ube2v1 expression and the clinicopathological
characteristics of the patients with CRC. Fisher's exact test was
used when >20% of the cells in the contingency table had an
expected count of ≤5 individuals. P<0.05 was considered to
indicate a statistically significant difference.
Results
Ube2v1 expression is higher in CRC
tissues
The distributions of gene expression levels are
displayed using box plots in the TIMER database results, with the
statistical significance of differential expression evaluated using
the Wilcoxon test. The results showed that Ube2v1 expression was
significantly elevated in colon adenocarcinoma and rectum
adenocarcinoma tissues compared to normal tissues (Fig. 1).
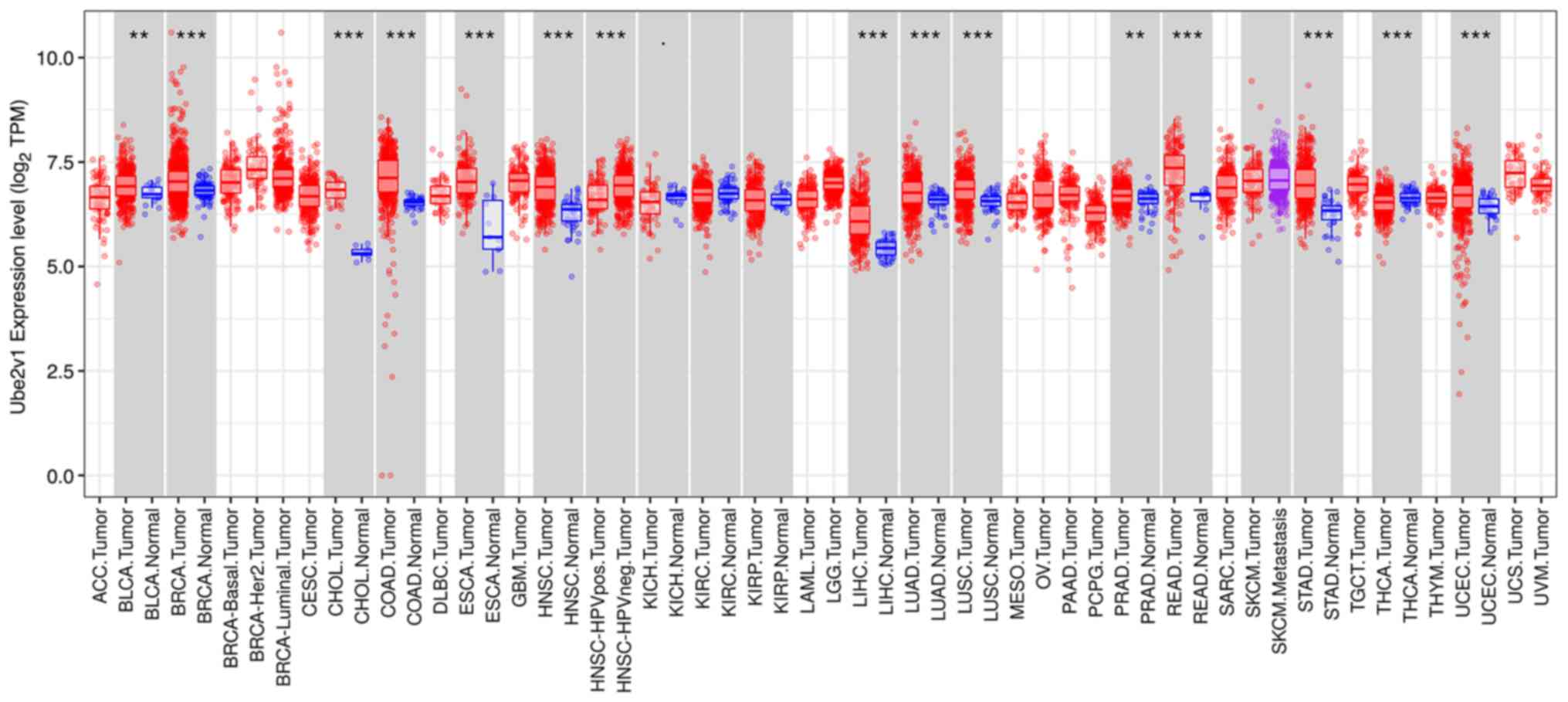 | Figure 1Expression of Ube2v1 in various types
of tumors. The analysis using the TIMER database revealed the
differential expression of Ube2v1 between various types of tumors,
including colorectal cancer and normal tissues.
**P<0.01 and ***P<0.001. Ube2v1,
ubiquitin-conjugating enzyme E2 variant 1; ACC, adrenocortical
carcinoma; BLCA, bladder Urothelial Carcinoma; BRCA, breast
invasive carcinoma; CESC, cervical squamous cell carcinoma and
endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon
adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell
lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma; HNSC, head
and neck squamous cell carcinoma; HPV, human papillomavirus; KICH,
kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP,
kidney renal papillary cell carcinoma; LAML, acute myeloid
leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular
carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell
carcinoma; MESO, mesothelioma; OV, ovarian serous
cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG,
pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma;
READ, rectal adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous
melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell
tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine
corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM,
uveal melanoma. |
Associations between Ube2v1 expression
and clinical pathological features of patients with CRC
Ube2v1 expression in CRC tissues was evaluated using
immunohistochemistry, and the associations between Ube2v1
expression and the clinical features of patients with CRC,
including sex, age, tumor location, tumor size, degree of
differentiation, depth of tumor, vascular invasion, perineural
invasion and lymph node metastasis were analyzed.
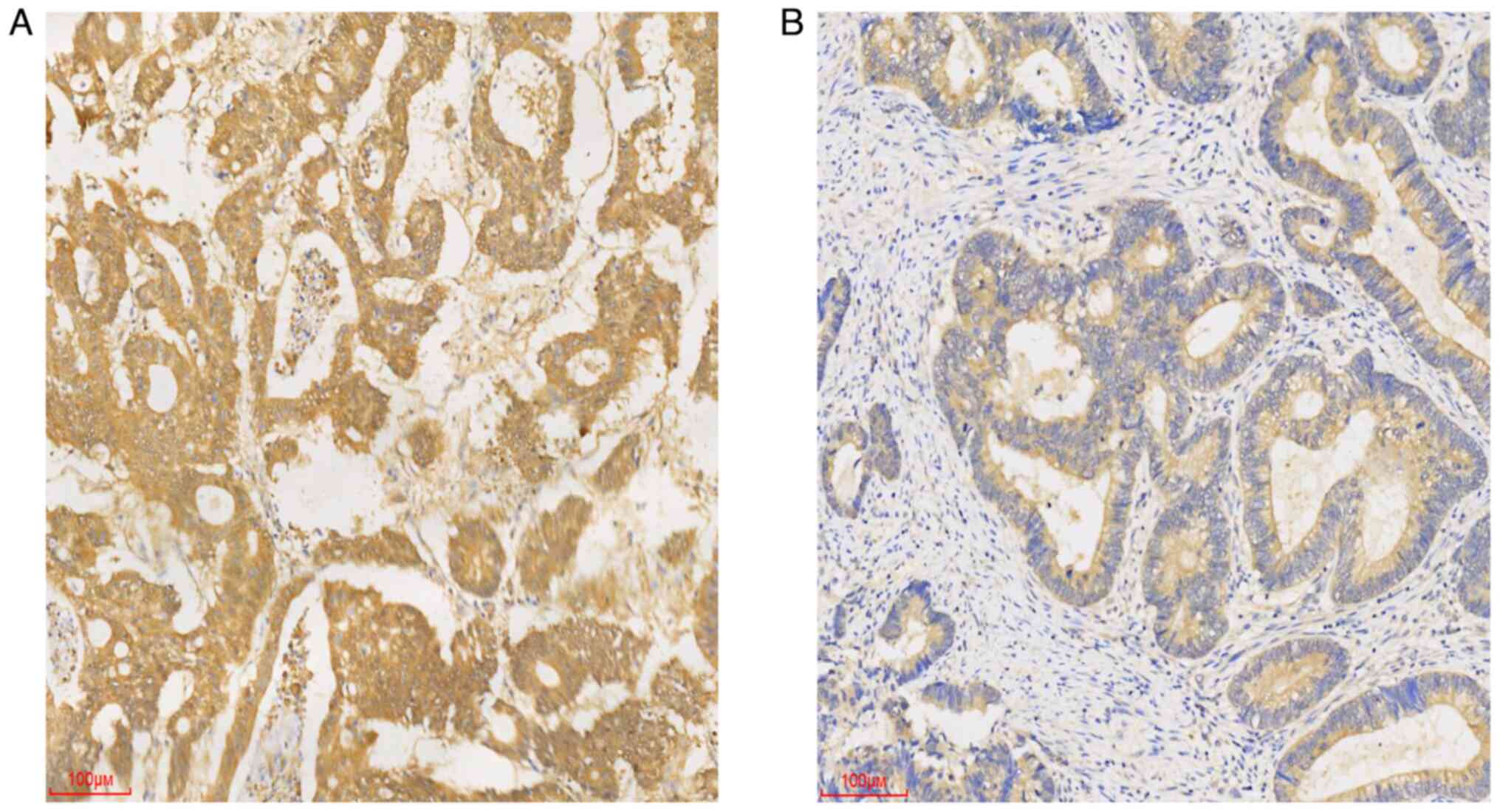
Immunohistochemical staining revealed that Ube2v1 expression in CRC
tissues localized to the cytoplasm (Fig.
2). In addition, the Ube2v1 expression level was closely
associated with the presence of lymph node metastasis and exhibited
a trend towards stage/invasion association (pT stage) (Table I).
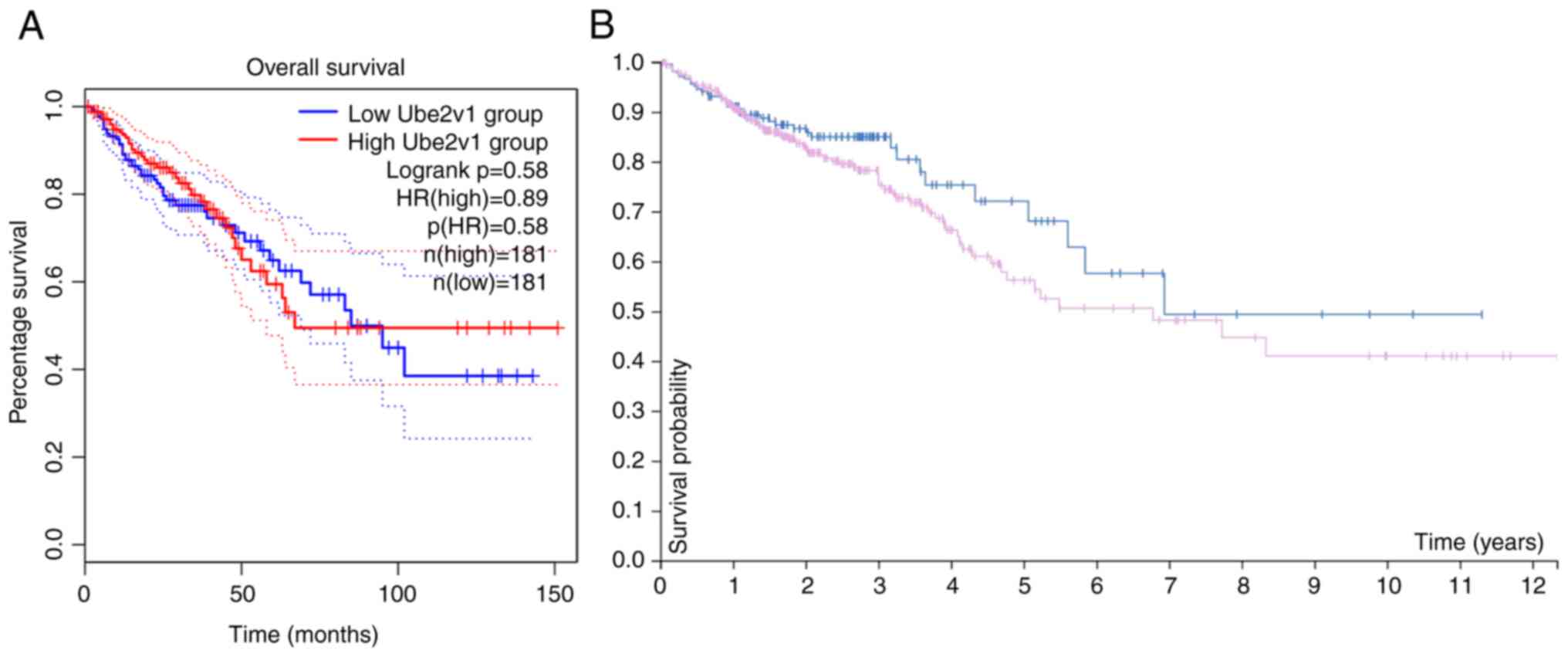
Ube2v1 expression is not associated
with the prognosis of patients with CRC
The association between Ube2v1 expression level and
patient prognosis was assessed using log-rank tests in both the
GEPIA2 and HPA databases. The analysis of the GEPIA2 database
(P=0.58, Fig. 3A) and the HPA
database (P=0.15, Fig. 3B) did not
reveal any significant association between the expression level of
Ube2v1 and the prognosis/survival of patients with CRC.
Discussion
Ube2v1 is an E2 variant and the corresponding gene
is located on chromosome 20q13.2(15). Ube2v1 was initially identified as an
activator of the trans-activating factor c-fos, and it is the
mammalian homolog of yeast MMS2(16). However, Ube2v1 and MMS2 have
different functions. For example, MMS2 forms a Ubc13-MMS2 complex
necessary for DNA damage repair but not for NF-κB activation,
whereas Ubc13-Ube2v1 is involved in NF-κB activation, but not DNA
repair (17). Ube2v1 interacts with
Ubc13 through non-covalent binding to mediate formation of
K63-linked polyubiquitin chains (K63Ub chains), which activate the
NF-κB pathway to regulate inflammation and cancer occurrence
(6,16,18).
Previous research has demonstrated that Ube2v1-regulated matrix
metalloproteinase-1 expression plays a key role in breast cancer
cell invasion and metastasis, which requires NF-κB activation
(16), and that Ube2v1 promotes CRC
metastasis by mediating Sirt1 ubiquitination, which suppresses
autophagy epigenetically (6).
In addition to the evidence provided by Shen et
al (6), who demonstrated that
Ube2v1 promoted CRC metastasis, and the evidence from the study by
Wu et al (19), which
demonstrated that Ube2v1 promoted breast cancer metastasis, Ren
et al (8) found that a high
expression of Ube2v1 was associated with the poor prognosis of
patients with cervical cancer, and Dikshit et al (20) demonstrated that the silencing of
Ube2v1 reduced malignant melanoma growth. All these studies
indicate that there is an association between Ube2v1 expression and
the prognosis of patients with various cancerous tumors. Due to
disagreements between two pathologists regarding the results of the
immunohistochemistry of CRC and normal tissue, the present study
decided to use TIMER and found that Ube2v1 expression was elevated
in colon cancer tissues when compared with normal colon tissues.
The results of immunohistochemistry revealed that Ube2v1 expression
was associated with lymph node metastasis, and a trend towards an
association with stage/invasion (pT stage) was observed.
Furthermore, due to the fact that some patients refused the
follow-up for various reasons during the process, the remaining
number of patients who continued to be followed-up was insufficient
to ensure the accuracy of the statistical results. Therefore, the
method of bioinformatics was adopted to investigate the association
between the expression level of UBE2V1 and the prognosis of
patients with CRC. Despite multiple studies suggesting that a high
expression of Ube2v1 promotes CRC metastasis and affects patient
prognosis, the bioinformatics analysis in the present study
demonstrated that Ube2v1 expression was not associated with the
prognosis/survival of patients with CRC. It was hypothesized that
this may be due to different data processing and analysis methods,
different sample selection and processing procedures, experimental
biases, and errors in handling samples. A recent study (21) proposed that cancer is a complex
multidimensional spatiotemporal ‘unity of ecology and evolution’
pathological ecosystem. Therefore, the aforementioned
contradictions may not be due to a few specific reasons and may
need to be comprehensively analyzed from multiple angles. In
summary, further experiments are required to validate the
association between Ube2v1 and the prognosis of patients with
CRC.
In conclusion, the present study found that Ube2v1
expression was higher in CRC tissues than in normal tissues, and
that Ube2v1 expression was associated with lymph node metastasis.
Due to the limited sample size, further studies with larger sample
sizes are warranted in order to validate and improve the accuracy
of the results.
Supplementary Material
Subsequent procedures used for
immunohistochemistry.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Research
Projects of Weifang Municipal Health Committee (grant no.
WFWSJK-2022-239) and the Sunshine Union Hospital Research Project
(grant no. 2022YGRH043).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QM and LG drafted the manuscript and conceived the
study. JB, NS, XY, LL, YC and WG performed the research and
analyzed the data. QM wrote the manuscript. LG and NS revised the
manuscript. QM and LG confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Sunshine Union Hospital (Approval no. 2022-04-0043).
The patients provided written informed consent prior to
participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fang Y, Yan C, Zhao Q, Xu J, Liu Z, Gao J,
Zhu H, Dai Z, Wang D and Tang D: The roles of microbial products in
the development of colorectal cancer: A review. Bioengineered.
12:720–735. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Dharwadkar P, Zaki TA and Murphy CC:
Colorectal cancer in younger adults. Hematol Oncol Clin North Am.
36:449–470. 2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fan A, Wang B, Wang X, Nie Y, Fan D, Zhao
X and Lu Y: Immunotherapy in colorectal cancer: Current
achievements and future perspective. Int J Biol Sci. 17:3837–3849.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Roberts JZ, Crawford N and Longley DB: The
role of Ubiquitination in Apoptosis and Necroptosis. Cell Death
Differ. 29:272–284. 2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
do Patrocinio AB, Rodrigues V and
Magalhães LG: P53: Stability from the ubiquitin-proteasome system
and specific 26S proteasome inhibitors. ACS Omega. 7:3836–3843.
2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Shen T, Cai LD, Liu YH, Li S, Gan WJ, Li
XM, Wang JR, Guo PD, Zhou Q, Lu XX, et al: Ube2v1-mediated
ubiquitination and degradation of Sirt1 promotes metastasis of
colorectal cancer by epigenetically suppressing autophagy. J
Hematol Oncol. 11(95)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Finley D: Recognition and processing of
ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem.
78:477–513. 2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Haglund K and Dikic I: Ubiquitylation and
cell signaling. EMBO J. 24:3353–3359. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Takahashi Y, Ishii Y, Nishida Y, Ikarashi
M, Nagata T, Nakamura T, Yamamori S and Asai S: Detection of
aberrations of ubiquitin-conjugating enzyme E2C gene (UBE2C) in
advanced colon cancer with liver metastases by DNA microarray and
two-color FISH. Cancer Genet Cytogenet. 168:30–35. 2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Xu N, Gulick J, Osinska H, Yu Y, McLendon
PM, Shay-Winkler K, Robbins J and Yutzey KE: Ube2v1 positively
regulates protein aggregation by modulating ubiquitin proteasome
system performance partially through K63 ubiquitination. Circ Res.
126:907–922. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ren Z, Liu Z, Ma S, Yue J, Yang J, Wang R,
Gao Y and Guo Y: Expression and clinical significance of UBE2V1 in
cervical cancer. Biochem Biophys Rep. 28(101108)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang Q, Lenardo MJ and Baltimore D: 30
years of NF-κB: A blossoming of relevance to human pathobiology.
Cell. 168:37–57. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu Z, Neufeld H, Torlakovic E and Xiao W:
Uev1A-Ubc13 promotes colorectal cancer metastasis through
regulating CXCL1 expression via NF-κB activation. Oncotarget.
9:15952–15967. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Niu T, Wu Z and Xiao W: Uev1A promotes
breast cancer cell migration by up-regulating CT45A expression via
the AKT pathway. BMC Cancer. 21(1012)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Syed NA, Andersen PL, Warrington RC and
Xiao W: Uev1A, a ubiquitin conjugating enzyme variant, inhibits
stress-induced apoptosis through NF-kappaB activation. Apoptosis.
11:2147–2157. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Andersen PL, Zhou H, Pastushok L, Moraes
T, McKenna S, Ziola B, Ellison MJ, Dixit VM and Xiao W: Distinct
regulation of Ubc13 functions by the two ubiquitin-conjugating
enzyme variants Mms2 and Uev1A. J Cell Biol. 170:745–755.
2005.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang Y, Li Y, Yang X, Wang J, Wang R,
Qian X, Zhang W and Xiao W: Uev1A-Ubc13 catalyzes K63-linked
ubiquitination of RHBDF2 to promote TACE maturation. Cell Signal.
42:155–164. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu Z, Shen S, Zhang Z, Zhang W and Xiao W:
Ubiquitin-conjugating enzyme complex Uev1A-Ubc13 promotes breast
cancer metastasis through nuclear factor-кB mediated matrix
metalloproteinase-1 gene regulation. Breast Cancer Res.
16(R75)2014.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Dikshit A, Jin YJ, Degan S, Hwang J,
Foster MW, Li CY and Zhang JY: UBE2N promotes melanoma growth via
MEK/FRA1/SOX10 signaling. Cancer Res. 78:6462–6472. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Luo W: Nasopharyngeal carcinoma ecology
theory: Cancer as multidimensional spatiotemporal ‘unity of ecology
and evolution’ pathological ecosystem. Theranostics. 13:1607–1631.
2023.PubMed/NCBI View Article : Google Scholar
|