1. Introduction
Lung cancer is the leading cause of cancer-related
mortality in the world, and 85% of cases are caused by tobacco
smoke (1). Other established risk
factors for lung cancer are exposure to second-hand cigarette
smoke, occupational exposure to agents such as asbestos, nickel,
chromium or arsenic, exposure to radiation, including radon gas in
homes, and exposure to air pollution (2).
The International Agency for Research on Cancer
(IARC) estimated in 2008 (3) that
the crude incidence and mortality of cancer was 12.7 and 7.6
million cases, respectively, of which 56% of new cancer cases and
63% of cancer deaths occur in the less developed regions of the
world. The most commonly diagnosed (N, proportion of total) cancers
worldwide are lung (1.61 million, 12.7%), breast (1.38 million,
10.9%) and colorectal cancers (1.23 million, 9.7%). Furthermore,
the most common causes (N, proportion of total) of cancer mortality
are lung (1.38 million, 18.2%), stomach (738,000, 9.7%) and liver
cancer (696,000, 9.2%).
In Korea, according to the 2008 annual report of
cancer statistics (4), the number
of new cancer cases was 178,816, a 7.8% increase compared to that
of 2007. The crude incidence rate of cancer in 2008 was 361.9 per
100,000, a 26% increase compared to that of 2001. Stomach cancer
was the most common form, while lung cancer was the 4th most
common. Furthermore, lung cancer is the most frequent cancer in
males over 65 years of age. The 5-year survival rate of lung cancer
between 2004 and 2008 was 17.5%, an increase of 4.8% compared to
that of the period between 1996 and 2000 (4).
In the United States, 75% of patients with lung
cancer present symptoms of advanced incurable disease (5). Despite advances in the treatment of
lung cancer, the 5-year survival rate for all stages combined is
approximately 16% (6). Patients
diagnosed at earlier stages inevitably have a significantly
improved 5-year survival rate: 60–75% for stage I disease (7). An efficacious screening test that
could result in early detection and reduced mortality would thus
represent a major advance in dealing with lung cancer
mortality.
Early detection would be a significant step towards
reducing lung cancer incidence and mortality. However, conventional
diagnostic methods for lung cancer are unsuitable for widespread
screening, as they are commonly expensive and occasionally miss
tumors or invasive cancers (8–11).
Computed tomography (CT) is widely used for early screening of lung
cancer, although it often produces high false-positive rates
(12,13). Better diagnostic methods are
urgently required to improve the detection of lung cancer. Tissue
and blood have been used extensively for the early detection of
lung cancer (14–16). The detection of aberrant promoter
methylation or metal ions in sputum has also been used (17–19).
Monoclonal antibody detection, fluorescence bronchoscopy and
low-dose spiral CT increase diagnostic sensitivity and improve the
ability to localize early-stage lesions (20), but the screening methods are
limited by the sample availability or composition variability.
Recently, biomarker discovery and their clinical use
have been accelerated by the completion of the human genome project
and the progress of techniques in proteomics (21). Numerous potential DNA biomarkers
have been discovered as lung cancer biomarkers (22). Progress has consequently been made
in early diagnosis, therapy guidance and prognosis monitoring of
cancers.
In this review, we discuss various molecular
biomarkers of lung cancer, and how reported individual biomarkers
could be used for the early diagnosis of lung cancer.
2. Biomarkers of lung cancer
Due to the vast development of knowledge over the
past several decades, different methods have been suggested to
classify lung cancer biomarkers. However, these classifications
should be considered in context, as identification of lung cancer
biomarkers is one of the major multidisciplinary areas in the
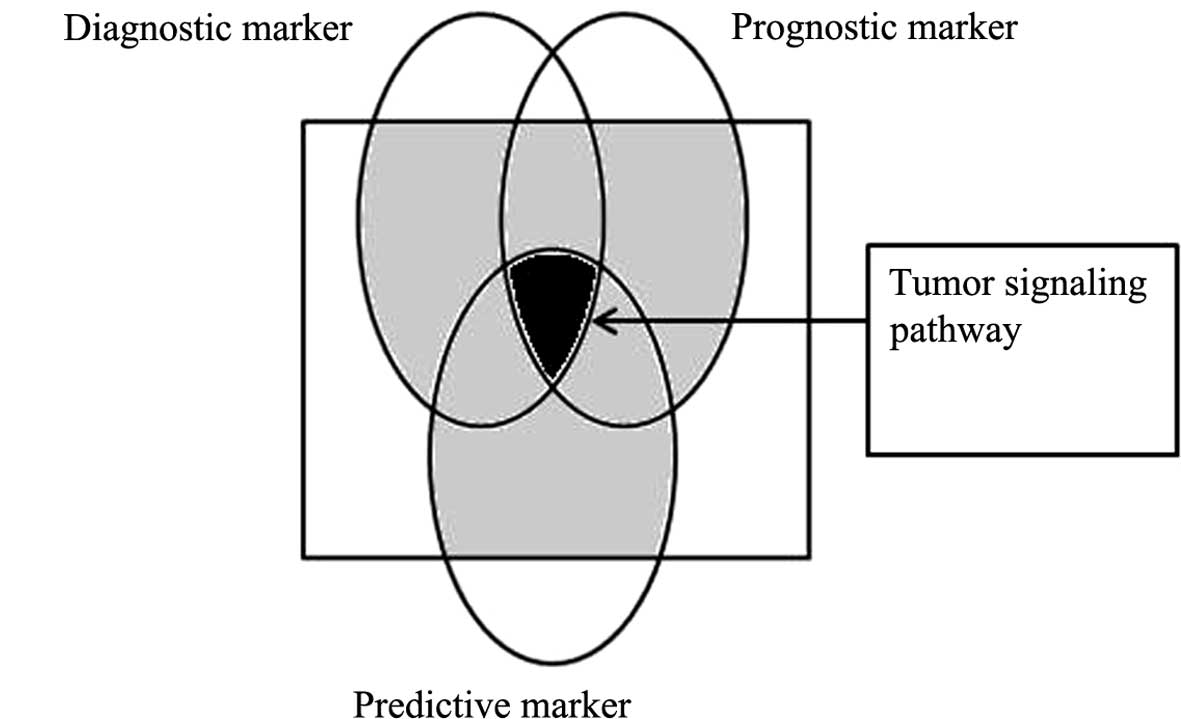
biomedical field. A schematic for the classification of biomarkers
is shown in Fig. 1.
Predictive, diagnostic and prognostic
biomarkers of lung cancer
Prognostic biomarkers are based on the
distinguishing features between benign and malignant tumors. They
could be selected based on the differentiation status of tumors,
which may affect clinicians’ decisions related to treatment
modalities. For example, the lung metagene model predicted
recurrence for individual patients significantly better than
clinical prognostic factors and was consistent across all early
stages of lung cancer (23,24).
Such markers are also important from the point of view of
predicting relapse of lung cancer.
Predictive biomarkers, occasionally referred to as
response markers, are utilized exclusively in assessing the effect
of administration of a specific drug. These biomarkers allow
clinicians to select a set of chemotherapeutic agents that will
work best for an individual patient. For example, gefitinib is
useful in non-small cell lung cancer (NSCLC) showing only epidermal
growth factor receptor (EGFR) mutation (25,26).
Consequently, EGFR mutation is a predictive lung cancer
biomarker.
Diagnostic markers may be present in any stage
during cancer development (27,28).
Carcinoembryonic antigen (CEA) in NSCLC is an example of a
diagnostic marker present in the early stages of lung cancer.
Moreover, a diagnostic cancer marker could be specific to stage,
tissue, relapse, follow-up and age.
Lung cancer biomarkers on the basis of
biomolecules
Lung carcinogenesis is a multi-step process
resulting from the accumulation of altered molecules generated from
genetic and epigenetic abnormalities of genes that are involved in
cell cycle, senescence, apoptosis, repair, differentiation and cell
migration controls (9,29). There are several distinct types of
cancer biomarkers based on different areas: genetics, epigenetics,
proteomics and metabolomics (22).
Genetics-based cancer biomarkers utilize functions such as DNA
arrays, polymerase chain reaction (PCR), reverse transcriptase
polymerase chain reaction (RT-PCR), DNA sequencing and fluorescent
in situ hybridization (FISH), to detect the genetic
alterations occurring in the cancerous state. Recent development of
epigenetic modification analyses has also improved tools for cancer
biomarkers. Epigenetic modification usually occurs in the CpG
island of the gene regulatory regions, which results in the
downregulation of the gene expression (30,31).
Proteomics includes techniques such as mass spectrometry (MS),
enzyme-linked immunosorbent assay (ELISA) and immunohistochemistry,
and it utilizes these tools to discover novel cancer biomarkers and
validate them in clinical trials. Other than using macromolecules
such as proteins and DNAs, metabolomics is concerned with the study
of low molecular weight molecules or metabolites such as amino
acids, peptides, lipids and carbohydrates (22,32).
DNA
Single nucleotide polymorphisms (SNPs) in many genes
are major DNA markers including XRCC1, ATM and
p53 (33). Other major DNA
markers include loss of heterozygosity (LOH), variation in copy
number of genes, chromosomal aberrations at a gross cytogenetic
level, such as translocation/fusion (BCR-ABL, PML-RARA
translocation in leukemias), micro-satellite instability (MSI) and
epigenetic modifications (27,34–36).
In many cases the inactivation is induced by loss of
DNA or accidental chromosomal rearrangement during cell division.
The most well-known, frequently-occurring abnormality is deletion
of the short arm of chromosome 3 (3p), where several tumor
suppressor genes (TSG) are present (37,38).
Loss of chromosomal material has also been detected in metaplastic
epithelium tissues of current-or ex-smokers. The loss of one allele
or LOH indicates a predisposing potential to lung cancers (39,40).
Notably, besides nuclear aberrations, alterations in
mitochondrial DNA (mtDNA) molecules are suggested as clear
biomarkers for lung cancers (41,42).
Epigenetic modification of nucleic acids and associated proteins
(histones and non-histones) are important in carcinogenesis
(35,43,44).
Histone de-acetylation, lysine-specific histone-H3 methylation and
promoter region CpG methylation modulate transcription of
tumor-suppressor genes (CDKN2A, TP53, APC, BRCA1) and DNA
mismatch-repair genes (MLH1 or the
O6-methyl-guanine-DNAmethyltransferase gene, MGMT). Gene silencing
by CpG methylation is one of the best-characterized epigenetic
modifications to date (35,41,44).
The degree of methylation in sputum/serum from patients with lung
cancer is directly implicated in the severity of the lesions.
RNA
Messenger RNAs (mRNAs) are promising biomarkers, and
microarrays represent a powerful approach for their discovery in
blood. More recent microarray studies have identified additional
early detection signatures. Some of the methods used to detect
cancer biomarkers at the RNA expression level include quantitative
reverse transcription polymerase chain reaction (RT-qPCR), serial
analysis of gene expression (SAGE), differential display,
bead-based methods and microfluid card and micro-array analysis
(45). A study of peripheral blood
mononuclear cells (PBMCs) from lung cancer patients identified a
signature of 29 genes that distinguished patients from controls
(46). Panels of mRNA biomarkers
for early detection have also been identified in bladder, breast
and renal carcinomas (47–49). These studies suggest that
blood-based mRNA signatures may be potentially useful tools for the
early detection of lung cancer.
The potential use of microRNAs (miRNAs) as
biomarkers for lung cancer has also been demonstrated. Several
studies have identified pathognomonic or tissue-specific miRNA
expression profiles in lung and other types of cancer (50–52).
There is sufficient evidence indicating that miRNA expression
profiles could be used to classify lung cancer, which also suggests
a correlation between disease prognosis and therapeutic outcome.
miRNA could act as a tumor suppressor, as well as an oncogene
(53). For example, in lung
cancer, let-7 is a suppressor for RAS. mir17 and mir21
clusters are oncogenic and modulate PTEN and TGFβ-RII (52). These observations emphasize the
potential application of miRNAs as biomarkers for diagnosis,
prognosis, stage, risk stratification and prediction and
drug-responses in patients with cancer.
Protein markers
Several proteins are currently in use for the
detection of lung cancer (Table
I). An oncofetal protein, CEA and cytokeratin 19-fragment
(CYFRA 21–1) proteins have been reported as potential indices for
monitoring response to treatment among advanced NSCLC patients
(5,54). Neuron-specific enolase (NSE) and
progastrin-releasing peptide (ProGRP) are also available to use as
lung cancer biomarkers for SCLC (55,56).
However, these protein biomarkers also usually lack lung-cancer
specificity.
 | Table ICurrently available protein-based
biomarkers in the detection of lung cancer. |
Table I
Currently available protein-based
biomarkers in the detection of lung cancer.
| Biomarkers | Diagnosis | Therapy | Prognosis
monitoring | Ontology
monitoring | Details |
|---|
| CEA | AdenoCA, LCLC
(>10 μg/l) | AdenoCA, Advanced
NSCLC | AdenoCA, NSCLC | Cellular
component
Cell membrane: lipid anchor Immunoglobulin superfamily | In combination with
CYFRA
Often elevated in smokers |
| CYFRA21-1 | NSCLC (23–70%), SCC
(no., sensitivity) | Advanced NSCLC | NSCLC, SCC | Structural
constitutent of cytoskeleton | Often elevated in
patients with benign lung diseases |
| TPA | NSCLC, SCC | - | NSCLC | | - |
| ProGRP | SCLC (47–86%)
(>200 ng/l, highly suspicious) (no., sensitivity) | SCLC | - | Neuropeptide
hormone activity | Increased in renal
failure and certain benign lung diseases In combination with
NSE |
| NSE | SCLC (>100 μg/l,
high probability) (sensitivity for SCLC, high 74%) | SCLC | SCLC | Phosphoglycerate
dehydrogenase activity
Subcellular location (cytoplasm) | In combination with
ProGRP
May correlate with short survival Increased in inflammatory
diseases |
| Tumor M2- pyruvate
kinase | AdenoCA (sensitivty
for SCLC, 50–71%) | - | AdenoCA | Pyruvate kinase
activity
Glycolysis
Cytoplasm | Increased in
multiple malignant and certain inflammatory diseases |
There are other potential lung cancer biomarker
molecules that are still not available for clinical use. The
potency of serum amyloid A (57,58)
and haptoglobin-α2 (59,60) as lung cancer biomarkers require
more clinical validation before they are approved for clinical use.
Plasma granulocyte colony-stimulating factor (G-CSF) levels were
significantly increased in patients with lung cancer, particularly
in the advanced TNM stages (61,62).
These results suggest that plasma G-CSF could be used to support
the diagnostic process of lung cancer staging and as an indicator
of metastasis.
Lung cancer and proteomics
Comprehensive and in-depth discovery of the disease
proteome is an important issue in recent proteomic developments.
Improvement in sample preparation tools will reduce the intrinsic
limitations in biological samples, such as variation among
individuals, differences in genetic make-up and non-specific
changes (63,64).
Protein-based lung cancer biomarkers are derived
from the techniques of classical 2-dimensional (2-D) fluorescence
difference gel electrophoresis (DIGE), polyacrylamide gel
electrophoresis (PAGE), mass spectroscopy, matrix-associated laser
absorption desorption ionization time-of-flight (MALDI-TOF),
surface-enhanced laser absorption desorption ionization
time-of-flight (SELDITOF) and reverse phase microarray (65–67).
Quantum dots and nanoparticles are recent additions
to the technologies available to assess the potential of protein
molecules as cancer biomarkers (68). Quantitative proteomics have been
utilized to discover biomarkers in lung cancer, such as stable
isotope labeling with amino acids in cell culture (SILAC), iTRAQ
and liquid chromatography-MS/MS (LC-MS/MS) (69–71).
Circulating tumor cells
Circulating tumor cells (CTCs) are indicators of
cancer and have been extensively reviewed (72,73).
For the early detection of cancer, one challenge is to find highly
specific markers that are able to address extremely low
signal-to-noise ratio. Detection of CTCs currently relies on a
single marker, the epithelial cell surface epitope EpCAM. Although
EpCAM is an excellent epithelial cell marker, it is not expressed
on all cancer cells (74).
Emerging approaches include the use of gene mutations, antibody
cocktails, negative selections and filtrations on the basis of cell
size or density (73).
3. Conclusion
The application of biomarkers for the early
detection of all types of cancer is of significant potential value
and deserves a similarly significant share of funding. However, in
lung cancer, there are no sensitive and specific biomarkers such as
prostate specific antigen for prostate cancer. Several biomarkers
will probably have to be used together, including DNA- and
RNA-based biomarkers, protein biomarkers, proteomics and CTCs.
Assay sensitivity and specificity need to be
improved, techniques must be standardized and validated, and
legislation on biomarkers needs to be regulated more closely. We
may be confident that general progress and marked new discoveries
will continue, as techniques for earlier detection are being
developed, which could have major impacts. The progress of clinical
medicine quite significantly depends on the testing of novel
theories.
References
|
1
|
Lee YS, Oh YM, Shim TS, Kim WS, An JS,
Choi CM and Jang SH: The clinical outcomes of photodynamic therapy
in early lung cancer patients. Tuberc Respir Dis. 71:266–270. 2011.
View Article : Google Scholar
|
|
2
|
Alberg AJ, Ford JG and Samet JM; American
College of Chest Physicians. Epidemiology of lung cancer: ACCP
evidence-based clinical practice guidelines (2nd edition). Chest.
132:29S–55S. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ministry for Health, Welfare and Family
Affairs. 2008 Annual Report of Cancer Statistics in Korea. Ministry
for Health, Welfare and Family Affairs; Seoul: 2010
|
|
5
|
Molina R, Filella X, Auge JM, et al: Tumor
markers (CEA, CA 125, CYFRA 21-1, SCC, and NSE) in patients with
non-small cell lung cancer as an aid in histological diagnosis, and
prognosis. Comparison with the main clinical and pathological
prognostic factors. Tumour Biol. 24:209–218. 2003. View Article : Google Scholar
|
|
6
|
American Cancer Society. Cancer Prevention
& Early Detection Facts & Figures, 2011. American Cancer
Society; Atlanta: 2011
|
|
7
|
Scott WJ, Howington J, Feigenberg S,
Movsas B and Pisters K; American College of Chest Physicians.
Treatment of non-small cell lung cancer stage I and stage II: ACCP
evidence-based clinical practice guidelines (2nd edition). Chest.
132:234S–242S. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hirsch FR, Merrick DT and Franklin WA:
Role of biomarkers for early detection of lung cancer and
chemoprevention. Eur Respir J. 19:1151–1158. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brambilla C, Fievet F, Jeanmart M, de
Fraipont F, Lantuejoul S, Frappat V, Ferretti G, Brichon PY and
Moro-Sibilot D: Early detection of lung cancer: role of biomarkers.
Eur Respir J Suppl. 39:36S–44S. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brower V: Biomarker studies abound for
early detection of lung cancer. J Natl Cancer Inst. 101:11–13.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Peng G, Tisch U, Adams O, Hakim M, Shehada
N, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A and Haick H:
Diagnosing lung cancer in exhaled breath using gold nanoparticles.
Nat Nanotechnol. 4:669–673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
van Klaveren RJ, Oudkerk M, Prokop M, et
al: Management of lung nodules detected by volume CT scanning. N
Engl J Med. 361:2221–2229. 2009.
|
|
13
|
Yau G, Lock M and Rodrigues G: Systematic
review of baseline low-dose CT lung cancer screening. Lung Cancer.
58:161–170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ilie M, Mazure NM, Hofman V, Ammadi RE,
Ortholan C, Bonnetaud C, Havet K, Venissac N, Mograbi B, Mouroux J,
Pouysségur J and Hofman P: High levels of carbonic anhydrase IX in
tumour tissue and plasma are biomarkers of poor prognostic in
patients with non-small cell lung cancer. Br J Cancer.
102:1627–1635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yee J, Sadar MD, Sin DD, Kuzyk M, Xing L,
Kondra J, McWilliams A, Man SF and Lam S: Connective
tissue-activating peptide III: a novel blood biomarker for early
lung cancer detection. J Clin Oncol. 27:2787–2792. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Patz EF, Campa MJ, Gottlin EB, Kusmartseva
I, Guan XR and Herndon JE II: Panel of serum biomarkers for the
diagnosis of lung cancer. J Clin Oncol. 25:5578–5583. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Palmisano WA, Divine KK, Saccomanno G,
Gilliland FD, Baylin SB, Herman JG and Belinsky SA: Predicting lung
cancer by detecting aberrant promoter methylation in sputum. Cancer
Res. 60:5954–5958. 2000.PubMed/NCBI
|
|
18
|
Belinsky SA, Klinge DM, Dekker JD, Smith
MW, Bocklage TJ, Gilliland FD, Crowell RE, Karp DD, Stidley CA and
Picchi MA: Gene promoter methylation in plasma and sputum increases
with lung cancer risk. Clin Cancer Res. 11:6505–6511. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gray RD, Duncan A, Noble D, Imrie M,
O’Reilly DS, Innes JA, Porteous DJ, Greening AP and Boyd AC: Sputum
trace metals are biomarkers of inflammatory and suppurative lung
disease. Chest. 137:635–641. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kennedy TC, Miller Y and Prindiville S:
Screening for lung cancer revisited and the role of sputum cytology
and fluorescence bronchoscopy in a high-risk group. Chest.
117:72S–79S. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hirsch J, Hansen KC, Burlingame AL and
Matthay MA: Proteomics: current techniques and potential
applications to lung disease. Am J Physiol Lung Cell Mol Physiol.
287:L1–L23. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sung HJ and Cho JY: Biomarkers for the
lung cancer diagnosis and their advances in proteomics. BMB Rep.
41:615–625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Potti A, Mukherjee S, Petersen R, et al: A
genomic strategy to refine prognosis in early-stage non-small-cell
lung cancer. N Engl J Med. 355:570–580. 2006. View Article : Google Scholar
|
|
24
|
Pleasance ED, Stephens PJ, O’Meara S, et
al: A small-cell lung cancer genome with complex signatures of
tobacco exposure. Nature. 463:184–190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mitsudomi T and Yatabe Y: Mutations of the
epidermal growth factor receptor gene and related genes as
determinants of epidermal growth factor receptor tyrosine kinase
inhibitors sensitivity in lung cancer. Cancer Sci. 98:1817–1824.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McDermott U, Downing JR and Stratton MR:
Genomics and the continuum of cancer care. N Engl J Med.
364:340–350. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Verma M and Manne U: Genetic and
epigenetic biomarkers in cancer diagnosis and identifying high risk
populations. Crit Rev Oncol Hematol. 60:9–18. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Habis AH, Vernon SD, Lee DR, Verma M and
Unger U: Molecular quality of exfoliated cervical cells:
implications for molecular epidemiology and biomarker discovery.
Cancer Epidemiol Biomarkers Prev. 13:492–496. 2004.PubMed/NCBI
|
|
29
|
Martin KJ, Fournier MV, Reddy GP and
Pardee AB: A need for basic research on fluid-based early detection
biomarkers. Cancer Res. 70:5203–5206. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2(Suppl 1): S4–S11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Belinsky SA: Gene-promoter
hypermethylation as a biomarker in lung cancer. Nat Rev Cancer.
4:707–717. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Parker CE, Pearson TW, Anderson NL and
Borchers CH: Mass-spectrometry-based clinical proteomics–a review
and prospective. Analyst. 135:1830–1838. 2010.
|
|
33
|
Qian B, Zhang H, Zhang L, Zhou X, Yu H and
Chen K: Association of genetic polymorphisms in DNA repair pathway
genes with non-small cell lung cancer risk. Lung Cancer.
73:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chatterjee SK and Zetter BR: Cancer
biomarkers: knowing the present and predicting the future. Future
Oncol. 1:37–50. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Verma M: Human epigenome and cancer. Human
Genome Epidemiology. Khoury M, Bedrosian SR, Gwinn M, Higgins JPT,
Ioannidis JPA and Little J: 2nd edition. Oxford University Press;
London: pp. 551–558. 2009, View Article : Google Scholar
|
|
36
|
Sidransky D: Emerging molecular markers of
cancer. Nat Rev Cancer. 2:210–219. 2002. View Article : Google Scholar
|
|
37
|
Zabarovsky ER, Lerman MI and Minna JD:
Tumor suppressor genes on chromosome 3p involved in the
pathogenesis of lung and other cancers. Oncogene. 21:6915–6935.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xue X, Zhu YM and Woll PJ: Circulating DNA
and lung cancer. Ann NY Acad Sci. 1075:154–164. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mao L, Lee JS, Kurie JM, et al: Clonal
genetic alterations in the lungs of current and former smokers. J
Natl Cancer Inst. 89:857–862. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wistuba II, Behrens C, Milchgrub S, Bryant
D, Hung J, Minna JD and Gazdar AF: Sequential molecular
abnormalities are involved in the multistage development of
squamous cell lung carcinoma. Oncogene. 18:643–650. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Verma M and Srivastava S: Epigenetics in
cancer: implications for early detection and prevention. Lancet
Oncol. 3:755–763. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Verma M and Kumar D: Application of
mitochondrial genome information in cancer epidemiology. Clin Chim
Acta. 383:41–50. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Enokida H, Shiina H, Igawa M, et al: CpG
hypermethylation of MDR1 gene contributes to the pathogenesis and
progression of human prostate cancer. Cancer Res. 64:5956–5962.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kaneuchi M, Sasaki M, Tanaka Y, Shiina H,
Verma M, Ebina Y, Nomura E, Yamamoto R, Sakuragi N and Dahiya R:
Expression and methylation status of 14-3-3 sigma gene can
characterize the different histological features of ovarian cancer.
Biochem Biophys Res Commun. 316:1156–1162. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Velculescu VE, Zhang L, Vogelstein B and
Kinzler KW: Serial analysis of gene expression. Science.
270:484–487. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Showe MK, Vachani A, Kossenkov AV, et al:
Gene expression profiles in peripheral blood mononuclear cells can
distinguish patients with non-small cell lung cancer from patients
with nonmalignant lung disease. Cancer Res. 69:9202–9210. 2009.
View Article : Google Scholar
|
|
47
|
McConkey DJ, Lee S, Choi W, Tran M,
Majewski T, Lee S, Siefker-Radtke A, Dinney C and Czerniak B:
Molecular genetics of bladder cancer: emerging mechanisms of tumor
initiation and progression. Urol Oncol. 28:429–440. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hennig G, Gehrmann M, Stropp U, Brauch H,
Fritz P, Eichelbaum M, Schwab M and Schroth W: Automated extraction
of DNA and RNA from a single formalin-fixed paraffin-embedded
tissue section for analysis of both single-nucleotide polymorphisms
and mRNA expression. Clin Chem. 56:1845–1853. 2010. View Article : Google Scholar
|
|
49
|
Hoffmann NE, Sheinin Y, Lohse CM, Parker
AS, Leibovich BC, Jiang Z and Kwon ED: External validation of IMP3
expression as an independent prognostic marker for metastatic
progression and death for patients with clear cell renal cell
carcinoma. Cancer. 112:1471–1479. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yanagihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rosenfeld N, Aharonov R, Meiri E, et al:
MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol.
26:462–469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang W, Winder T, Ning Y, et al: A let-7
microRNA-binding site polymorphism in 3′-untranslated region of
KRAS gene predicts response in wild-type KRAS patients with
metastatic colorectal cancer treated with cetuximab monotherapy.
Ann Oncol. 22:104–109. 2011.
|
|
53
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kulpa J, Wojcik E, Radkowski A,
Kolodziejski L and Stasik Z: CYFRA 21-1, TPA-M, TPS, SCC-Ag, and
CEA in patients with squamous cell lung cancer, and in chemical
industry workers as a reference group. Anticancer Res.
20:5035–5040. 2000.PubMed/NCBI
|
|
55
|
Lamy P, Grenier J, Kramar A and Pujol JL:
Pro-gastrin-releasing peptide, neuron specific enolase, and
chromogranin A as serum markers of small cell lung cancer. Lung
Cancer. 29:197–203. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Schneider J, Philipp M, Velcovsky HG, Morr
H and Katz N: Pro-gastrin-releasing peptide (ProGRP), neuron
specific enolase (NSE), carcinoembryonic antigen (CEA), and
cytokeratin 19-fragments (CYFRA 21-1) in patients with lung cancer
in comparison to other lung diseases. Anticancer Res. 23:885–893.
2003.
|
|
57
|
Cho WC, Yip TT, Cheng WW and Au JS: Serum
amyloid A is elevated in the serum of lung cancer patients with
poor prognosis. Br J Cancer. 102:1731–1735. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cremona M, Calabrò E, Randi G, De Bortoli
M, Mondellini P, Verri C, Sozzi G, Pierotti MA, La Vecchia C,
Pastorino U and Bongarzone I: Elevated levels of the acute-phase
serum amyloid are associated with heightened lung cancer risk.
Cancer. 116:1326–1335. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Abdullah M, Marwitz S, Kähler D, Schultz
H, Kugler C, Zabel P, Vollmer E and Goldmann T: Pulmonary
haptoglobin: a new marker for adenocarcinomas of the lung?
Pathology. 43:70–72. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Shah A, Singh H, Sachdev V, Lee J,
Yotsukura S, Salgia R and Bharti A: Differential serum level of
specific haptoglobin isoforms in small cell lung cancer. Curr
Proteomics. 7:49–65. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Coward JI, Nathavitharana R and Popat S:
True hypoglycaemia secondary to treatment with granulocyte colony
stimulating factor (G-CSF) in a diabetic patient with non-small
cell lung cancer. Lung Cancer. 75:133–135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Song JS, Kim SY, Jo HJ, et al: The role
and significance of biomarker for plasma G-CSF in patients with
primary lung cancer. Tuber Respir Dis. 66:444–450. 2009. View Article : Google Scholar
|
|
63
|
Hanash SM, Pitteri SJ and Faca VM: Mining
the plasma proteome for cancer biomarkers. Nature. 452:571–579.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Taguchi A, Politi K, Pitteri SJ, et al:
Lung cancer signatures in plasma based on proteome profiling of
mouse tumor models. Cancer Cell. 20:289–299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jeong HC, Kim GI, Cho SH, Lee KH, Ko JJ,
Yang JH and Chung KH: Proteomic analysis of human small cell lung
cancer tissues: up-regulation of coactosin-like protein-1. J
Proteome Res. 10:269–276. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Du J, Yang S, Lin X, Bu L, Nan Y, Huo S
and Shang W: Use of anchorchip-time-of-flight spectrometry
technology to screen tumor biomarker proteins in serum for small
cell lung cancer. Diagn Pathol. 5:602010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Rahman SM, Gonzalez AL, Li M, et al: Lung
cancer diagnosis from proteomic analysis of preinvasive lesions.
Cancer Res. 71:3009–3017. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Seydel C: Quantum dots get wet. Science.
300:80–81. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Guha U, Chaerkady R, Marimuthu A, et al:
Comparisons of tyrosine phosphorylated proteins in cells expressing
lung cancer-specific alleles of EGFR and KRAS. Proc Natl Acad Sci
USA. 105:14112–14117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Haura EB, Müller A, Breitwieser FP, Li J,
Grebien F, Colinge J and Bennett KL: Using iTRAQ combined with
tandem affinity purification to enhance low-abundance proteins
associated with somatically mutated EGFR core complexes in lung
cancer. J Proteome Res. 10:182–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zeng X, Hood BL, Sun M, Conrads TP, Day
RS, Weissfeld JL, Siegfried JM and Bigbee WL: Lung cancer serum
biomarker discovery using glycoprotein capture and liquid
chromatography mass spectrometry. J Proteome Res. 9:6440–6449.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Taniguchi K, Uchida J, Nishino K, Kumagai
T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F and Kato
K: Quantitative detection of EGFR mutations in circulating tumor
DNA derived from lung adenocarcinomas. Clin Cancer Res.
17:7808–7815. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kaiser J: Medicine. Cancer’s circulation
problem. Science. 327:1072–1074. 2010.
|
|
74
|
Pantel K, Alix-Panabieres C and Riethdorf
S: Cancer micrometastases. Nat Rev Clin Oncol. 6:339–351. 2009.
View Article : Google Scholar : PubMed/NCBI
|