Introduction
Osteoarthritis (OA) is a group of states associated
with defective articular cartilage and changes in the underlying
bone.OA is divided into erosive or non-erosive. Erosive OA is more
abrupt and commonly exhibits subchondral bone erosions (1). Pathological changes in articular
cartilage and subchondral bone result from chondrocyte imbalance in
the extracellular matrix. Although numerous studies have reported
probable chemical or mechanical causes of cartilage destruction
(2,3), this area of research requires more
detailed investigation.
Rheumatoid arthritis (RA) is a complex, chronic
multisystemic autoimmune disease, which affects the synovial
membranes of multiple joints, cartilage and bone as well as bursa
and tendon sheaths (4). RA is a
prevalent chronic inflammatory joint disease affecting 0.5–1% of
the world’s population (5). RA
leads to severe morbidity and disability if incorrectly treated,
imposing a substantial economic burden on the affected individuals
and society. The inflammatory process associated with RA is
primarily observed in the synovial tissue. Synovial hyperplasia
results from synovial outgrowths or synovial villi, comprised of
macrophages, synovial lining cells, lymphocytes and blood vessels
(6). Joint destruction occurs when
the synovial pannus produces enzymes resulting in cartilage
penetration, cartilage damage and joint erosion (7).
Although RA and OA share similar symptoms, it has
been demonstrated that RA follows an alternative inflammatory
pathway of pathogenesis to OA. Diagnosis and assessment of RA and
OA is largely based on semi-quantitative methods of diagnosis,
including symptoms, joint damage and physical function (8). At present, no cure exists for RA and
OA and the management of these diseases depends upon early
detection and aggressive treatment. Therefore, it is increasingly
important to explore the molecular mechanisms of these diseases and
analyze the associated signaling pathways, in order to uncover an
effective therapeutic approach. In this study, we analyzed gene
expression profiles of OA and RA cells to determine differentially
expressed genes (DEGs) in the two forms of arthritis. Furthermore,
through comparison we determined changed metabolic and
non-metabolic pathways, small bioactive molecules and SNP
corresponding genes associated with RA and OA.
Materials and methods
Gene expression profiles of synovial
tissue samples from RA and OA patients and normal donors
The transcription profile of GSE1919 was obtained
from the National Center for Biotechnology Information Gene
Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and was based on the
Affymetrix Human Genome U95A Array (Santa Clara, CA, USA). A total
of 15 chips were used in this study, including 5 OA tissue chips, 5
RA tissue chips and 5 normal donor (ND) tissue chips (9). The study was approved by the Ethics
Committee of the Charité Universitätsmedizin Berlin and has been
performed in accordance with the ethical standards laid down in the
1964 Declaration of Helsinki. All patients gave their informed
consent prior to their inclusion in the study.
Analysis of DEGs
Raw data were normalized using the robust multichip
average method (10) with the
default settings implemented in the R affy package (version
2.13.0). The Limma (linear models for microarray data) method was
used to identify DEGs (11). The
original expression datasets from all conditions were extracted
into expression estimates and used to construct the linear model.
Significance of gene expression differences between OA, RA and ND
cells were tested by classical t-test and P-values were adjusted
for multiple comparisons using the false discovery rate (FDR) of
Benjamini and Hochberg (12).
FDR-corrected P<0.05 was considered to indicate a statistically
significant difference.
Pathway analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway database is a collection of manually drawn pathway maps of
the molecular interaction and reaction networks. A total of 130
pathways, involving 2,287 genes, were collected from KEGG (updated
2011/06). The Database for Annotation, Visualization and Integrated
Discovery (DAVID), a high-throughput and integrated data-mining
environment, analyzed gene lists derived from high-throughput
genomic experiments. The DAVID (13,14)
was used to identify over-represented pathways based on the
hypergeometric distribution. Pathways with P<0.05 and count
>2 were considered to be significant.
Small molecule expression analysis
CMap (the connectivity map) is a collection of gene
expression profiles from cultured human cells treated with
bioactive small molecules. The database contains 6,100 bioactive
small molecule-interfering tests and 7,056 corresponding gene
expression profiles (15). DEGs
were analyzed through the CMap database to identify small
bio-active molecules which resulted in similar or adverse gene
expression. Probes of DEGs were converted to the accession number
of GeneBank and then probe numbers for use in the CMap.
RA- and OA-related SNP analysis
RA- and OA- related SNPs were obtained following a
dbSNP database search (http://www.ncbi.nlm.nih.gov/projects/SNP/) using the
keywords ‘osteoarthritis’ and ‘osteoporosis’. SNPs of RA and OA,
which were suitable for probing, were acquired through the
comparison of their corresponding genes with DEGs under
pathological conditions.
Results
Recognition of DEGs in different
samples
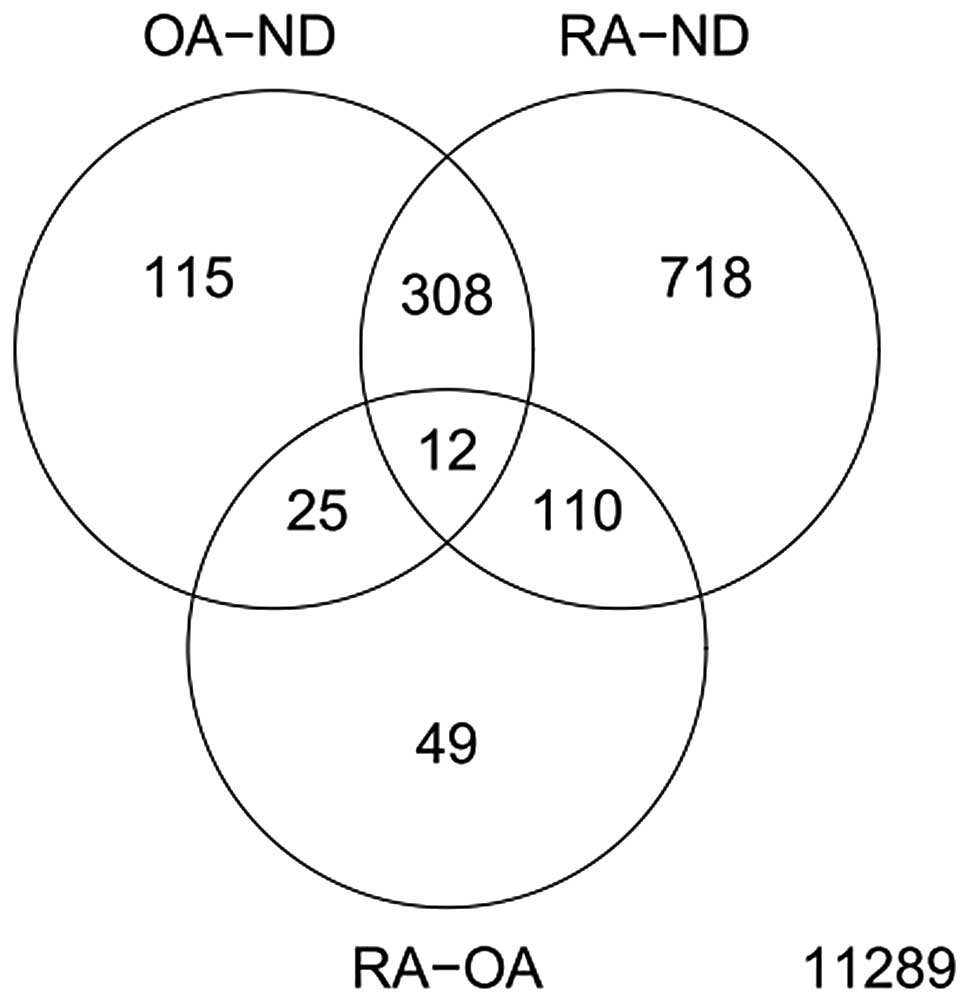
Analysis of GSE1919 using the Limma method
identified a total of 460 DEGs in OA tissues compared with normal
tissues. In RAt issues 1,148 DEGs were identified in comparison
with ND (P<0.05; Fig. 1). Only
196 DEGs were identified when we compared the gene expression in OA
tissues with that of RA. These results indicate similarities in the
molecular mechanisms of OA and RA.
Analysis of pathways induced by
arthritis
In arthritic tissues, gene expression profiles were
significantly changed compared with the normal tissues. Therefore,
DEGs were adopted to perform KEGG sub-pathway enrichment analysis.
Significantly changed pathways with ≥2 genes included and P<0.05
in arthritis were obtained. It was demonstrated that the majority
of pathways were involved in metabolic and non-metabolic processes,
indicative of a large number of changes in arthritic tissues
compared with normal tissues (Table
I). These results are likely to be important for drug discovery
for the treatment of arthritis.
 | Table IDifference in signaling pathways
between normal and arthritic tissues. |
Table I
Difference in signaling pathways
between normal and arthritic tissues.
| Arthritic tissue | Term | P-value |
|---|
| OA-ND | hsa04612:Antigen
processing and presentation | 7.07E-04 |
| hsa05322:Systemic
lupus erythematosus | 7.72E-04 |
| hsa04910:Insulin
signaling pathway | 0.001034 |
|
hsa05332:Graft-versus-host disease | 0.002426 |
| hsa03320:PPAR
signaling pathway | 0.003083 |
| hsa04940:Type I
diabetes mellitus | 0.003567 |
| hsa05416:Viral
myocarditis | 0.00369 |
| hsa05330:Allograft
rejection | 0.008703 |
|
hsa04640:Hematopoietic cell lineage | 0.011659 |
| hsa04010:MAPK
signaling pathway | 0.014961 |
| hsa05310:Asthma | 0.019068 |
| hsa04514:Cell
adhesion molecules | 0.019897 |
|
hsa04142:Lysosome | 0.024332 |
| hsa04672:Intestinal
immune network for IgA production | 0.030296 |
|
hsa04920:Adipocytokine signaling
pathway | 0.032485 |
| hsa05320:Autoimmune
thyroid disease | 0.035251 |
| RA-OA |
hsa04060:Cytokine-cytokine receptor
interaction | 2.55E-06 |
| hsa05340:Primary
immunodeficiency | 1.52E-05 |
| hsa04062:Chemokine
signaling pathway | 0.002685 |
| hsa04630:Jak-STAT
signaling pathway | 0.003031 |
| hsa04650:Natural
killer cell-mediated cytotoxicity | 0.004972 |
| hsa04660:T cell
receptor signaling pathway | 0.007133 |
| hsa04672:Intestinal
immune network for IgA production | 0.00735 |
| hsa02010:ABC
transporters | 0.032296 |
| hsa04510:Focal
adhesion | 0.040002 |
|
hsa04640:Hematopoietic cell lineage | 0.04746 |
| RA-ND | hsa04666:Fcγ
R-mediated phagocytosis | 7.49E-10 |
| hsa04612:Antigen
processing and presentation | 2.56E-07 |
| hsa04062:Chemokine
signaling pathway | 8.32E-07 |
| hsa04672:Intestinal
immune network for IgA production | 8.82E-07 |
| hsa05330:Allograft
rejection | 1.71E-06 |
| hsa04514:Cell
adhesion molecules | 2.55E-06 |
| hsa04940:Type I
diabetes mellitus | 2.58E-06 |
| hsa05416:Viral
myocarditis | 4.06E-06 |
|
hsa05332:Graft-versus-host disease | 5.22E-06 |
| hsa05322:Systemic
lupus erythematosus | 8.12E-06 |
| hsa04650:Natural
killer cell-mediated cytotoxicity | 2.57E-05 |
|
hsa05310:Asthma | 3.00E-05 |
| hsa04662:B cell
receptor signaling pathway | 3.72E-05 |
| hsa04660:T cell
receptor signaling pathway | 3.87E-05 |
| hsa05340:Primary
immunodeficiency | 4.24E-05 |
| hsa04670:Leukocyte
transendothelial migration | 1.71E-04 |
|
hsa04640:Hematopoietic cell lineage | 2.68E-04 |
| hsa04620:Toll-like
receptor signaling pathway | 3.16E-04 |
| hsa05320:Autoimmune
thyroid disease | 5.90E-04 |
| hsa04664:Fcɛ RI
signaling pathway | 6.59E-04 |
| hsa04010:MAPK
signaling pathway | 0.001477 |
|
hsa04060:Cytokine-cytokine receptor
interaction | 0.001906 |
| hsa04520:Adherens
junction | 0.004307 |
|
hsa04142:Lysosome | 0.005237 |
|
hsa04722:Neurotrophin signaling
pathway | 0.019966 |
| hsa05210:Colorectal
cancer | 0.022059 |
|
hsa04512:ECM-receptor interaction | 0.022059 |
| hsa04910:Insulin
signaling pathway | 0.02415 |
| hsa04510:Focal
adhesion | 0.031928 |
| hsa05221:Acute
myeloid leukemia | 0.039772 |
| hsa04810:Regulation
of actin cytoskeleton | 0.04152 |
| hsa05120:Epithelial
cell signaling in Helicobacter pylori infection | 0.048088 |
Analysis of small molecules resulting in
RA and OA
DEGs were first divided into upregulated and
downregulated genes and then enriched with significantly changed
genes obtained from treatment of small molecules from the CMap
database. Targeted molecules observed to induce similar effects to
arthritis were selected and the 20 targeted molecules with the
lowest P-values were enumerated (Table II). Parthenolide and
alsterpaullone were identified in OA and RA tissue analysis and are
highly relevent molecules.
 | Table IIIntersection of gene expressions
between small bio-active molecules and the differentially expressed
genes of arthritis. |
Table II
Intersection of gene expressions
between small bio-active molecules and the differentially expressed
genes of arthritis.
| Arthritic
tissue | CMap name | P-value |
|---|
| OA vs. ND | Doxorubicin | 0 |
| H-7 | 0 |
| Alsterpaullone | 0 |
| GW-8510 | 0 |
| Anisomycin | 0 |
| Thapsigargin | 0 |
| MG-262 | 0 |
| Parthenolide | 0 |
| Withaferin A | 0 |
| Cephaeline | 0 |
| 15-delta
prostaglandin J2 | 0 |
| Mitoxantrone | 0.00002 |
| Valinomycin | 0.00006 |
| Disulfiram | 0.00016 |
| Lomustine | 0.00024 |
| Terfenadine | 0.00026 |
| Lanatoside C | 0.00026 |
| Gossypol | 0.00052 |
| 5224221 | 0.00056 |
| 5194442 | 0.00062 |
| RA vs. ND | Thapsigargin | 0 |
| Parthenolide | 0 |
| Niclosamide | 0 |
| Alsterpaullone | 0.00002 |
| Helveticoside | 0.00016 |
| Valinomycin | 0.0003 |
| Fluticasone | 0.00097 |
| 5194442 | 0.00105 |
| Cephaeline | 0.00134 |
|
Diphenylpyraline | 0.00179 |
| Tiapride | 0.00192 |
|
Methylergometrine | 0.00219 |
| Metixene | 0.00292 |
| Methyldopate | 0.00294 |
| Lanatoside C | 0.00326 |
| CP-320650-01 | 0.00329 |
| Enoxacin | 0.00394 |
| Procainamide | 0.004 |
| CP-690334-01 | 0.00445 |
|
Tranylcypromine | 0.00449 |
Analysis of RA- and OA-related SNPs
A total of 10 SNPs were obtained from the dbSNP
database with the keyword ‘osteoarthritis’ and 15 were obtained
with the keyword ‘osteoporosis’. Following comparison of these
acquired SNP corresponding genes with DEGs, we revealed that no
arthritis-related SNP corresponding genes were the same as the
previously identified DEGs and three osteoporosis-related SNP
corresponding genes were identified in the DEGs (Table III).
 | Table IIICorresponding differently expressed
genes of disease-related SNPs. |
Table III
Corresponding differently expressed
genes of disease-related SNPs.
| Arthritic
tissue | Gene | SNP ID |
|---|
| OA-ND | IGF1 | 121912430 |
| COL1A2 | 72658152 |
| RA-ND | IGF1 | 1E+08 |
| SATB2 | 1E+08 |
Discussion
During the development of OA and RA, significant
changes in gene expression occur. The present study demonstrated
that more than 320 genes changed in OA and RA. Study of these
common DEGs may help identify potential broad-spectrum
anti-arthritis drugs. Furthermore, only 196 DEGs were identified
between OA and RA, including interleukin 3 receptor α
(IL3RA), transforming growth factor β receptor III and
CRYAB, indicating that the two diseases are correlated and
drugs that simultaneously treat these diseases may exist.
Cluster analysis of DEGs demonstrated several common
pathways associated with these diseases, including the classic
mitogen-activated protein kinase (MAPK) signaling pathway and the
insulin signaling pathway. The signaling pathways leading to MAPK
activation have been linked to various catabolic responses in
diseases, including arthritis (16,17).
Two immune pathways, antigen processing and presentation and
intestinal immune network for IgA production, were also activated
in the arthritic tissues, indicating that the immune response is
involved in these diseases. Inflammation and cytokines play
significant roles in RA and in certain cases of OA (17). Furthermore, changes of several cell
adhesion molecules, including integrin β2 (ITGB2) (18) and protein tyrosine phosphatase
receptor type c (PTPRC) (19) and lysosome-related molecules,
including phospholipase A2 group XV (PLA2G15) (20) and adaptor-related protein complex
1β (AP1B1) (21) in OA and
RA cells demonstrated that the two diseases altered their
microenvironment and removed the exogenous substances by the
lysosome. In addition, graft-versus-host (22) and autoimmune thyroid
disease-related pathways were also activated in OA and RA (23). Further investigation of these
pathways is likely to be shed light the network of signal pathways
under OA and RA. More studies on the DEGs between OA and RA may
provide useful information to differentiate the molecular
mechanisms associated with OA and RA.
Based on the DEGs and data from the CMap database,
we acquired a series of small molecules. Two of these small
molecules, parthenolide and alsterpaullone, demonstrated
significant similarity in OA and RA tissues (P<0.05) and
required additional analysis to determine their suitability as
broad spectrum anti-arthritis drugs.
Parthenolide is a sesquiterpene lactone of the
germacranolide class which occurs naturally in the plant feverfew
(Tanacetum parthenium). Parthenolide modulates the
NF-κB-mediated inflammatory responses in experimental
atherosclerosis (24) and blocks
lipopolysaccharide-induced osteolysis through suppression of NF-κB
activity (25). Parthenolide
induces apoptosis in acute myelogenous leukemia cells, leaving
normal bone marrow cells relatively unscathed (26). Pharthenolide also exhibits
microtubule-interfering activity (27), anti-inflammatory and
anti-hyperalgesic effects (28)
and activity against the parasite Leishmania
amazonensis(29). Since
numerous cases of OA and RA result from the interruption of immune
responses, including inflammation, it is probable that parthenolide
is a suitable therapeutic for these ailments.
Alsterpaullone is a potent, ATP-competitive
inhibitor of the cell cycle regulating cyclin-dependent kinases
CDK1/cyclin B (IC50 = 0.035 μM) and an inhibitor of
GSK-3β and the neuronal CDK5/p25 (30). In addition, alsterpaullone induces
apoptosis by activation of caspase-8 and -9 followed by disruption
of mitochondrial potential (31).
Through analysis of RA- and OA-related SNPs, we
identified that three osteoporosis-related SNP corresponding genes
(IGF1, COL1A2 and SATB2) were differentially
expressed. Insulin-like growth factor 1 (IGF-1), also called
somatomedin C, is a protein encoded by the IGF1 gene in humans
(32,33). IGF-1 is expressed and produced by
chondrocytes and is one of the anabolic growth factors associated
with cartilage (34) and thus is
involved in arthritis. Collagen α2(I) chain is a protein encoded by
the COL1A2 gene in humans (35,36).
Mutations in this gene are associated with osteogenesis imperfecta,
Ehlers-Danlos syndrome, idiopathic osteoporosis and atypical Marfan
syndrome. Special AT-rich sequence-binding protein 2 (SATB2), also
known as DNA-binding protein SATB2, is a human protein encoded by
the SATB2 gene (37). SATB2 has
been identified to be disrupted in two unrelated cases with de
novo apparently balanced chromosome translocations associated
with cleft palate and Pierre Robin Sequence (38). The present study also demonstrated
that these DEGs were often mutated under arthritis and thereby more
studies should focus on their roles in OA and RA.
The present findings shed new light on the molecular
mechanisms of OA and RA. Results revealed more than 320 DEGs in
both diseases which may be involved in OA and RA development via
MAPK and insulin signaling pathways, antigen processing and
presentation, intestinal immune network, graft-versus-host disease
and autoimmune thyroid disease-related pathways. Notably,
parthenolide and alsterpaullone were identified as important small
molecules involved in the induction of anti-inflammatory and
apoptosis-related gene expression and thus we suggest that these
molecules may be suitable anti-arthritis drugs for OA and RA.
Furthermore, mutations of IGF1, COL1A2 and SATB2
genes were critical for the pathogenesis of OA and RA. However,
there are specific limitations in our study. The pathway enrichment
was only based on the connection between genes and therefore genes
without strong neighbors are likely to be excluded from the
analysis (13). In addition,
further experimental analysis is required to confirm the
conclusions of the present study.
References
|
1
|
Roach HI: The complex pathology of
osteoarthritis: even mitochondria are involved. Arthritis Rheum.
58:2217–2218. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Berenbaum F: New horizons and perspectives
in the treatment of osteoarthritis. Arthritis Res Ther. 10(Suppl
2): S12008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dieppe P: Osteoarthritis of the knee in
primary care. BMJ. 336:105–106. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gay S, Gay RE and Koopman WJ: Molecular
and cellular mechanisms of joint destruction in rheumatoid
arthritis: two cellular mechanisms explain joint destruction? Ann
Rheum Dis. 52(Suppl 1): S39–S47. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kvien TK: Epidemiology and burden of
illness of rheumatoid arthritis. Pharmacoeconomics. 22:1–12. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Beasley J: Osteoarthritis and rheumatoid
arthritis: conservative therapeutic management. J Hand Ther.
25:163–171. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scott DL and Symmons DP: The role of
specialists in managing established rheumatoid arthritis.
Rheumatology (Oxford). 47:237–238. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Biswas S, Manikandan J and Pushparaj PN:
Decoding the differential biomarkers of Rheumatoid arthritis and
Osteoarthritis: A functional genomics paradigm to design disease
specific therapeutics. Bioinformation. 6:153–157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ungethuem U, Haeupl T, Witt H, et al:
Molecular signatures and new candidates to target the pathogenesis
of rheumatoid arthritis. Physiol Genomics. 42A:267–282. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Irizarry RA, Hobbs B, Collin F, et al:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar
|
|
11
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:32004.PubMed/NCBI
|
|
12
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 57:289–300.
1995.
|
|
13
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
14
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009.PubMed/NCBI
|
|
15
|
Lamb J, Crawford ED, Peck D, et al: The
connectivity map: using gene-expression signatures to connect small
molecules, genes and disease. Science. 313:1929–1935. 2006.
View Article : Google Scholar
|
|
16
|
Sondergaard BC, Schultz N, Madsen SH,
Bay-Jensen AC, Kassem M and Karsdal MA: MAPKs are essential
upstream signaling pathways in proteolytic cartilage
degradation-divergence in pathways leading to aggrecanase and
MMP-mediated articular cartilage degradation. Osteoarthritis
Cartilage. 18:279–288. 2010. View Article : Google Scholar
|
|
17
|
Lawrence MC, Jivan A, Shao C, et al: The
roles of MAPKs in disease. Cell Res. 18:436–442. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kotovuori A, Pessa-Morikawa T, Kotovuori
P, Nortamo P and Gahmberg CG: ICAM-2 and a peptide from its binding
domain are efficient activators of leukocyte adhesion and integrin
affinity. J Immunol. 162:6613–6620. 1999.PubMed/NCBI
|
|
19
|
Kaplan R, Morse B, Huebner K, et al:
Cloning of three human tyrosine phosphatases reveals a multigene
family of receptor-linked protein-tyrosine-phosphatases expressed
in brain. Proc Natl Acad Sci U S A. 87:7000–7004. 1990. View Article : Google Scholar
|
|
20
|
Wang A and Dennis EA: Mammalian
lysophospholipases. Biochim Biophys Acta. 1439:1–16. 1999.
View Article : Google Scholar
|
|
21
|
Peyrard M, Fransson I, Xie YG, et al:
Characterization of a new member of the human beta-adaptin gene
family from chromosome 22q12, a candidate meningioma gene. Hum Mol
Genet. 3:1393–1399. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ferrara JL, Levine JE, Reddy P and Holler
E: Graft-versus-host disease. Lancet. 373:1550–1561. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weetman AP and McGregor AM: Autoimmune
thyroid disease: further developments in our understanding. Endocr
Rev. 15:788–830. 1994.PubMed/NCBI
|
|
24
|
López-Franco O, Hernández-Vargas P,
Ortiz-Muñoz G, et al: Parthenolide modulates the NF-kappaB-mediated
inflammatory responses in experimental atherosclerosis.
Arterioscler Thromb Vasc Biol. 26:1864–1870. 2006.PubMed/NCBI
|
|
25
|
Yip KH, Zheng MH, Feng HT, Steer JH, Joyce
DA and Xu J: Sesquiterpene lactone parthenolide blocks
lipopolysaccharide-induced osteolysis through the suppression of
NF-kappaB activity. J Bone Miner Res. 19:1905–1916. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guzman ML, Rossi RM, Karnischky L, et al:
The sesquiterpene lactone parthenolide induces apoptosis of human
acute myelogenous leukemia stem and progenitor cells. Blood.
105:4163–4169. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miglietta A, Bozzo F, Gabriel L and Bocca
C: Microtubule-interfering activity of parthenolide. Chem Biol
Interact. 149:165–173. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feltenstein MW, Schuhly W, Warnick JE,
Fischer NH and Sufka KJ: Anti-inflammatory and anti-hyperalgesic
effects of sesquiterpene lactones from Magnolia and Bear’s foot.
Pharmacol Biochem Behav. 79:299–302. 2004.PubMed/NCBI
|
|
29
|
Tiuman TS, Ueda-Nakamura T, Garcia Cortez
DA, et al: Antileishmanial activity of parthenolide, a
sesquiterpene lactone isolated from Tanacetum parthenium.
Antimicrob Agents Chemother. 49:176–182. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leost M, Schultz C, Link A, et al:
Paullones are potent inhibitors of glycogen synthase kinase-3beta
and cyclin-dependent kinase 5/p25. Eur J Biochem. 267:5983–5994.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lahusen T, De Siervi A, Kunick C and
Senderowicz AM: Alsterpaullone, a novel cyclin-dependent kinase
inhibitor, induces apoptosis by activation of caspase-9 due to
perturbation in mitochondrial membrane potential. Mol Carcinog.
36:183–194. 2003. View
Article : Google Scholar
|
|
32
|
Hoppener JW, de Pagter-Holthuizen P,
Geurts van Kessel AH, et al: The human gene encoding insulin-like
growth factor I is located on chromosome 12. Hum Genet. 69:157–160.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jansen M, van Schaik FM, Ricker AT, et al:
Sequence of cDNA encoding human insulin-like growth factor I
precursor. Nature. 306:609–611. 1983. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Studer RK: Nitric oxide decreases IGF-1
receptor function in vitro; glutathione depletion enhances this
effect in vivo. Osteoarthritis Cartilage. 12:863–869. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Retief E, Parker MI and Retief AE:
Regional chromosome mapping of human collagen genes alpha 2(I) and
alpha 1(I) (COLIA2 and COLIA1). Hum Genet. 69:304–308. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wenstrup RJ, Cohn DH, Cohen T and Byers
PH: Arginine for glycine substitution in the triple-helical domain
of the products of one alpha 2(I) collagen allele (COL1A2) produces
the osteogenesis imperfecta type IV phenotype. J Biol Chem.
263:7734–7740. 1988.
|
|
37
|
Kikuno R, Nagase T, Ishikawa K, et al:
Prediction of the coding sequences of unidentified human genes. XIV
The complete sequences of 100 new cDNA clones from brain which code
for large proteins in vitro. DNA Res. 6:197–205. 1999. View Article : Google Scholar
|
|
38
|
FitzPatrick DR, Carr IM, McLaren L, et al:
Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum
Mol Genet. 12:2491–2501. 2003. View Article : Google Scholar : PubMed/NCBI
|