Introduction
Bone is a dynamic tissue that constantly undergoes
remodeling in which a coupled process of bone formation and
resorption continues throughout life. This remodeling is necessary
to maintain the structural integrity of the skeleton under
conditions of changing mechanical forces. Over the past few
decades, investigators have expounded mechanisms for the adaptive
response of bone to mechanical stimuli, including the most
well-known theory of mechanostat originally proposed by Frost
(1,2). It has been reported that disuse
activates remodeling, but inhibits modeling, leading to bone loss,
whereas overload inhibits remodeling and activates formation mode
modeling, leading to bone gain. Furthermore, strain above 1,500 μɛ
evokes bone increase (a positive adaptive response) and a strain
below 100 μɛ causes a loss of bone (a negative adaptive response),
while a strain ranging from 100 to 1,500 μɛ evokes no response.
Bone tissue functionally adjusts its mass and architecture to
mechanical stimuli, mainly depending on 2 cell types involved in
remodeling, one of which is osteoblasts, engaged in bone formation,
and the other is osteoclasts, mainly responsible for bone
resorption. Excessive osteoclast bone resorption leads to bone loss
resulting in skeletal pathologies, such as rheumatoid arthritis,
periodontal disease, postmenopausal osteoporosis, implant
osteolysis and tumor-associated bone loss (3). However, the understanding of the
cellular mechanisms producing such a mechanically meaningful
structure remains poor.
The osteoclast is a macrophage polykaryon developed
from the differentiation and fusion of hematopoietic precursors at
or near the bone surface in response to the essential tumor
necrosis factor (TNF) family-related signal molecule receptor
activator of NF-κB (RANK) ligand (RANKL) and macrophage
colony-stimulating factor (M-CSF) (4,5).
RANKL directly engages a membrane receptor, RANK, on osteoclast
precursors and mature osteoclasts to trigger multiple intracellular
signaling cascades that stimulate osteoclast gene expression,
development, function and survival (6). Mice deficient in RANKL or RANK have
severe osteopetrosis due to osteoclastogenesis (7–10).
Bone tissue is sensitive to mechanical strain. In a
previous study, in the establishment of a mechanobiology model of
bone and functional adaptation, the ulna was subjected to peak
strains of 2,000 and 3,000 μɛ, suggesting a dose-dependent
adaptation of bone to mechanical stimuli (11). However, to date, little is known
about the mechanisms invovled in the dose-response relationship
between mechanical stimuli and osteoclasts in vitro.
Therefore, in the present study, the cells were stretched with
cyclic predominantly uniaxial strain by substrate movement along a
given axis in order to mimic the mechanical stimuli within or
beyond physiological load. Strain magnitudes ranging from 0 to
5,000 μɛ were applied over a period of 3 days at a constant cycle
number and frequency to explore the effect of mechanical strain
magnitude on osteoclast fusion and activation.
Materials and methods
Cell culture
The RAW264.7 murine monocyte/macrophage cell line
(obtained from the School of Basic Medicine of the Peking Union
Medical College, Beijing, China) was used as an osteoclast
precursor, that has been shown to differentiate into
osteoclast-like cells in the presence of M-CSF and soluble RANKL
(PeproTech Inc., Rocky Hill, NJ, USA) over a period of 4–5 days
(12–14). The cells were cultured in α-minimal
essential medium (α-MEM; Gibco BRL, Rockville, MD, USA)
supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco), 1%
(v/v) penicillin-streptomycin solution (Gibco), 10 mM HEPES at 37°C
in a humidified atmosphere of 95% air and 5% CO2. For
treatment with mechanical stretching, the cells were seeded onto
34.8-mm cell culture plates at a density of 5×106
cells/cm2. After overnight incubation, the cells were
maintained in α-MEM containing 10% FBS, 1% (v/v)
penicillin-streptomycin solution, 10 mM HEPES, M-CSF (40 ng/ml) and
RANKL (40 ng/ml) for 7 days. The medium was changed every 3
days.
Application of mechanical strain to
cultured cells
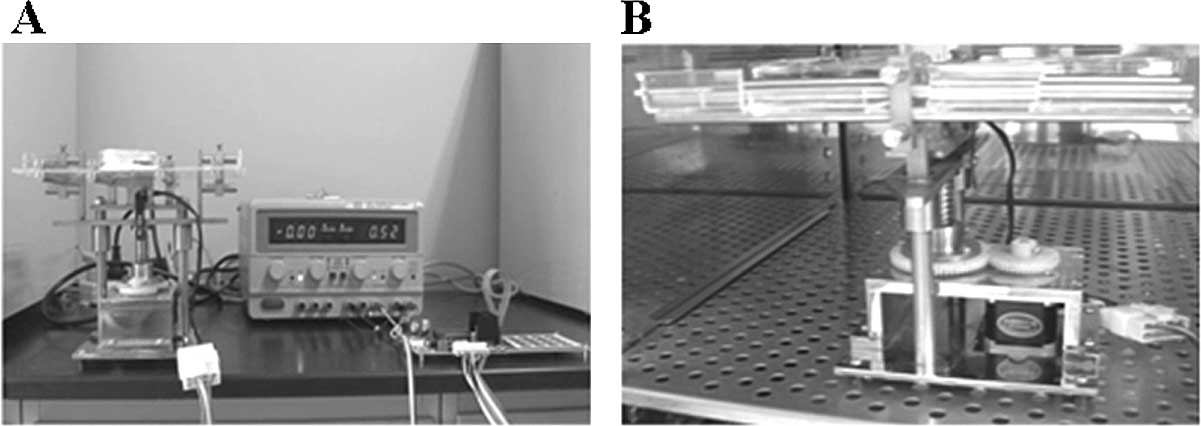
A 4-point bending system (invented by the Institute
of Medical Equipment, Academy of Military Medical Science, Tianjin,
China), composed of a cell culture unit, loading unit and circuit
controller, was used to apply uniaxial and homogeneous mechanical
strain, as described previously (Fig.
1) (15). Strain magnitudes
(substrate stretching) ranged from 1,000 to 5,000 μɛ, with a strain
frequency fixed at 0.5 Hz. After treatment with RANKL and M-CSF for
3 days, the cells were divided into 6 groups (0, 1,000, 1,500,
2,000, 2,500 and 5,000 μɛ) at random, and subjected to substrate
stretching for 3 days for 1 h per day, keeping the original culture
condition unchanged (16). After 3
days of substrate stretching, the cells were harvested for the
following experiments. The control culture was grown under the same
condition without mechanical strain.
Morphological observation and
tartrate-resistant acid phosphatase (TRAP) staining
After substrate stretching, the cells from each
group were washed twice with phosphate-buffered saline (PBS) and
stained with TRAP, using a leukocyte acid phosphatase kit
(Institute of Hematology and Blood Diseases Hospital Chinese
Academy of Medical Sciences, Tianjin, China). The TRAP+
multinucleated cells (≥5 nuclei) in 3 representative fields were
manually enumerated as osteoclasts under a microscope.
Immunocytochemistry
The cells were washed 3 times with PBS after
substrate stretching and then fixed in 4% (v/v) paraformaldehyde.
Permeabilization was performed with 0.2% (v/v) Triton X-100 (Sigma
Aldrich Chemical Co., St. Louis, MO, USA), then the cells were
incubated in 3% H2O2 to quench the endogenous
peroxidase activity. After washing with PBS, the samples were
incubated with anti-RANK polyclonal antibody (Santa Cruz
Biotechnology, Santa Cruz, CA, USA; working dilution, 1:200) at 4°C
overnight using the peroxidase-conjugated mouse IgG SABC kit (Wuhan
Boster Biological Technology, Ltd., Wuhan, China). After the
peroxidase detection, cells were washed with PBS 4 times using the
DAB Chromogenic kit (Wuhan Boster Biological Technology, Ltd.).
Seal slide with balsam neutral and average optical density of RANK
was calculated. The control samples were incubated in the same way
with 0.01 M PBS alone, instead of primary antibody.
Apoptosis of osteoclasts
The cells from each group were washed twice with PBS
and then fixed in 4% (v/v) paraformaldehyde. The fixed cells were
stained with 1 μg/ml DAPI for 20 min at room temperature. After
washing with PBS, the nuclear morphology of the cells was observed
by fluorescence microscopy. Triplicate samples were prepared for
each group and cells with condensed nuclei were counted in a
selected field of each sample.
RT-PCR
The cells from each group were harvested as
described above followed by RNA extraction using TRIzol reagent
(Invitrogen) according to the manufacturer’s instructions. cDNA was
synthesized with 3 μg of total RNA, oligo(dT) primer and the
TIANScript RT kit (Tiangen Biotech, Beijing, China). The following
primers were used: RANK, TRAP, matrix metalloproteinase-9 (MMP-9),
cathepsin K and carbonic anhydrase II (CAII). PCRs were conducted
by initial denaturation at 94°C for 3 min 30 sec, with 1 μl of
reverse transcriptase product added. The sequences of the used
primers are shown in Table I. PCR
products were separated by 1.5% agarose gel electrophoresis,
stained with ethidium bromide, photographed using Gel-Doc (Bio-Rad)
and quantified by density determination using Quantity One image
analysis software (Bio-Rad). The results were normalized to β-actin
signals determined in parallel for each sample, and the data
expressed as a ratio of the target gene to β-actin. All amplicons
were of the expected size (Table
I), and products were directly sequenced to confirm identities
by comparison with sequences published using computation performed
at NCBI and the BLAST network service.
 | Table INucleotide sequences of the used
primers. |
Table I
Nucleotide sequences of the used
primers.
| Gene | GeneBank™ accession
no. | Product size
(bp) | Temperature cycling
(cycle no.) | Forward (F) and
reverse (R) primer sequences (5′-3′) |
|---|
| TRAP | NM_007388 | 465 | 94°C, 25 sec; 57°C,
30 sec; 68°C, 35 sec (32) |
ACACAGTGATGCTGTGTGGCAACTC (F)
CCAGAGGCTTCCACATATATGATGG (R) |
| MMP-9 | NM_013599 | 354 | 94°C, 25 sec; 64°C,
30 sec; 68°C, 35 sec (32) |
CGAGTGGACGCGACCGTAGTTGG (F)
CAGGCTTAGAGCCACGACCATACAG (R) |
| RANK | NM_009399 | 351 | 94°C, 25 sec; 57°C,
30 sec; 68°C, 35 sec (32) | ACCTCCAGTCAGCAAGAAGT
(F)
TCACAGCCCTCAGAATCCAC (R) |
| CAII | NM_009801 | 407 | 94°C, 25 sec; 54°C,
30 sec; 68°C, 35 sec (32) | CTTCAGGACAATGCAGTGC
(F)
ATCCAGGTCACACATTCCAGC (R) |
| Cath K | NM_007802 | 364 | 94°C, 25 sec; 56°C,
30 sec; 68°C, 35 sec (32) |
CTGAAGATGCTTTCCCATATGTGGG (F)
GCAGGCGTTGTTCTTATTCCGAG (R) |
| β-actin | NM_007393 | 306 | 94°C, 25 sec; 55°C,
30 sec; 68°C, 35 sec (32) | GAAGAGCTATGAGCTGCCTG
(F)
CACAGAGTACTTGCGCTCAG (R) |
Statistical analysis
Data are presented as the means ± SE, typically from
2 to 3 independent trials, each with 3 replicates. Statistical
comparisons between the treatment groups were performed using
one-way analysis of variance. For simultaneous comparisons between
multiple treatments, significant differences were determined using
Bonferroni’s post hoc analysis of variance test; p<0.05 denoted
a statistically significant difference.
Results
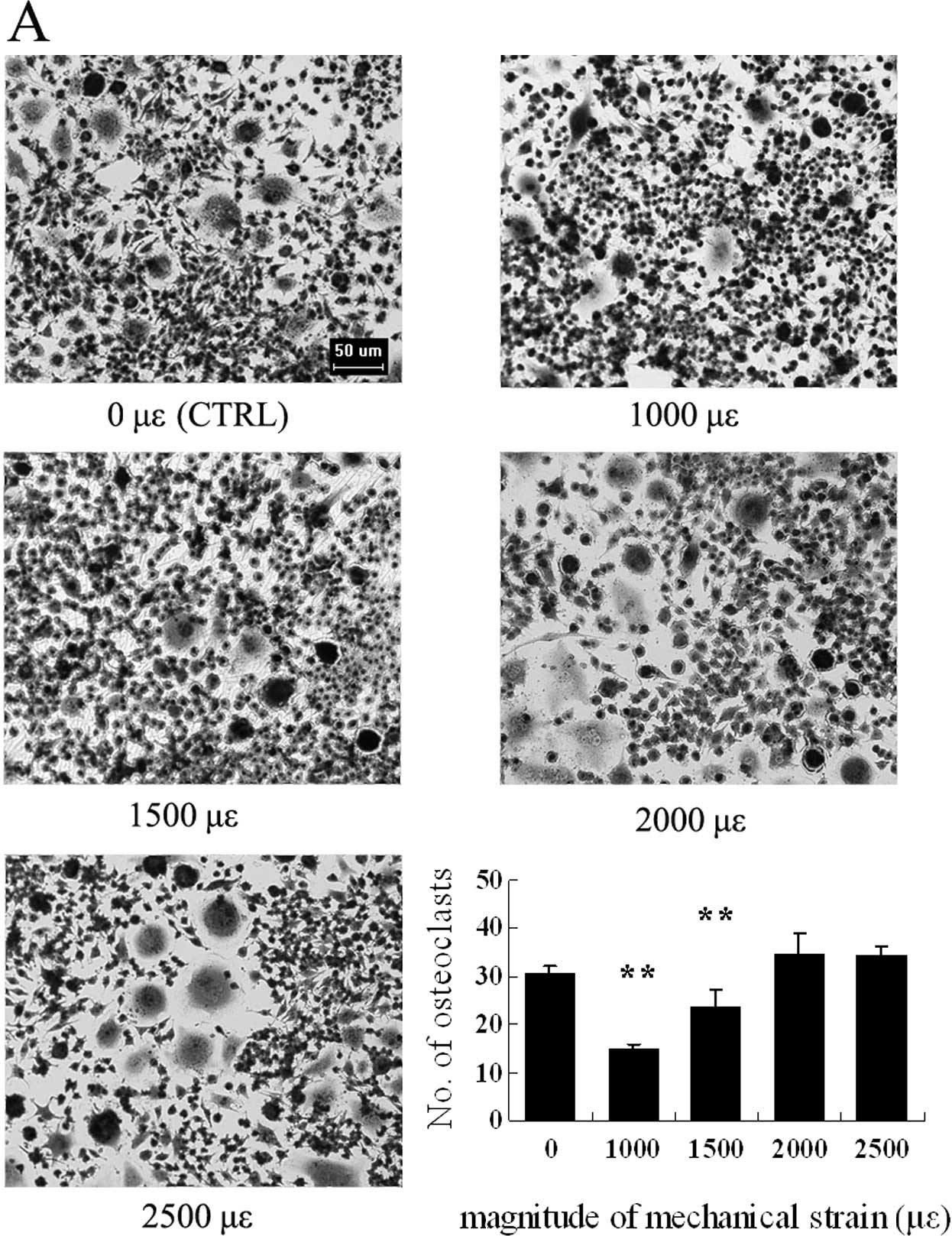
Osteoclasts change morphologically and
the number of TRAP-positive multinucleated osteoclasts vary
depending on the magnitude of mechanical strain
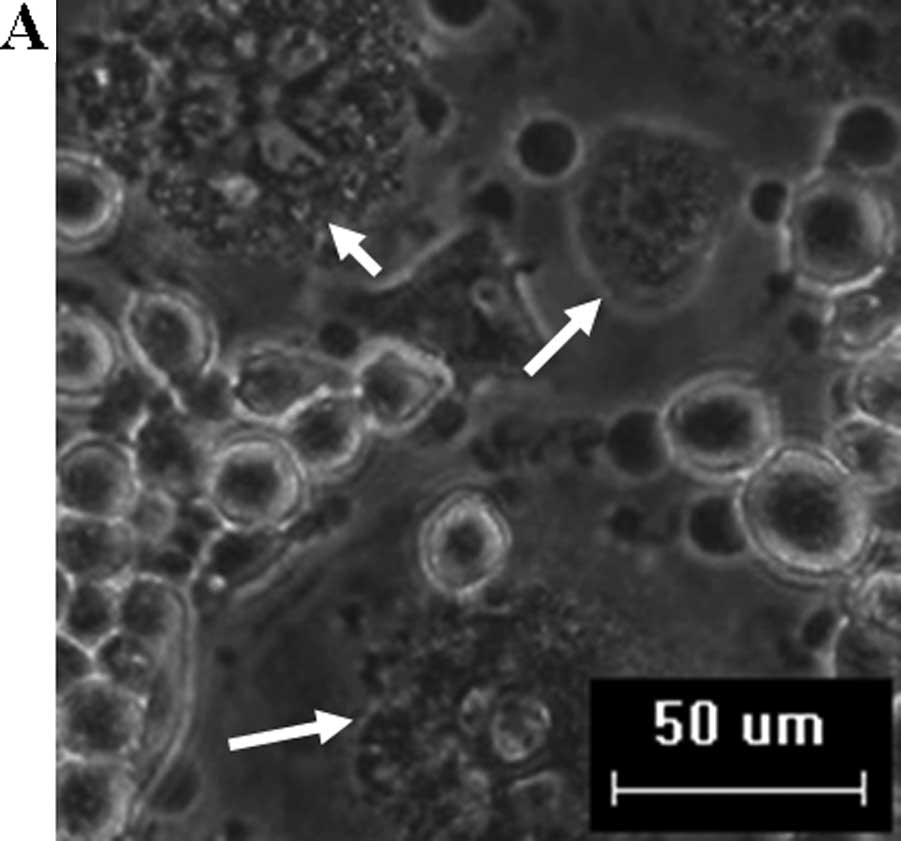
Osteoclasts in culture with M-CSF and RANKL were
observed with representative typical multinuclei. After substrate
stretching, the number of TRAP+ multinucleated
osteoclasts significantly decreased in the groups subjected to a
strain of 1,000 and 1,500 μɛ as compared to the control, whereas
there was no significant difference in the groups subjected to a
strain of 2,000 and 2,500 μɛ compared to the control (Figs. 2A and B, and 3A).
Expression of RANK increases under high
mechanical strain within physiological load
RANK is the sole signaling receptor for RANKL in the
process of inducing the development and activation of osteoclasts.
In immunocytochemistry staining for RANK, the groups subjected to a
strain of 2,000 and 2,500 μɛ showed stronger positive staining than
the control after substrate stretching (Fig. 3B).
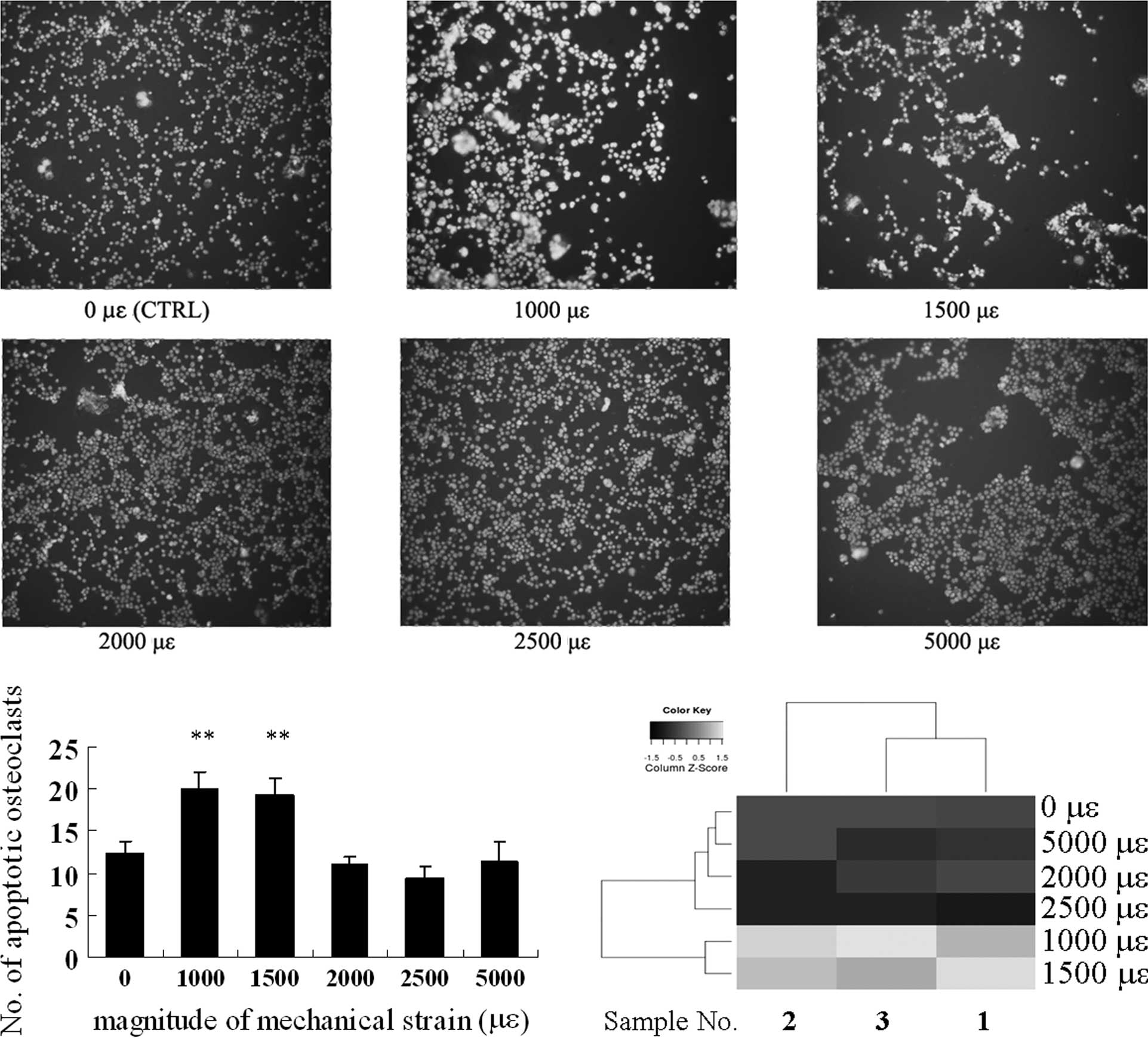
Osteoclast apoptosis is induced by
substrate stretching of low magnitude
The number of apoptotic osteoclasts increased with
the mechanical strain of 1,000 and 1,500 μɛ compared to the
control, whereas no significant stimulation was observed in the
groups subjected to a strain of 2,000, 2,500 and 5,000 μɛ (Fig. 4A and B).
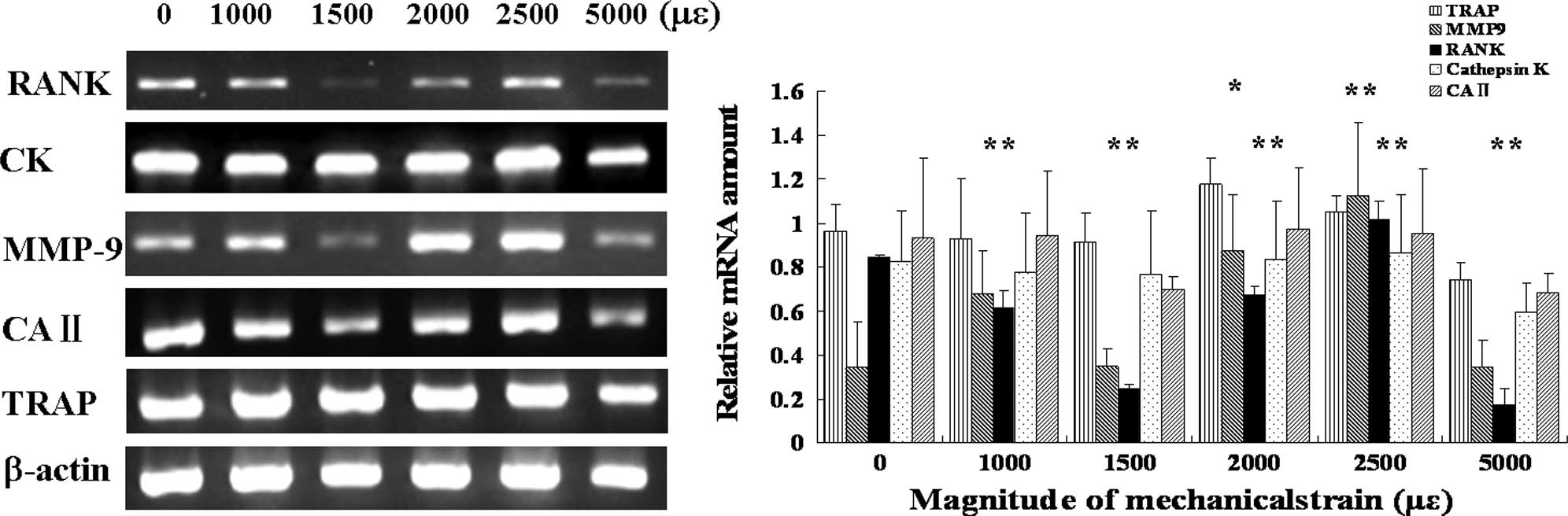
Expression of RANK and MMP-9 genes
differs depending on different magnitudes of mechanical strain
The expression of RANK mRNA increased in the groups
subjected to a strain of 2,000 and 2,500 μɛ, while it decreased in
the groups subjected to a strain of 1,000, 1,500 and 5,000 μɛ. The
expression of MMP9 mRNA increased in the groups subjected to a
strain of 2,000 and 2,500 μɛ, while there were no significant
differences in the groups subjected to a strain of 1,000, 1,500 and
5,000 μɛ. There were no significant differences in the expression
of TRAP, cathepsin K and CAII mRNA under mechanical strain compared
to the control (Fig. 5).
Discussion
Since the pioneering work of Frost, it has become
generally accepted that bone tissue maintains its structure
throughout life by the coupled activities of bone-forming
osteoblasts and bone-resorbing osteoclasts. The skeleton is able to
continually adapt to the mechanical environment by adding new bone
to withstand increased amounts of loading, and by removing bone in
response to unloading or disuse. Furthermore, most adult skeletal
diseases are due to excess osteoclastic activity. Studies have
shown that mechanical strain inhibits osteoclast differentiation
indirectly by osteoblasts and stromal cells (17,18).
Other reports have shown that compressive mechanical stress
promoted osteoclast formation through RANKL expression in synovial
cells (19). Ceratin studies have
shown that mechanical strain also directly suppresses osteoclast
differentiation (13,20,21);
others have proven that mechanical stimuli enhance osteoclast
differentiation and activities (22–24).
However, little is known about the direct effects of different
magnitudes of mechanical strain on osteoclast differentiation.
The amount of nuclei per osteoclast decreased
significantly with a mechanical strain of 1,000 and 1,500 μɛ, while
the number of osteoclasts with 5 or more nuclei increased when the
loading magnitude was tuned to 2,000 and 2,500 μɛ compared to the
control with no strain (0 μɛ) (Fig.
2). When TRAP staining was performed, the number of osteoclasts
was found to be downregulated by low mechanical strain (Fig. 3A). It is well known that RANK
signaling is essential for osteoclast differentiation, activation
and survival. In immunocytochemistry staining of osteoclasts for
RANK, the groups subjected to a strain of 2,000 and 2,500 μɛ showed
stronger positive staining than the control after 3 days of
substrate stretching (Fig. 3B).
These findings indicate that a mechanical strain of low magnitude
within physiological load inhibits osteoclast differentiation, but
promotes the fusion of mononuclear osteoclasts related to high
magnitude strain. A strain of 5,000 μɛ regarded as high magnitude
beyond physiological load had little effect (data not shown).
In RT-PCR analysis (Fig. 5), RANK mRNA expression decreased
with the mechanical strain of 1,000, 1,500 and 5,000 μɛ, while both
RANK and MMP-9 increased with the mechanical strain of 2,000 and
2,500 μɛ compared to the control. RANK mediated the ability of
precursor cells to undergo differentiation. The selective
inhibition of RANK with RANK:Fc or RANK receptor inhibitor has been
shown to block osteoclast maturation and function in vivo or
in vitro (25–27). MMP-9 has been proven to be
indispensable for the migration of osteoclasts through collagen,
both in the periosteum and developing marrow cavity of primitive
long bones (28,29). MMP-9 antisense oligonucleotides
exert an inhibitory effect on osteoclastic bone resorption by
suppressing cell migration (30).
Bone resorption is specifically reduced by the chemical inhibition
of MMP-9 (31,32). The results of this study suggest
the involvement of both RANK and MMP-9 expression depending on the
mechanical strain magnitude, thus indicating that low-magnitude
strain suppresses osteoclast differentiation, while high-magnitude
strain within physiological load stimulates osteoclast fusion and
activation.
We examined the apoptotic osteoclasts under
mechanical strain. Compared to the control, apoptotic osteoclasts
increased with the mechanical strain of 1,000 and 1,500 μɛ, whereas
there was no significant stimulation was observed with a strain of
2,000, 2,500 and 5,000 μɛ (Fig.
4), suggesting that apoptosis occurs when a mechanical strain
of low magnitude is applied. A heatmap produced by the heatmap
function from R intuitively indicated the diverse trends of cell
survival between the groups of different mechanical strain
magnitudes as well.
Currently, mechanical exercise appears to be a
concern for clinicians as a co-ordinated treatment. Little is known
about how different magnitudes of mechanical strain exert an effect
on the differentiation and fusion of osteoclasts. The data from our
study provide a further understanding of the diverse regulation by
different magnitudes of mechanical strain, leading to the
development of therapeutics optimized for diseases related to bone
loss. We found that mechanical strain turned out to be inhibitory
towards RAW264.7 cell differentiation at a low magnitude, but
stimulatory at a high magnitude within physiological load. The
osteoclasts morphologically changed depending on the different
magnitudes mechanical strain. The expression of RANK and related
genes also changed depending on different magnitutes of strain.
TRAP is often used as one of the macrophage/osteoclast lineage
markers (33,34). However, the results of the present
study on mRNA expression are in disagreement with those from the
study of Fujisaki et al, who observed that the expression of
CAII and cathepsin K induced by RANKL was increased according to
the maturity and differentiation of the RAW264.7 cells (35). No significant differences were
observed in the expression of TRAP, cathepsin K and CAII mRNA under
mechanical strain compared to the control. Therefore,
RANKL/osteoprotegerin-dependent signal transduction pathways are
possibly more active during late differentiation than
TRAP-dependent pathways. However, further study of other
co-stimulators and/or mechanisms unknown during various stages of
osteoclast development differentially regulated by mechanical
strain is warranted.
Acknowledgements
The authors are grateful to their colleagues at the
Institute of Medical Equipment, Academy of Military Medical
Science, for their tremendous support. The study was supported by
the National Natural Science Foundation Key Program of China (no.
10832012).
References
|
1
|
Frost HM: Bone Remodeling Dynamics. Thomas
CC: Springfield, IL: 1963
|
|
2
|
Frost HM: Bone ‘mass’ and the
‘mechanostat’: a proposal. Anat Rec. 219:1–9. 1987.
|
|
3
|
Teitelbaum SL: Osteoclasts: what do they
do and how do they do it? Am J Pathol. 170:427–435. 2007.
|
|
4
|
Kodama H, Nose M, Niida S and Yamasaki A:
Essential role of macrophage colony-stimulating factor in the
osteoclast differentiation supported by stromal cells. J Exp Med.
173:1291–1294. 1991.
|
|
5
|
Yasuda H, Shima N, Nakagawa N, et al:
Identity of osteoclastogenesis inhibitory factor (OCIF) and
osteoprotegerin (OPG): a mechanism by which OPG/OCIF inhibits
osteoclastogenesis in vitro. Endocrinology. 139:1329–1337.
1998.
|
|
6
|
Mochizuki A, Takami M, Kawawa T, et al:
Identification and characterization of the precursors committed to
osteoclasts induced by TNF-related activation-induced
cytokine/receptor activator of NF-kappa B ligand. J Immunol.
177:4360–4368. 2006.
|
|
7
|
Kong YY, Yoshida H, Sarosi I, et al: OPGL
is a key regulator of osteoclastogenesis, lymphocyte development
and lymph-node organogenesis. Nature. 397:315–323. 1999.
|
|
8
|
Kim N, Odgren PR, Kim DK, Marks SC Jr and
Choi Y: Diverse roles of the tumor necrosis factor family member
TRANCE in skeletal physiology revealed by TRANCE deficiency and
partial rescue by a lymphocyte-expressed TRANCE transgene. Proc Nat
Acad Sci USA. 97:10905–10910. 2000.
|
|
9
|
Dougall WC, Glaccum M, Charrier K, et al:
RANK is essential for osteoclast and lymph node development. Genes
Dev. 13:2412–2424. 1999.
|
|
10
|
Li J, Sarosi I, Yan XQ, et al: RANK is the
intrinsic hematopoietic cell surface receptor that controls
osteoclastogenesis and regulation of bone mass and calcium
metabolism. Proc Nat Acad Sci USA. 97:1566–1571. 2000.
|
|
11
|
Chen XY, Zhang XZ, Guo Y, Li RX, Lin JJ
and Wei Y: The establishment of a mechanobiology model of bone and
functional adaptation in response to mechanical loading. Clin
Biomech (Bristol, Avon). 23(Suppl 1): 88–95. 2008.
|
|
12
|
Suda T, Takahashi N, Udagawa N, Jimi E,
Gillespie MT and Martin TJ: Modulation of osteoclast
differentiation and function by the new members of the tumor
necrosis factor receptor and ligand families. Endoc Rev.
20:345–357. 1999.
|
|
13
|
Suzuki N, Yoshimura Y, Deyama Y, Suzuki K
and Kitagawa Y: Mechanical stress directly suppresses osteoclast
differentiation in RAW264.7 cells. Int J Mol Med. 21:291–296.
2008.
|
|
14
|
Takahashi N, Udagawa N, Tanaka S and Suda
T: Generating murine osteoclasts from bone marrow. Methods Mol Med.
80:129–144. 2003.
|
|
15
|
Tang LL, Wang YL, Pan J and Cai SX: The
effect of step-wise increased stretching on rat calvarial
osteoblast collagen production. J Biomech. 37:157–161. 2004.
|
|
16
|
Shibata K, Yoshimura Y, Kikuiri T, et al:
Effect of the release from mechanical stress on osteoclastogenesis
in RAW264.7 cells. Int J Mol Med. 28:73–79. 2011.
|
|
17
|
Kreja L, Liedert A, Hasni S, Claes L and
Ignatius A: Mechanical regulation of osteoclastic genes in human
osteoblasts. Biochem Biophys Res Commun. 368:582–587. 2008.
|
|
18
|
Rubin J, Murphy T, Nanes MS and Fan X:
Mechanical strain inhibits expression of osteoclast differentiation
factor by murine stromal cells. Am J Physiol. 278:C1126–1132.
2000.
|
|
19
|
Ichimiya H, Takahashi T, Ariyoshi W,
Takano H, Matayoshi T and Nishihara T: Compressive mechanical
stress promotes osteoclast formation through RANKL expression on
synovial cells. Oral Surg Oral Med Oral Pathol Oral Radiol Endod.
103:334–341. 2007.
|
|
20
|
Rubin J, Fan X, Biskobing DM, Taylor WR
and Rubin CT: Osteoclastogenesis is repressed by mechanical strain
in an in vitro model. J Orthop Res. 17:639–645. 1999.
|
|
21
|
Burger EH, Klein-Nulend J and Smit TH:
Strain-derived canalicular fluid flow regulates osteoclast activity
in a remodelling osteon – a proposal. J Biomech. 36:1453–1459.
2003.
|
|
22
|
McAllister TN, Du T and Frangos JA: Fluid
shear stress stimulates prostaglandin and nitric oxide release in
bone marrow-derived preosteoclast-like cells. Biochem Biophys Res
Commun. 270:643–648. 2000.
|
|
23
|
Kurata K, Uemura T, Nemoto A, et al:
Mechanical strain effect on bone-resorbing activity and messenger
RNA expressions of marker enzymes in isolated osteoclast culture. J
Bone Miner Res. 16:722–730. 2001.
|
|
24
|
Zhang Q, Liang X, Zhu B, et al: Effects of
fluid shear stress on mRNA expression of carbonic anhydrase II in
polarized rat osteoclasts. Cell Biol Int. 30:714–720. 2006.
|
|
25
|
Childs LM, Paschalis EP, Xing L, et al: In
vivo RANK signaling blockade using the receptor activator of
NF-κB:Fc effectively prevents and ameliorates wear debris-induced
osteolysis via osteoclast depletion without inhibiting
osteogenesis. J Bone Miner Res. 17:192–199. 2002.
|
|
26
|
Feeley BT, Liu NQ, Conduah AH, et al:
Mixed metastatic lung cancer lesions in bone are inhibited by
noggin overexpression and Rank:Fc administration. J Bone Miner Res.
21:1571–1580. 2006.
|
|
27
|
Kim H, Choi HK, Shin JH, et al: Selective
inhibition of RANK blocks osteoclast maturation and function and
prevents bone loss in mice. J Clin Invest. 119:813–825. 2009.
|
|
28
|
Blavier L and Delaisse JM: Matrix
metalloproteinases are obligatory for the migration of
preosteoclasts to the developing marrow cavity of primitive long
bones. J Cell Sci. 108:3649–3659. 1995.
|
|
29
|
Sato T, Foged NT and Delaissé JM: The
migration of purified osteoclasts through collagen is inhibited by
matrix metalloproteinase inhibitors. J Bone Miner Res. 13:59–66.
1998.
|
|
30
|
Ishibashi O, Niwa S, Kadoyama K and Inui
T: MMP-9 antisense oligodeoxynucleotide exerts an inhibitory effect
on osteoclastic bone resorption by suppressing cell migration. Life
Sci. 79:1657–1660. 2006.
|
|
31
|
Hill PA, Murphy G, Docherty AJ, et al: The
effects of selective inhibitors of matrix metalloproteinases (MMPs)
on bone resorption and the identification of MMPs and TIMP-1 in
isolated osteoclasts. J Cell Sci. 107(Pt 11): 3055–3064. 1994.
|
|
32
|
Spessotto P, Rossi FM, Degan M, et al:
Hyaluronan-CD44 interaction hampers migration of osteoclast-like
cells by down-regulating MMP-9. J Cell Biol. 158:1133–1144.
2002.
|
|
33
|
Alatalo SL, Halleen JM, Hentunen TA,
Monkkonen J and Vaananen HK: Rapid screening method for osteoclast
differentiation in vitro that measures tartrate-resistant acid
phosphatase 5b activity secreted into the culture medium. Clin
Chem. 46:1751–1754. 2000.
|
|
34
|
Rissanen JP, Suominen MI, Peng Z and
Halleen JM: Secreted tartrate-resistant acid phosphatase 5b is a
Marker of osteoclast number in human osteoclast cultures and the
rat ovariectomy model. Calcif Tissue Int. 82:108–115. 2008.
|
|
35
|
Fujisaki K, Tanabe N, Suzuki N, et al:
Receptor activator of NF-kappaB ligand induces the expression of
carbonic anhydrase II, cathepsin K, and matrix metalloproteinase-9
in osteoclast precursor RAW264.7 cells. Life Sci. 80:1311–1318.
2007.
|