Introduction
Glioma, which incurs high mortality due to its
fast-growing and invasive nature, is the most frequently
encountered intracranial tumor (1). The pathogenesis of glioma is complex
and involves the aberrant activation of proto-oncogenes (e.g.,
EGFR and IDH1/2) and inactivation of tumor suppressor
genes (e.g., TP53 and PTEN) (2–4). The
causes and consequences of intracellular signaling network
dysregulation in the development of glioma, however, have not yet
been fully elucidated. A number of treatment modalities, such as
neurosurgery, chemotherapy and radiotherapy, have been devised for
gliomas (5). Nevertheless, the
prognosis of patients with this malignancy remains dismal, with a
median survival time of 9–12 months after diagnosis. This
highlights the need to develop novel agents for the treatment of
this highly aggressive disease.
microRNA (miRNA) is a class of non-coding RNA. These
RNAs, which are of 20–25 nucleotides in length, carry out their
biological functions by binding to the 3′ untranslated regions
(UTRs) of their target mRNAs, thereby repressing the translation of
target mRNAs into proteins and/or directly inducing the degradation
of target mRNAs (6,7). miRNA genes are first transcribed to
primary miRNAs, which are then processed by Drosha into precursor
miRNAs. After exportation from the nucleus, precursor miRNAs are
further processed by the RNase Dicer to produce
miRNA:miRNA* duplexes. The mature miRNA strand then
guides the RNA-induced silencing complex to the target mRNA to
repress its expression (7,8). miRNA is emerging as a novel player in
tumorigenesis. In this regard, miRNA expression is dysregulated in
most, if not all, types of cancer. In glioma tissues, the aberrant
upregulation of miR-15b (9),
miR-21 (10), miR-221/222
(11) and miR-296 (6), and the downregulation of miR-7
(12), miR-124 (13), miR-128 (14), miR-137 (15) and miR-181a/b/c (16,17)
have been reported. Notably, the restoration of these dysregulated
miRNAs to normal expression levels has been shown to impair the
growth and survival of glioma cells. These findings support that
miRNAs play important functional roles in cancer development. Thus,
miRNA represents a novel target for the treatment of glioma.
In the present study, we employed a novel miRNA
targeting approach using a lentiviral vector to deliver anti-miRNAs
into glioma cells. This vector was constructed to produce short
hairpin RNAs, which eventually give rise to short, single-stranded
anti-miRNAs that competitively bind to and inhibit endogenous
miR-27a, a miRNA that displays oncogenic properties in many types
of solid tumors, including breast (18), gastric (19), colon (20) and pancreatic cancers (21). miR-27a has also been shown to be
expressed in glioma cells and is detectable in glioma-secreted
exosomes (22). In this study, we
provide evidence that targeting miR-27a by the lentiviral
expression of anti-miRNAs substantially impairs the malignant
phenotypes of glioma cells. Moreover, coupled with computational
prediction and proteomic analysis, we successfully identified the
targets specifically repressed by miR-27a in glioma cells.
Materials and methods
Construction of short hairpin-expressing
lentiviral vector
The short hairpin RNAs targeting miR-27a were cloned
into the pGreenPuro™ shRNA expression lentivector (SBI). The
hairpins were rationally designed for asymmetry such that the upper
strand of the hairpin did not contain the miR-27a sequence, whereas
the lower strand was fully complementary to miR-27a. The expression
of the short hairpin was driven by the H1 promoter. This vector
also encoded cop green fluorescence protein (copGFP) for the
selection of stably transfected clones. Successful cloning was
confirmed by sequencing. The pPACK-H1 Lentivector Packaging System
(SBI) and the 293TN cell line (SBI) were used for the production of
pseudoviral particles according to the manufacturer’s instructions.
U87 cells were then transduced at a multiplicity of infection (MOI)
of 3.
Cell culture and proliferation assay
The U87 human glioma cell line was obtained from the
American Type Culture Collection (Manassas, VA, USA). The cells
were maintained in DMEM (Invitrogen, Carlsbad, CA, USA),
supplemented with 10% fetal bovine serum (Thermo Scientific), 100
U/ml penicillin and 100 μg/ml streptomycin (Invitrogen) at 37°C in
a humidified atmosphere of 5% CO2 and 95% air. Cell
proliferation was measured by the colorimetric Cell Counting kit-8
(CCK-8) assay (Dojindo). The transfected cells were plated at a
density of 5,000 cells/well in 96-well plates. After incubation for
2 h to allow cell attachment to the bottom of the well, 10 μl CCK-8
solution were added to each well at 0, 24 and 48 h, and the plates
were incubated for another 2 h. The optical density was then
determined at 450 nm using a microplate reader.
Cell cycle analysis
U87 cells were fixed with ice-cold 70% ethanol in
phosphate-buffered saline, followed by incubation with 50 μg/ml
propidium iodide, 3.8 mmol/l sodium citrate and 0.5 μg/ml RNase A
at 4°C for 3 h and analyzed by flow cytometry.
Cell invasion assay
The invasive capacity of cells was determined using
the BD BioCoat Matrigel invasion chambers (8-μm pores) (BD
Biosciences). The transfected cells were seeded on the top chamber
of each insert with complete medium added to the bottom chamber.
After 48 h, cells on the membrane were wiped off with a cotton
swab. Fixed and stained with H&E, cells on the underside of the
membrane were counted from 4 microscope fields (magnification,
×200). The mean number of invading or migrating cells was expressed
as a percentage relative to the control.
Quantitation of miR-27a expression
Total RNA was isolated using TRIzol (Invitrogen) and
cDNA was synthesized using the QuantiMir kit (SBI) following the
manufacturer’s instructions. Real-time PCR was performed with
miR-27a-specific forward primer and universal reverse primer.
Conditions for real-time PCR were 50°C for 2 min, 95°C for 10 min,
40 cycles of 95°C for 15 sec and 60°C for 1 min. Quantitative PCR
was carried out using SYBR-Green JumpStart Taq ReadyMix (Sigma) and
the 7300 Real-Time PCR Detection System (ABI). The results were
analyzed using the comparative threshold cycle (CT) method.
Two-dimensional (2D) electrophoresis
The immobilized pH gradient (IPG) strip (pH 3–10,
length 13 cm; Bio-Rad, Hercules, CA, USA) was rehydrated with 1,500
μg protein in 450 ml rehydration buffer containing 7 M urea, 2 M
thiourea, 4% CHAPS, 65 mM DTT, 20 mM Trizma base, 1% IPG buffer and
0.002% bromophenol blue for 14 h at room temperature. Isoelectric
focusing (IEF) was performed using the Protean IEF System (Bio-Rad)
for a total of 70 kVh. The strip was then subjected to two-step
equilibration in a buffer containing 6 M urea, 20% glycerol, 2% SDS
and 50 mM Tris-HCl (pH 8.8), with 2% w/v DTT for the first step and
2.5% w/v iodoacetamide for the second step. The second-dimension
SDS-PAGE (12% T, 260×200×1.5 mm3) was carried out using
a Mini-Protean 3 system (Amersham Biosciences, Piscataway, NJ, USA)
according to the following procedures: 45 min at a constant power
of 5 W, followed by 20 W/gel until the bromophenol blue front
reached the bottom of the gel. Subsequently, the proteins in the
gels were visualized using the Dodeca silver staining kit
(Bio-Rad). The silver-stained protein 2D gels were scanned using an
Amersham Biosciences Imagescanner and analyzed using ImageMaster 2D
Elite v.6.0 (Amersham Biosciences).
In-gel digestion and matrix-assisted
laser desorption/ionisation-time-of-flightmass spectrometry
(MALDI-TOF-MS) identification
Protein spots were excised from gel with an
operating knife blade and equilibrated in 50 mM
NH4HCO3 to pH 8.0. After dehydrating with ACN
and drying in N2 at 37°C for 20 min, the gel pieces were
rehydrated in 10 μl trypsin solution (12.5 ng/μl in 50 mM
NH4HCO3) at 4°C for 30 min and incubated at
37°C overnight. Peptides were extracted twice using 0.1% TFA in 30%
CAN. The peptides were analyzed using Ultraflex II MALDI-TOF/TOF.
Mass spectra were internally and externally calibrated with
self-digested fragments of trypsin and standard peptides (Bruker,
USA), respectively.
Protein identification and database
searching
Protein identification using peptide mass
fingerprinting (PMF) was performed by the Mascot search engine
(http://www.matrixscience.com/;
MatrixScience Ltd., London, UK) against the SwissProt protein
database. The errors in peptide mass were in the range of 100 ppm.
One missed tryptic cleavage site per peptide was allowed during the
search. Proteins matching more than 4 peptides and with a Mascot
score of >64 were considered significant (P<0.05).
MH+ was selected as the modification. Protein
identification results were filtered with the GPS software.
miRNA target prediction
In order to define the potential targets of miR-27a,
4 publicly available computational algorithms, miRanda, miRWalk,
RNA22 and TargetScan, were used. Targets commonly predicted by 2 or
more algorithms were considered as putative targets of miR-27a.
Statistical analysis
The results are representative of multiple
experiments and expressed as the means ± SEM. Statistical analysis
was performed with an analysis of variance (ANOVA) followed by the
Turkey’s t-test. P-values <0.05 denoted statistically
significant differences.
Results
Stable lentiviral transduction of
anti-miRNAs targeting miR-27a in U87 glioma cells
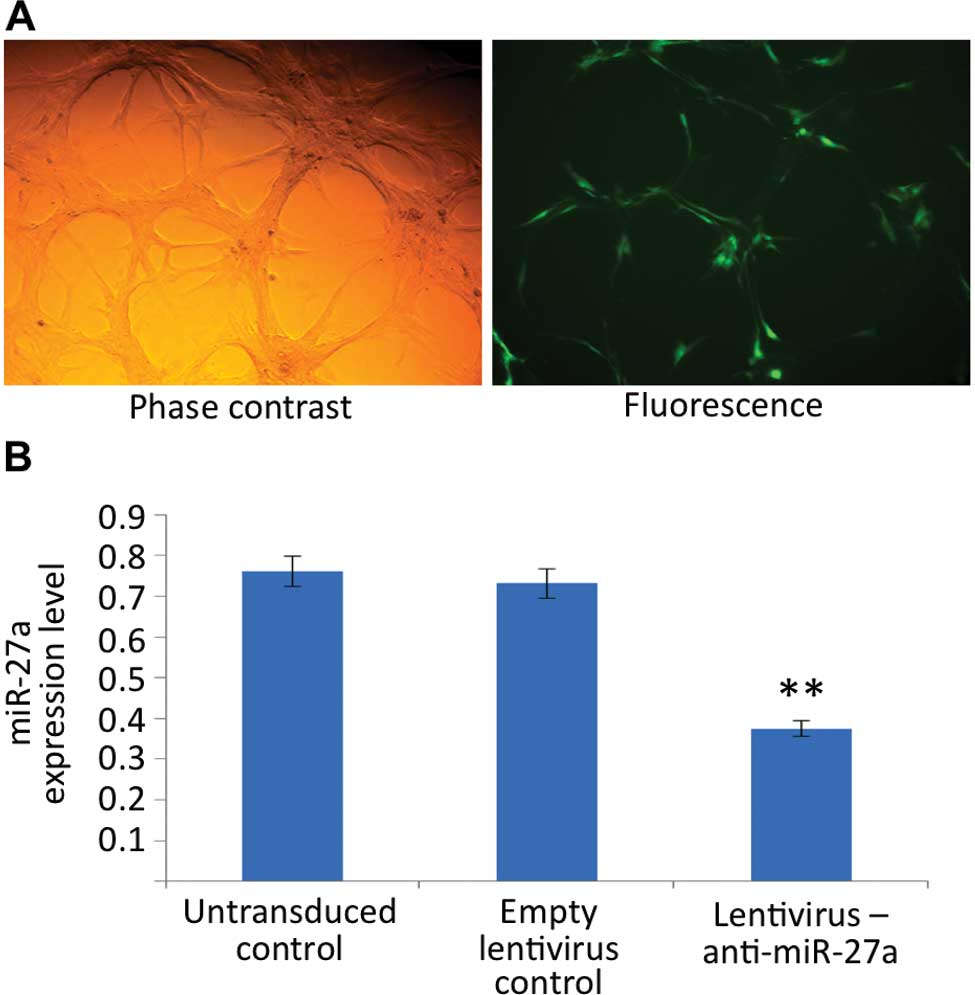
The lentivirus transduction efficiency of U87 glioma
cells was determined by the detection of GFP signals by
fluorescence microscopy at 72 h after transduction and confirmed to
be >80% (Fig. 1A). To select
stably transduced cells, fluorescence-activated cell sorting was
performed. After cell sorting, the miR-27a expression in stably
transduced U87 cells was measured by real-time PCR. Fig. 1B demonstrates that the levels of
miR-27a were significantly repressed in cells transduced with
lentivirus stably expressing anti-miR-27a when compared to the
control lentivirus-transduced cells or the untransduced cells.
Anti-miR-27a reduces the viability and
increases apoptosis of U87 cells
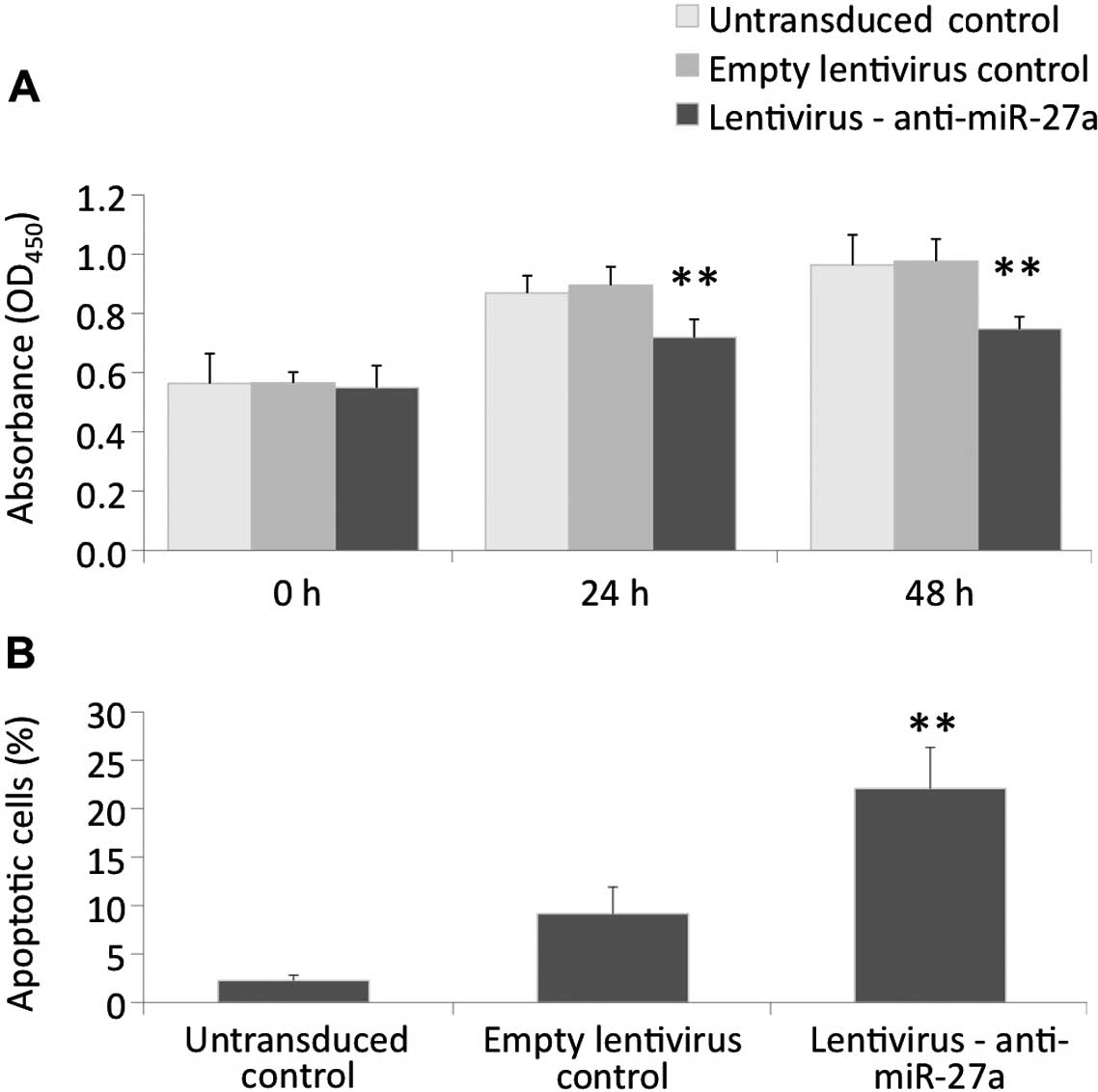
To study the effect of the blockade of miR-27a on
the proliferation of glioma cells, we examined changes in viable
cell number by CCK-8 assays after transduction with
anti-miR-27a-encoding lentiviral particles in U87 cells. The stable
expression of anti-miR-27a significantly impaired the proliferation
of U87 cells as indicated by the reduced viable cell number at the
24- and 48-h time-points when compared to the untransduced control
or cells transduced with the control lentiviral particles (Fig. 2A). At the 48-h time-point, the
viable cell number was significantly reduced by 16.5% when compared
to the control lentivirus-transduced group. To further confirm the
anti-proliferative action of anti-miR-27a, cell cycle analysis by
flow cytometry was performed. The results showed that the stable
expression of anti-miR-27a induced a substantial accumulation of
U87 cells at the sub-G1 phase without affecting the distribution of
other phases of the cell cycle, suggesting that miR-27a inhibition
may cause apoptotic cell death in U87 cells (Fig. 2B).
Anti-miR-27a reduces invasiveness of U87
cells
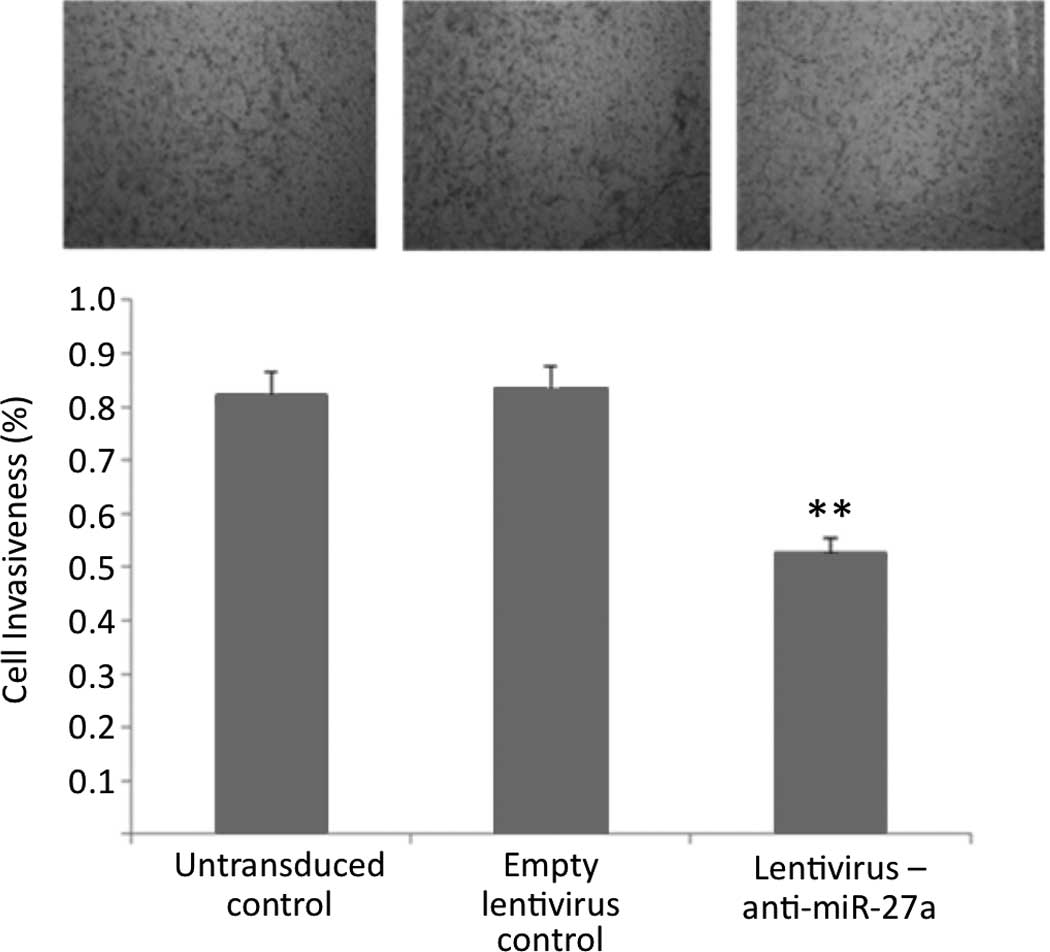
To investigate the effect of anti-miR-27a expression
on cell invasiveness, Transwell invasion assay was performed using
a chamber coated with a thin layer of extracellular matrix. The
results showed that the stable expression of anti-miR-27a
substantially reduced the invasiveness of U87 glioma cells, as
indicated by a marked decrease in the number of invaded cells
(Fig. 3).
Target identification by 2D-gel
electrophoresis/mass spectrometry
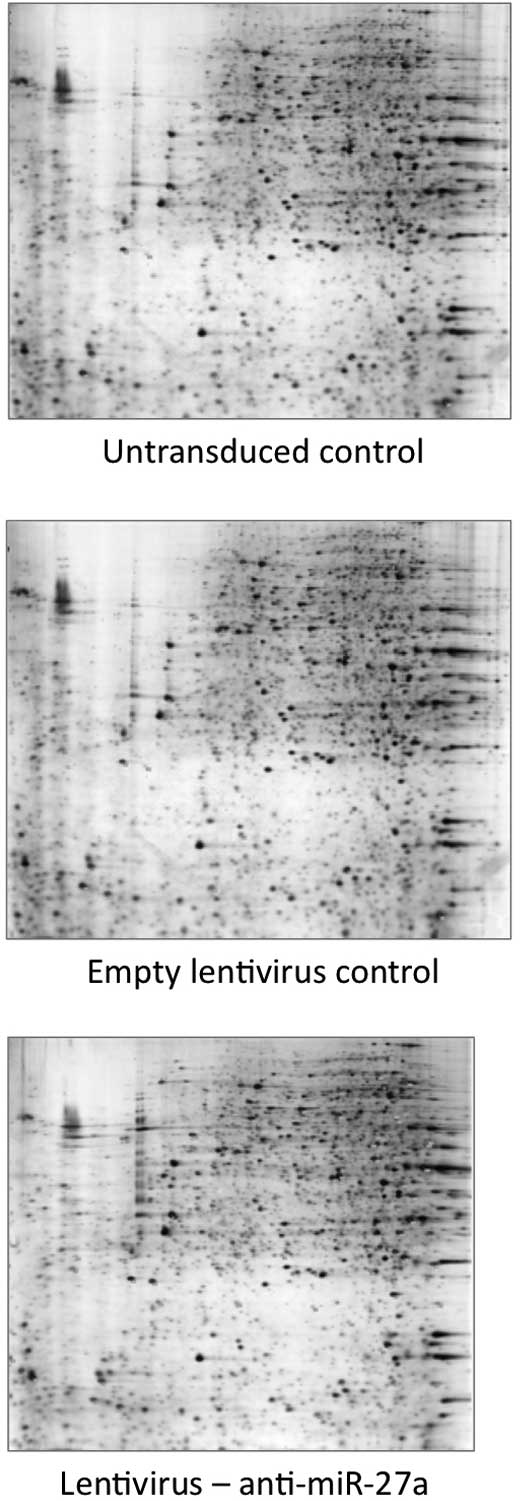
To determine the change in protein profile in
response to miR-27a inhibition, gel-based comparative proteomic
analysis was performed. As shown in Fig. 4, 29 protein spots were found to be
significantly altered in the cells stably expressing anti-miR-27a
as compared to the control lentivirus-transduced group, among which
13 and 16 proteins were found to be significantly downregulated and
upregulated, respectively. Most of the protein spots of interest
were successfully identified by MALDI-TOF MS and by subsequent
comparative sequence search in the Mascot database (Table I). Among the 16 upregulated
proteins, the mRNAs of 8 proteins were predicted by at least 2 out
of 4 computational algorithms to be the direct targets of
miR-27a.
 | Table IProteins showing a 10-fold up- or
downregulation by lentiviral transduction of anti-miR-27a in U87
cells. |
Table I
Proteins showing a 10-fold up- or
downregulation by lentiviral transduction of anti-miR-27a in U87
cells.
| Spot no. | Serial no. | Sequence
coverage | Score |
Down-/up-regulation | Calculated PI
value/nominal mass | Protein
description | Function | Algorithms that
predicted the protein as target of miR-27a |
|---|
| 276 | gi|14277700 | 67% | 104 | Down | 6.81/14905 | 40S ribosomal
protein S12 | Ribosomal
proteins | |
| 363 | gi|149242397 | 50% | 105 | Down | 8.62/15837 | Chain A, crystal
structure of RhoGDI E155h, E157h mutant | Negative regulator
of Cdc42 activation | |
| 381 | gi|62702115 | 59% | 139 | Down | 6.12/25320 | Unknown | Unknown | |
| 392 | gi|4506181 | 59% | 118 | Down | 6.92/25996 | Proteasome subunit
α type-2 | Processing of class
I MHC peptides | |
| 400 | gi|5453549 | 55% | 109 | Down | 5.86/30749 |
Peroxiredoxin-4 | Activator of the
transcription factor NF-κB | |
| 408 | gi|6912328 | 38% | 89 | Down | 5.53/31444 |
N(G),N(G)-dimethylarginine
dimethylaminohydrolase 1 isoform 1 | Nitric oxide
generation | |
| 422 | gi|119603918 | 49% | 93 | Down | 6.46/31898 | Capping protein
(actin filament) muscle Z-line, α 2, isoform CRA_a | Regulation of
growth of the actin filament by capping the barbed end of growing
actin filaments | |
| 428 | gi|4506667 | 57% | 141 | Down | 5.71/34423 | 60S acidic
ribosomal protein P0 | Component of the
60S subunit ribosomal protein | |
| 431 | gi|4758638 | 30% | 72 | Down | 6.00/25133 |
Peroxiredoxin-6 | Regulation of
phospholipid turnover, protection against oxidative injury | |
| 446 | gi|229359377 | 61% | 159 | Down | 8.75/27690 |
ADP-ribosyltransferase 4 (Dombrock blood
group) | Catalysis of
ADP-ribosylation | |
| 457 | gi|92911770 | 62% | 189 | Down | 6.42/23896 |
XTP3TPA-transactivated protein 1 |
Transactivation | |
| 768 | gi|113411427 | 37% | 78 | Up | 12.05/29451 | PREDICTED:
hypothetical protein LOC642441 | Unknown | |
| 846 | gi|32364694 | 71% | 173 | Up | 5.44/41317 | p40 | Lysosomal membrane
proteins | |
| 850 | gi|225939 | 32% | 76 | Up | 6.34/36674 | Aldehyde
reductase | Reversible
conversion of an aldose to an alditol | |
| 909 | gi|77736367 | 51% | 99 | Up | 6.60/37177 | Poly(rC)-binding
protein 2 | | |
| 1108 | gi|4502101 | 33% | 69 | Up | 6.57/38918 | Annexin A1 | Potential
anti-inflammatory activity | |
| 1135 | gi|12056473 | 38% | 85 | Up | 6.29/40738 | Sialic acid
synthase | Generating
phosphorylated forms of Neu5Ac and KDN | |
| 1279 | gi|15277503 | 61% | 202 | Down | 5.55/40536 | ACTB protein | Cytoskeletal
protein; associated with asthenospermia | |
| 1291 | gi|13938339 | 31% | 113 | Up | 9.42/44476 | ATP5A1 protein | Regulation of
apoptosis and possible involvement in colorectal cancers | |
| 1308 | gi|203282367 | 55% | 225 | Up | 6.99/47350 | Chain A, crystal
structure of human enolase 1 | Catalysis of the
conversion of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate
(PEP) | miRanda,
miRWalk |
| 1335 | gi|119626209 | 48% | 194 | Up | 6.50/49260 | Septin 11, isoform
CRA_b | Regulator of tumor
progression in human malignancies | miRanda, miRWalk,
TargetScan |
| 1357 | gi|1706611 | 46% | 175 | Up | 7.26/49852 | Elongation factor
Tu, mitochondrial | Promotion of
GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes
during protein biosynthesis | |
| 1393 | gi|7657381 | 26% | 79 | Up | 6.14/55603 | Pre-mRNA-processing
factor 19 | Cell survival and
DNA repair | miRanda, miRWalk,
TargetScan |
| 1416 | gi|27436946 | 42% | 204 | Up | 6.57/74380 | Prelamin-A/C
isoform 1 precursor | Nuclear stability,
chromatin structure and gene expression | miRanda, RNA22 |
| 1557 | gi|169404695 | 20% | 77 | Down | 8.00/57091 | Chain A, pyruvate
kinase M2 is a phosphotyrosine binding protein |
Phosphotyrosine-binding protein | |
| 1727 | gi|21493039 | 30% | 177 | Up | 6.68/94811 | A-kinase anchor
protein 4 isoform 2 | Regulation of sperm
motility | miRanda, miRWalk,
TargetScan |
| 1888 | gi|308818195 | 36% | 192 | Up | 5.94/74027 |
Dihydropyrimidinase-related protein 2
isoform 1 | Neuronal
differentiation and axonal guidance | miRanda,
TargetScan |
| 1994 | gi|2687853 | 22% | 80 | Up | 6.19/155341 | RAD50 homologue
hsRAD50 | Component of the
MRN protein complex | miRanda, miRWalk,
TargetScan |
| 2055 | gi|1710248 | 24% | 88 | Up | 4.95/46512 | Protein disulfide
isomerase-related | With isomerase and
protein 5 chaperone activities | miRanda, miRWalk,
TargetScan |
Discussion
miRNA dysregulation plays an active role in cancer
development. In this regard, miR-27a, an oncogenic miRNA
overexpressed in many types of cancer, has been reported to promote
cell proliferation (19),
oncogene-induced transformation (23), metastasis (24) and multidrug resistance (25). miR-27a has also been implicated in
the regulation of apoptosis (26),
angiogenesis (18) and hormone
sensitivity (27). The antagonism
of miR-27a function thus represents a novel approach for the
treatment of cancer. In the present study, we demonstrate that the
inhibition of miR-27a by the stable expression of anti-miRNA
significantly suppresses the proliferation and invasiveness of U87
glioma cells. Although miR-27a has been shown to be abundantly
expressed in glioma tissues (22),
this is the first study to demonstrate the influence of miR-27a on
the malignant phenotypes of glioma cells.
miRNA performs its biological functions by
repressing the protein translation and/or inducing the degradation
of its mRNA targets. To date, a number of genes, including Fas
associated protein with death domain (FADD) (26), zinc finger and BTB domain
containing 10 (ZBTB10; a Sp1 repressor) (28), Ring1 and YY1 binding protein (RYBP;
an apoptotic facilitator) (28),
Myt-1 (a cdc2 inhibitor) (18),
Forkhead box protein O1 (FOXO1) (29), homeodomain-interacting protein
kinase-2 (HIPK2) (30), Sprouty2
(21), prohibitin (19) and F-box/WD repeat-containing
protein 7 (Fbxw7) (23), have been
identified to be the targets of miR-27a. In the current study, we
hypthesized that the repressing effects of miR-27a on its targets
would be relieved through the stable expression of anti-miR-27a,
leading to the upregulation of its targets. By the gel-based
comparative proteomic approach, we identified 16 proteins that were
upregulated by more than 10-fold in anti-miR-27a-expressing U87
cells. Using multiple computational algorithms, 8 of these
upregulated proteins, including the RAD50 homolog, protein
disulfide isomerase family A member 5 (PDIA5),
dihydropyrimidinase-like 2 (DPYSL2), A kinase anchor protein 4
(AKAP4), lamin A, PRP19/PSO4 pre-mRNA processing factor 19 homolog
(PRPF19), septin 11 and enolase 1, were predicted to be the targets
of miR-27a in glioma cells. These findings suggest that the
combined use of bioinformatic and proteomic approaches may be an
efficient method for the identification of novel miRNA targets.
Some of the newly identified proteins putatively
targeted by miR-27a have been implicated in cancer biology. For
instance, both the RAD50 homolog and PRPF19 have been shown to
mediate DNA repair and to have an impact on cell cycle and
apoptosis (31,32). The loss of lamin A has also
frequently been observed during tumor progression and may
contribute to the disruption of nuclear architecture and chromatin
structure, thereby increasing genetic instability (33). Moreover, the expression of DPYSL2
has been reported to be reduced in carcinogen-exposed murine lung
tissue (34). Septins, a family of
cytoskeleton-related proteins dysregulated in cancer, are also
involved in cytokinesis, chromosome segregation, DNA repair,
migration and apoptosis, all of which are important to cancer
development (35). In addition,
glioma tissues have been found to possess lower enolase activity
than normal brain tissues (36).
The mechanism by which the orchestrated expression of these
proteins mediates the anticancer effect of anti-miR-27a in glioma
cells, however, warrants further investigation.
Lentiviral vectors are promising for gene therapy
applications due to their ability to sustain long-term transgene
expression (37). In this study,
we demonstrate that the lentiviral vector may be used to deliver
specific anti-miRNAs to glioma cells and impair their growth and
invasiveness. Since 2002, a number of trials using lentiviral
vectors for the treatment of both infectious and genetic diseases
have been carried out (37). Our
study provides in vitro evidence that the lentiviral vector
may be used to stably express anti-miRNAs in glioma cells. With the
advance of tissue-specific expression control and further
understanding of miRNA dysregulation in gliomas, we anticipate that
the anti-miRNA-encoding lentivirus will become the latest addition
to the armamentarium to fight against gliomas in humans in the near
future.
Acknowledgements
This study was supported by the Jiangsu Natural
Scientific Fund (SBK200921106) and by the Yangtz River Scholar and
Innovation Research Team Development Program (Project no.
IRT0945).
References
|
1
|
Westphal M and Lamszus K: The neurobiology
of gliomas: from cell biology to the development of therapeutic
approaches. Nat Rev Neurosci. 12:495–508. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hatanpaa KJ, Burma S, Zhao D and Habib AA:
Epidermal growth factor receptor in glioma: signal transduction,
neuropathology, imaging, and radioresistance. Neoplasia.
12:675–684. 2010.PubMed/NCBI
|
|
3
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. New Eng J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Endersby R and Baker SJ: PTEN signaling in
brain: neuropathology and tumorigenesis. Oncogene. 27:5416–5430.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Giannopoulos S and Kyritsis AP: Diagnosis
and management of multifocal gliomas. Oncology. 79:306–312. 2010.
View Article : Google Scholar
|
|
6
|
Wurdinger T, Tannous BA, Saydam O, Skog J,
Grau S, Soutschek J, Weissleder R, Breakefield XO and Krichevsky
AM: miR-296 regulates growth factor receptor overexpression in
angiogenic endothelial cells. Cancer Cell. 14:382–393. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu WK, Law PT, Lee CW, Cho CH, Fan D, Wu
K, Yu J and Sung JJ: MicroRNA in colorectal cancer: from benchtop
to bedside. Carcinogenesis. 32:247–253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: a new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baraniskin A, Kuhnhenn J, Schlegel U,
Maghnouj A, Zollner H, Schmiegel W, Hahn S and Schroers R:
Identification of microRNAs in the cerebrospinal fluid as biomarker
for the diagnosis of glioma. Neuro Oncol. 14:29–33. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Quintavalle C, Garofalo M, Zanca C, Romano
G, Iaboni M, Del Basso De Caro M, Martinez-Montero JC, Incoronato
M, Nuovo G, Croce CM and Condorelli G: miR-221/222 overexpession in
human glioblastoma increases invasiveness by targeting the protein
phosphate PTPmu. Oncogene. 31:858–868. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kefas B, Godlewski J, Comeau L, Li Y,
Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S and
Purow B: microRNA-7 inhibits the epidermal growth factor receptor
and the Akt pathway and is down-regulated in glioblastoma. Cancer
Res. 68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li KK, Pang JC, Ching AK, Wong CK, Kong X,
Wang Y, Zhou L, Chen Z and Ng HK: miR-124 is frequently
down-regulated in medulloblastoma and is a negative regulator of
SLC16A1. Hum Pathol. 40:1234–1243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Godlewski J, Nowicki MO, Bronisz A,
Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca
EA and Lawler S: Targeting of the Bmi-1 oncogene/stem cell renewal
factor by microRNA-128 inhibits glioma proliferation and
self-renewal. Cancer Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Silber J, Lim DA, Petritsch C, Persson AI,
Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello
JF, et al: miR-124 and miR-137 inhibit proliferation of
glioblastoma multiforme cells and induce differentiation of brain
tumor stem cells. BMC Med. 6:142008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu
Z and You Y: hsa-mir-181a and hsa-mir-181b function as tumor
suppressors in human glioma cells. Brain Res. 1236:185–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ciafre SA, Galardi S, Mangiola A, Ferracin
M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM and Farace MG:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mertens-Talcott SU, Chintharlapalli S, Li
X and Safe S: The oncogenic microRNA-27a targets genes that
regulate specificity protein transcription factors and the G2-M
checkpoint in MDA-MB-231 breast cancer cells. Cancer Res.
67:11001–11011. 2007. View Article : Google Scholar
|
|
19
|
Liu T, Tang H, Lang Y, Liu M and Li X:
MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by
targeting prohibitin. Cancer Lett. 273:233–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chintharlapalli S, Papineni S, Abdelrahim
M, Abudayyeh A, Jutooru I, Chadalapaka G, Wu F, Mertens-Talcott S,
Vanderlaag K, Cho SD, Smith R III and Safe S: Oncogenic
microRNA-27a is a target for anticancer agent methyl
2-cyano-3,11-dioxo-18beta-olean-1,12-dien-30-oate in colon cancer
cells. Int J Cancer. 125:1965–1974. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma Y, Yu S, Zhao W, Lu Z and Chen J:
miR-27a regulates the growth, colony formation and migration of
pancreatic cancer cells by targeting Sprouty2. Cancer Lett.
298:150–158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Skog J, Wurdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Q, Li DC, Li ZF, Liu CX, Xiao YM,
Zhang B, Li XD, Zhao J, Chen LP, Xing XM, et al: Upregulation of
miR-27a contributes to the malignant transformation of human
bronchial epithelial cells induced by SV40 small T antigen.
Oncogene. 30:3875–3886. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: microRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
25
|
Zhu H, Wu H, Liu X, Evans BR, Medina DJ,
Liu CG and Yang JM: Role of MicroRNA miR-27a and miR-451 in the
regulation of MDR1/P-glycoprotein expression in human cancer cells.
Biochem Pharmacol. 76:582–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chhabra R, Adlakha YK, Hariharan M, Scaria
V and Saini N: Upregulation of miR-23a-27a-24-2 cluster induces
caspase-dependent and -independent apoptosis in human embryonic
kidney cells. PloS One. 4:e58482009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Mertens-Talcott SU, Zhang S, Kim K,
Ball J and Safe S: MicroRNA-27a indirectly regulates estrogen
receptor {alpha} expression and hormone responsiveness in MCF-7
breast cancer cells. Endocrinology. 151:2462–2473. 2010.PubMed/NCBI
|
|
28
|
Scott GK, Mattie MD, Berger CE, Benz SC
and Benz CC: Rapid alteration of microRNA levels by histone
deacetylase inhibition. Cancer Res. 66:1277–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast
cancer cells. J Biol Chem. 284:23204–23216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Hu S, Wang J, Cai J, Xiao L, Yu L
and Wang Z: MiR-27a modulates MDR1/P-glycoprotein expression by
targeting HIPK2 in human ovarian cancer cells. Gynecol Oncol.
119:125–130. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abuzeid WM, Jiang X, Shi G, Wang H,
Paulson D, Araki K, Jungreis D, Carney J, O’Malley BW Jr and Li D:
Molecular disruption of RAD50 sensitizes human tumor cells to
cisplatin-based chemotherapy. J Clin Invest. 119:1974–1985. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu X and Legerski RJ: The Prp19/Pso4 core
complex undergoes ubiquitylation and structural alterations in
response to DNA damage. Biochem Biophys Res Commun. 354:968–974.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gonzalez-Suarez I, Redwood AB and Gonzalo
S: Loss of A-type lamins and genomic instability. Cell Cycle.
8:3860–3865. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bortner JD Jr, Das A, Umstead TM, Freeman
WM, Somiari R, Aliaga C, Phelps DS and El-Bayoumy K:
Down-regulation of 14-3-3 isoforms and annexin A5 proteins in lung
adenocarcinoma induced by the tobacco-specific nitrosamine NNK in
the A/J mouse revealed by proteomic analysis. J Proteome Res.
8:4050–4061. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Connolly D, Abdesselam I, Verdier-Pinard P
and Montagna C: Septin roles in tumorigenesis. Biol Chem.
392:725–738. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Joseph J, Cruz-Sanchez FF and Carreras J:
Enolase activity and isoenzyme distribution in human brain regions
and tumors. J Neurochem. 66:2484–2490. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
D’Costa J, Mansfield SG and Humeau LM:
Lentiviral vectors in clinical trials: current status. Curr Opin
Mol Ther. 11:554–564. 2009.
|