Introduction
Hemangiopoietin (HAPO), a 294-amino acid protein,
stimulates the proliferation and hematopoiesis and/or endothelial
differentiation of human bone marrow mononuclear cells and of
purified CD34+, CD133+, kinase domain receptor-positive (KDR+) or
CD34+/KDR+ cell populations (1).
The native protein is one of the alternative splicing products of
proteoglycan 4 (PRG4), which consists of 12 exons. The coding
sequence of native HAPO includes part of exon 2, full-length exons
3 and 4 and part of PRG4 exon 6. Research performed to date
supports the assignment of at least three domains within HAPO: i)
The N-terminal domain encoded by PRG4 exons 2 and 3 consists of two
somatomedin B (SMB) homology domains (SMBrhHAPO), while
HAPO lacks N-terminal 15 amino acids in exon 2. SMB, with the
consensus sequence
X2CX6CX9CXCX3CX5CCX5CX5,
is homologous to the cysteine-rich region of vitronectin (Vn) and
the sequence identity is 45% (2).
All 14 cysteines of HAPO are distributed over the two SMB domains.
ii) The center region of HAPO encoded by PRG4 exon 4 is a lysine
(K)-rich region with the probable function of heparin binding and
is known as putative heparin-binding domain (pHBD) (2). The interaction between protein and
heparin is one of the most investigated topics in previous studies,
particularly the mechanism of promoting cell adhesion. iii) The
C-terminus encoded by PRG4 exon 6 is a mucin-like O-linked
oligosaccharide-rich repeat region composed of KEPATTT/P and XXTTTX
consensus sequences (2). These
repeats are highly diverse in length among species (3).
PRG4, an extracellular matrix (ECM) protein, has
been identified as megakaryocyte-stimulating factor (MSF) or
cartilage superficial zone proteoglycan (SZP) encoded by the
DOL54 gene. Two tissue-specific alternative splice variants
of PRG4 have been detected in human pathological tendon by
polymerase chain reaction (PCR) with forward primers specific for
exon 3 in combination with a reverse primer specific for exon 6.
One splice variant, similar to HAPO, lacks exon 5, while the other
lacks exons 4 and 5 (3–6).
The recombinant HAPO expressed in E. coli
supported the survival of MO7e cells through a PI3K-Akt pathway
following deprivation of granulocyte-macrophage colony stimulating
factor (7). The transfection of
the HESS-5 mouse bone marrow stromal cell line with a eukaryotic
HAPO-expressing vector supported the rapid generation of primitive
progenitor cells and maintains reconstitution of CD34+
hematopoietic stem cells in vitro(8).
HAPO has clinical potential in the management of
various cytopenias and radiation injury. However, there is limited
knowledge with regard to its biophysical characteristics. In this
study, we cloned and expressed the full-length protein and two
deletion mutants. Biophysical experiments were also performed in
order to gain knowledge of the protein structure.
Materials and methods
Expression and purification of rhHAPO and
the mutation variants
The complementary DNA (cDNA)-encoding full length
HAPO was amplified by PCR from human fetal liver cDNA and inserted
into plasmid pET22b (+) (Novagen, Madison, WI, USA) which had been
digested with NcoI and XhoI as previously described.
The human fetal liver was obtained from aborted fetuses of 17–22
weeks’ gestation after informed consent was obtained (1).
The protein was extracted using the one-step
extraction method. The cells were suspended into three volumes of
20% sucrose, 1 mM ethylenediaminetetraacetic acid (EDTA) and 20 mM
Tris-HCl (pH 7.9). Seven times volume ice water was then added.
Following centrifugation at 12,000 rpm for 30 min at 4°C, the
supernatant was loaded to a nickel-chelating column. The fraction
washed from the affinity column containing His-bind resin with 0.5
M NaCl, 200 mM imidazol and 20 mM Tris-HCl (pH 7.9), was dialyzed
into 20 mM citrate sodium buffer (pH 5.5) and then applied to a
fast flow SP Sepharose™ column. The target protein was eluted at
the same buffer with 0.5 M NaCl.
The cDNA of rhHAPOΔmucin and rhHAPOΔmucin-pHBD was
amplified from human fetal liver cDNA using the 5′
(5′-CATgCCATggATgCCACCTgCAACTgTgA-3′) and 3′
(5′-CTAgCTCgAgAgTTgTgACCTTgAAgTCAC-3′) oligonucleotide primers. The
amplified cDNA products were digested with NcoI and
XhoI. The rhHAPOΔmucin cDNA was inserted into pET22b (+) and
the rhHAPOΔmucin-pHBD cDNA was inserted into pET32c (+). The
purification of rhHAPOΔmucin mutant was performed as described
above for the full-length HAPO. For the purification of the
rhHAPOΔmucin-pHBD, the protein was eluted by 160 mM imidazol, 0.5 M
NaCl and 50 mM Tris-HCl (pH 8.0), dialyzed into 20 mM Tris-HCl (pH
8.0) and excised with enterokinase (3 U/mg protein; Invitrogen,
Carlsbad, CA, USA) at 4°C for 16 h. The peptide and the enzyme were
separated with Resource Q (GE Healthcare Bioscience Corp.,
Piscataway, NJ, USA) at 0–0.5 M NaCl. The protein concentration was
determined by measuring absorbance (A) at 280 nm with a U-3010
spectrophotometer (Hitachi High-Technologies Corporation, Tokyo,
Japan) and using a calculated extinction coefficient given by
ExPASy-ProtParam tool (http://kr.expasy.org/tools/protparam.html).
Size-exclusion chromatogaraphy
Following dialyzation into 20 mM Tris-HCl (pH 8.0),
rhHAPO eluted from Reasource Q was applied to a Superdex 200 10/300
GL column (Amersham Pharmasia Biotech, Piscataway, NJ, USA) at 0.5
ml/min. Purified rhHAPOΔmucin and rhHAPOΔmucin-pHBD were dialyzed
into phosphate-buffered saline (PBS) and applied to a Superdex 200
10/300 GL column. The column had a void volume (Vo) of 8.3 ml and
the standards IgG1 (150 kDa), bovine serum albumin (BSA; 67 kDa)
and lysozyme (14.4 kDa) eluted at 12.305, 14.345 and 17.147 ml
(Ve), respectively. The elution volumes of the standards were
divided by the elution volume of the thyroglobulin (Ve/Vo) and
plotted against the log of the molecular weights of the standards.
Then, the molecular masses of the peaks of rhHAPO could be
measured.
Western blot analysis
Western blot analysis was routinely performed.
Following electrophoresis and transfer, the blotted membrane was
blocked in Tris-buffered saline (TBS) buffer (PBS, 0.02% Tween-20)
containing 5% non-fat dried milk for 1 h at room temperature. The
membrane was then incubated with anti-rhHAPO MoAb prepared by the
laboratory of National Research Center for Stem Cell Engineering
and Technology in TBS buffer overnight at 4°C and horseradish
peroxidase-conjugated goat anti-mouse IgG (Beijing Zhongshan
Biotechnology Co., Beijing, China) for 1 h at room temperature.
Dynamic light-scattering (DLS)
analysis
HAPO and rhHAPOΔmucin-pHBD were dialyzed into PBS (1
mg/ml). DLS was carried out to characterize the aggregating with
Protein Solutions DynaPro (Protein Solutions, Charlottesville, VA,
USA). DLS was recorded on a DynaPro-801 (Protein Solutions) with a
temperature-controlled microsampler at 20°C. Twenty scans were
averaged for each measurement. Dynamics version 5.24.02 instrument
software was used to analyze the data.
Mass spectrum assay
The second fraction (1 mg/ml) of rhHAPO eluted from
Superdex 200 was dialyzed into water and matrix-assisted laser
desorption-ionization time-of-flight mass spectrometry
(MALDI-TOF-MS) was performed for molecular weight analysis
(National Center of Biomedical Analysis, Beijing, China).
Isoelectrofocusing
The pI values of rhHAPO and rhHAPOΔmucin-pHBD were
calculated using the ExPASy- ProtParam tool. Isoelectrofocusing was
performed to examine the purification and check their pI with the
method suggested by the manufacturer (Instruction 1818-A,
LKB-Produkter AB, Bromma, Sweden) using a 0.55-mm thin-layer
polyacrylamide gel, ampholine carrier ampholytes (pH 3.5–9.5;
LKB-Produkter AB). rhHAPO and pHBD were dialyzed into water and 20
μl 1 mg/ml protein was loaded into the loading filter paper. The
marker was broad pI calibration kit (pI 3.5–9.3; Amersham Pharmasia
Biotech). Following electrofocusing, the gel was stained with
Coomassie blue and the pI was monitored.
Cell adhesion assay
The murine bone marrow stromal cell line HESS-5 was
routinely cultured in Iscove’s modified Dulbecco’s medium (IMDM)
containing 10% fetal calf serum, 1% Gln, 1%
penicillin-streptomycin. Ninety-six-well plates were coated with
0.5% BSA at 4°C overnight and washed with PBS twice to prevent
non-specific adhesion. HESS-5 cells (5×104) were plated
into every well with IMDM medium containing 2% fetal calf serum, 1%
Gln and 1% penicillin-streptomycin. rhHAPO in dimeric or mulimeric
forms (1,000 ng/ml) was added. The recombinant proteins were
separated from rhHAPO by size-exclusion chromatogaraphy with
Superdex 200. Cells were cultured at 37°C for 2 h, the supernatant
was then removed and the cells were washed twice with PBS. The
crystal violet method was performed to measure A at 596 nm. Ratio
of relative adhesion = [(AHAPO−)/(APBS)]
×100.
Heparin binding assay
Heparin binding assay was performed as previously
described (9). Protein was applied
to a 1×5-cm heparin agarose column in 20 mM Tris-HCl buffer (pH
7.4) and eluted at a flow rate of 1 ml/min with a NaCl gradient in
the same buffer. A was monitored at 280 nm. NaCl concentration was
determined by conductivity.
Circular dichroism (CD)
The CD spectrum was performed on a Jasco J720
spectropolarimeter (Jasco, Easton, MD, USA). Far UV measurements
were taken in a 0.1-cm path length utensil with rhHAPO dialyzed
into PBS buffer. Time constants were 4 sec and 5 scans were
averaged for each measurement. CD was expressed in terms of
ellipticity [θ] in degree × cm2/dmol. Low molecular
weight (LMr) heparin was purchased from Sigma-Aldrich (St. Louis,
MO, USA). To determine whether rhHAPO binds to heparin, solutions
containing protein plus heparin were prepared at various
protein:heparin ratios (w/w). CD was performed to analyze the
conformational change. The K2D program (http://kal-el.ugr.es/k2d/spectra.html) was used for
the prediction of protein secondary structure from CD spectra.
Results and Discussion
rhHAPO is a stable tetramer
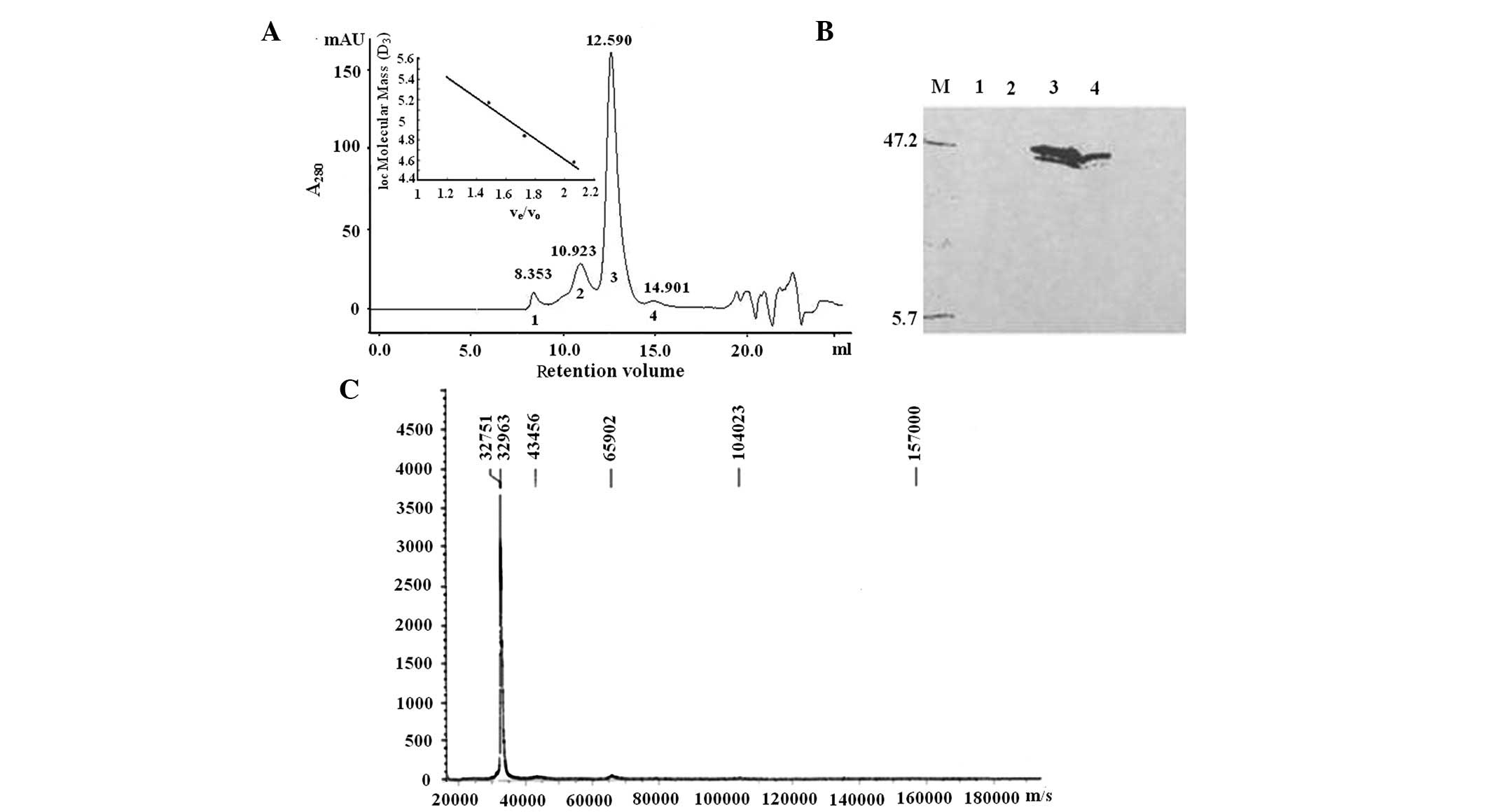
In the gel filtration chromatogram by Superdex 200
300/10 GL column chromatography, there were four A peaks of rhHAPO
purified from SPFF with the first peak eluted at void volume
suggesting matter with a high molecular weight (Fig. 1A). Western blot analysis with
anti-rhHAPO MoAb was performed to identify the peaks (Fig. 1B). The first two peaks did not
contain the protein recognized by the rhHAPO antibody and after the
endotoxin was removed from the sample, fraction 1 disappeared (data
not shown), suggesting that this peak was mostly composed of
endotoxin. According to the retention volume of standards, the
calculated molecular mass of fractions 3 (principal component) and
4 was 129.8 and 65.2 kDa (Fig.
1A), respectively. Since the theoretical molecular weight
deduced from amino acid sequence of rhHAPO with
(His)6-tag was 32,742.6 Da, rhHAPO is likely to be a
homotetramer with a trace amount of the dimeric form. Fraction 3
was expected to be a 137-kDa protein calculated by DLS. This was
consistent with the results from gel filtration, suggesting a
tetrameric form of rhHAPO.
MALDI-TOF MS was then used to obtain a more accurate
measurement of the mass weight of the protein in peak 3. The result
gave a perfect peak with a molecular weight of 32,751 Da (Fig. 1C), the mass weight of one rhHAPO
molecule. The electronic energy of MALDI-TOF MS changed the
multimeric conformation of rhHAPO into a monomer.
Multimeric rhHAPO is more potent in
promoting cell adhesion than the other form
HESS-5 cell adhesion assay was performed to
determine the different efficacy between the monomer, dimer and
tetramer structures of rhHAPO. HESS-5 is a murine bone marrow
stroma cell line that supports the reconstituting ability of ex
vivo-generated hematopoietic stem cells from human bone marrow
and cytokine-mobilized peripheral blood (10). When HESS-5 cells were cultured in
the presence of the tetrameric HAPO, increased adhesion was
observed compared with the presence of the dimeric HAPO (relative
adhesion value =54.3±11.1 vs. 33.2±12.1%; P=0.033). Platelet Vn is
another protein known to promote endothelial cell adhesion and the
conformationally altered multimeric Vn is more potent compared with
the monomeric form (11). Binding
efficiency of multimeric Vn with porcine endothelial cells
monolayer is 3–4 times higher compared with monomeric Vn (11). A possible explanation for this
different behavior is the steric influence due to aggregation. A
number of studies suggest that the SMB domain is indeed cryptic in
native monomeric Vn (12). Highly
sulfated glycosaminoglycans (GAGs) bind to native plasma Vn and
induce Vn multimerization at physiological ionic strength,
resulting in the exposure of the SMB domain. Although the principal
site for cell attachment in Vn is the Arginine-Glycine-Aspartic
(RGD) sequence locating in the SMB domain, it has been confirmed
that the purified recombinant SMB domain, which does not contain
the RGD sequence, is able to promote the attachment of HT-1080 and
U937 cells (13). Multimeric Vn is
able to bind the endothelial cells through the heparin-binding
domain, not the RGD sequence (14). Due to the high sequence homology
between Vn and HAPO, it is possible that dimeric HAPO
self-associates into the tetrmeric form, which may cause a
structural change and consequently SMB or heparin-binding domain
exposure, thus facilitating binding to their receptors.
Multimeric rhHAPO is more potent in
binding heparin compared with the monomeric rhHAPO in the absence
of significant conformational change
Heparin-binding peptides from ECM proteins have been
identified to promote cell adhesion (8,15).
Direct binding of the peptide to heparin may be assessed by
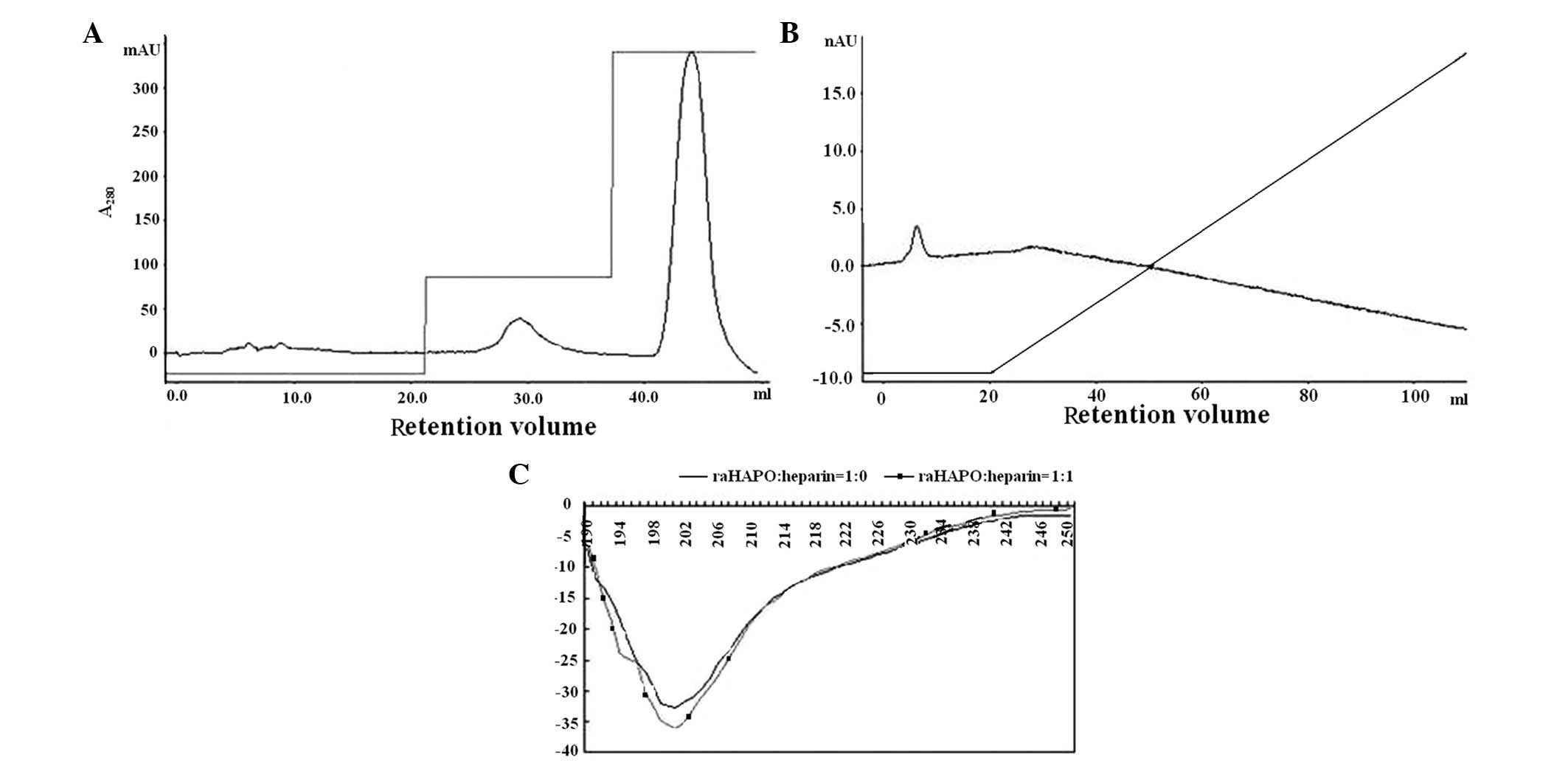
affinity chromatography on a heparin-agarose column. High binding
activity was evaluated from rhHAPO tetramer interactions with
heparin-agarose column at 1 M NaCl elution (Fig. 2A). However, during the elution
procedure with a linear salt gradient of 0–1.0 M NaCl, dimeric
rhHAPO was eluted with loading buffer and did not bind heparin
(Fig. 2B). There was a molecular
weight dependence of rhHAPO binding to heparin. This notable
observation was similar to Vn (16). Vn association into a multivalent
form contains proximal aligned heparin-binding sites exposed on its
surface and has been shown to have a stronger binding activity due
to the increased number of heparin-binding sites (17). A similar phenomenon may explain why
the tetrameric rhHAPO was more effective in promoting cell adhesion
compared with the monomeric rhHAPO.
The far-ultraviolet CD spectrum of multimeric rhHAPO
is shown in Fig. 3. There was no
significant structural change after binding with heparin (Fig. 2C). This spectrum differs
significantly from CD spectra of proteins known to assume
disordered structure. The spectrum gives a strong negative
ellipticity maximum at 200 nm, while weak positive ellipticities
are not observed at 218 nm. Although a negative signal near 200 nm
may be associated with disordered structure, the broad nature of
the band suggested some structural contribution. This spectrum was
characteristic of rhHAPO with predominant β-sheets and random
coils, with limited α-helical content (18). Analysis of this CD spectrum using
K2D online software indicated that rhHAPO is 61% random coils, 27%
α-helices and the remainder is β-sheets.
SMB and pHBD is the oligomerization
region of mulitimetic rhHAPO
Denaturation and renaturation of Vn under
physiological solution conditions is invariably accompanied by
self-association of the protein into a multimeric form (19). Intermolecular disulfide
cross-linking occurs primarily at the multimeric Vn. We predicted a
three-dimensional structure model of the first SMBrhHAPO
with swissmodel (http://swissmodel.expasy.org/) (20) to investigate the disulfide model.
The proposed disulfide linkage was Cys5-Cys9,
Cys19-Cys31,
Cys21-Cys32 and
Cys25-Cys39. In this pattern, the first two
cysteines form an independent disulfide bond. The first SMB domain
of the native HAPO purified from patients and rhHAPO lack the
N-terminal 15 amino acids containing these two cysteines. Thus,
absence of these 15 amino acids does not appear to form dissociated
cysteines in the molecule.
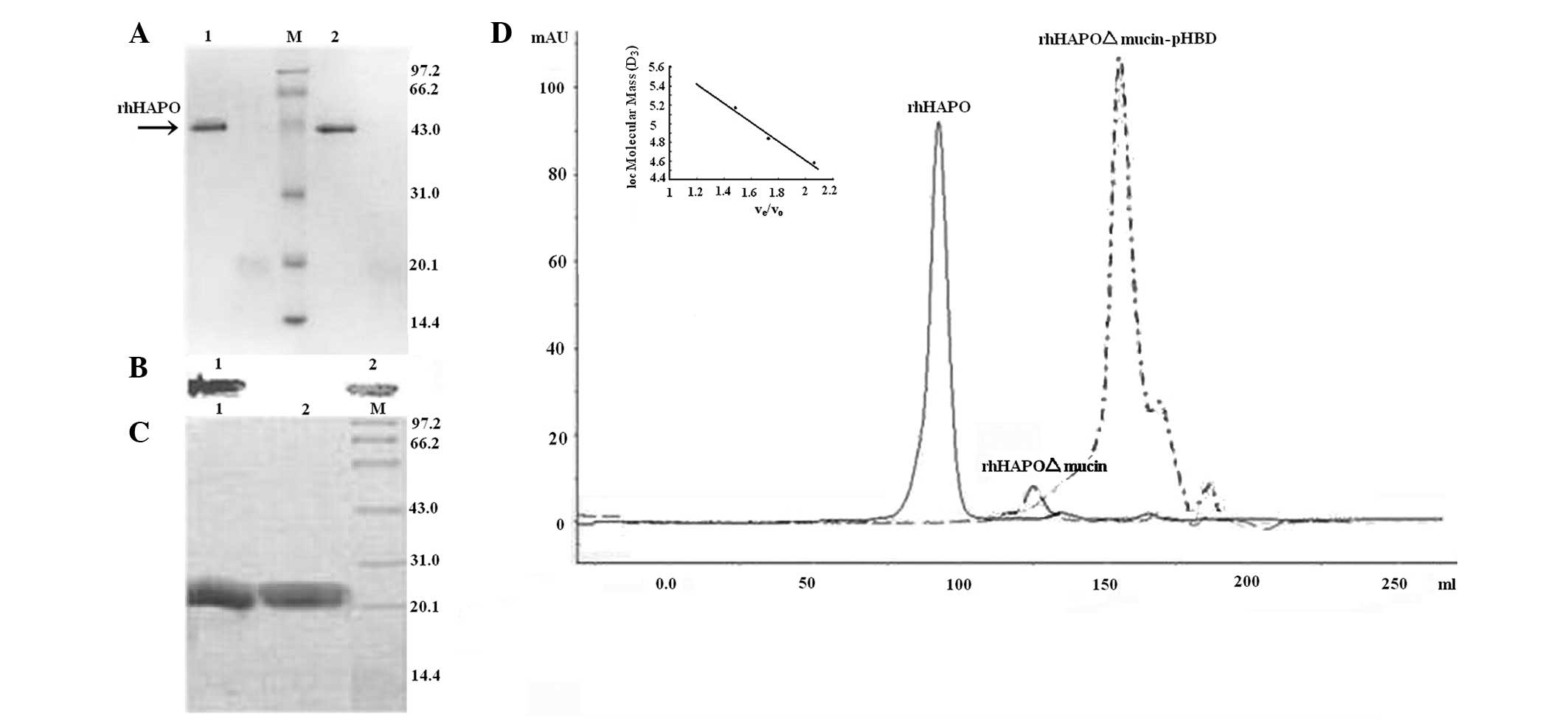
The results of reducing and non-reducing SDS-PAGE
further supported our hypothesis. On SDS-PAGE under reducing and
non-reducing conditions, the multimeric protein migrated as a
single band with a molecular mass of ~42 kDa (Fig. 3A and B). The results showed that
disulfide bond was not involved in the aggregation. There was no
free sulfhedryl in rhHAPO as well as the result of recombinant
SMBVn mensurated by Ellman’s method (21). Under non-reducing conditions, the
mobility of rhHAPO was slightly increased. This was caused by 14
cysteines being distributed over the SMB domain (22). The protein amount of positive
charges or cysteines does not have a lineal correlation with the
relative migratory ratio and molecular weight. Meanwhile, there was
a high content of positively charged amino acids throughout the
full length HAPO. Sequence analysis showed that is a K-rich protein
with 14.3% Lysine amino acid residues. The ratio of positively
charged residues (K+R) to negatively charged residues (D+E) was
50/38.
To identify the oligomerization sequence, we
investigated the consequences of the absence of the mucin-like
domain. According to the amino acid sequence, the predicted
molecular weight of rhHAPOΔmucin was 18.2 kDa, whereas the
calculated molecular weight was 72.4 kDa in Superdex 200
size-exclusion chromatogaraphy (Fig.
3D), suggesting that the protein was also a stable tetramer. We
found that deletion of the mucin-like repeat did not hamper the
ability of rhHAPO to form a tetramer. C-terminal mucin-like repeats
did not participate in the oligomerization.
Based on the alternative splicing of PRG4 mRNA, a
cDNA lacking the center region pHBD was amplified with the same
primers as those of rhHAPOΔmucin. In Superdex 200 300/10 GL column
chromatography, the retention volume of rhHAPOΔmucin-pHBD was 17.98
ml (Fig. 3D) with the calculated
molecular mass 28.2 kDa, validating the dimeric form of
rhHAPOΔmucin-pHBD. The recombinant SMBVn with C-terminal
of thioredoxin had monomeirc and dimeric form (23). The two N-terminal SMB domains of
nucleotide pyrophosphatases/phosphodiesterases 1 (NPP1) were
disulphide-linked homodimers and dimers could even be detected
after reducing SDS-PAGE (24).
However, we found that recombinant SMBHAPO was a
non-covalent dimer by reducing and non-reducing SDS-PAGE (Fig. 3C). Expression of this deletion
mutant protein resulted in structural alternations. Thus, SMB and
pHBD consisted of an intact tetramer. The absence of pHBD caused
the protein to be a dimer and changed certain physical
characteristics of the protein (Table
I). It has been proposed that the heparin-binding domain in Vn
mediates the protein association, but this conclusion has been
disproved in biochemical and biophysical studies (19). Intermolecular disulfide
cross-linking close to the C-terminal heparin-binding domain is the
oligomerization force of Vn. This study showed that pHBD mediated
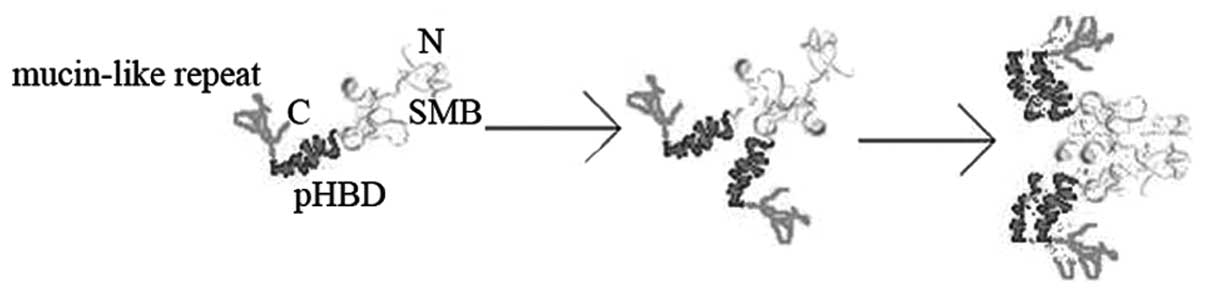
the association of the rhHAPO dimer into a tetramer. We proposed a
potential model to account for the oligomerization of rhHAPO
(Fig. 4). The subunits of rhHAPO
were assembled post-translationally into a dimer by interactions
between SMB domains, and this interaction caused a conformational
change of pHBD which mediated the association of the rhHAPO dimer
into a tetramer.
 | Table IComparison of characteristics of
rhHAPO and rhHAPOΔmucin-pHBD. |
Table I
Comparison of characteristics of
rhHAPO and rhHAPOΔmucin-pHBD.
| Protein | Monomer (kDa) | Oligomer (kDa) | pI | Predicted extinction
coefficientsa
(M−1cm−1) |
|---|
| rhHAPO | 32.7 | 129.8 | 8.52 | 5,960 |
|
rhHAPOΔmucin-pHBD | 12.9 | 28.6 | 6.39 | 6,835 |
Taken together, these observations demonstrate that
rhHAPO is a stable 129-kDa noncovalent homological oligomer.
Initially self-associated rhHAPO dimers are formed by interactions
between SMB domains, whereas the formation of a tetramer is a
secondary event mediated by pHBD sequences. Tetrameric rhHAPO is
more potent in promoting the adhension of HESS-5 cells compared
with dimeric rhHAPO. Dimeric rhHAPO does not bind heparin, while
tetrameric rhHAPO has a high affinity for heparin although there
are no clear conformational changes.
Acknowledgements
This study was supported by the National Natural
Science Foundation of Zhejiang, no. LQ12H16001 and Zhejiang
Provincial Program for the Cultivation of High-level Innovative
Health talents.
References
|
1
|
Liu YJ, Lu SH, Xu B, Yang RC, Ren Q, Liu
B, Li B, Lu M, Yan FY, Han ZB and Han ZC: Hemangiopoietin, a novel
human growth factor for the primitive cells of both hematopoietic
and endothelial cell lineages. Blood. 103:4449–4456. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Flannery CR, Hughes CE, Schumacher BL,
Tudor D, Aydelotte MB, Kuettner KE and Caterson B: Articular
cartilage superficial zone protein (SZP) is homologous to
megakaryocyte stimulating factor precursor and is a multifunctional
proteoglycan with potential growth-promoting, cytoprotective, and
lubricating properties in cartilage metabolism. Biochem Biophys Res
Commun. 254:535–541. 1999. View Article : Google Scholar
|
|
3
|
Ikegawa S, Sano M, Koshizuka Y and
Nakamura Y: Isolation, characterization and mapping of the mouse
and human PRG4 (proteoglycan 4) genes. Cytogenet Cell Genet.
90:291–297. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jay GD, Tantravahi U, Britt DE, Barrach HJ
and Cha CJ: Homology of lubricin and superficial zone protein
(SZP): products of megakaryocyte stimulating factor (MSF) gene
expression by human synovial fibroblasts and articular chondrocytes
localized to chromosome 1q25. J Orthop Res. 19:677–687. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rees SG, Davies JR, Tudor D, Flannery CR,
Hughes CE, Dent CM and Caterson B: Immunolocalisation and
expression of proteoglycan 4 (cartilage superficial zone
proteoglycan) in tendon. Matrix Biol. 21:593–602. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Panagopoulos I, Mertens F, Isaksson M and
Mandahl N: Expression of DOL54 is not restricted to myxoid
liposarcomas with the FUS-DDIT3 chimera but is found in various
sarcomas. Oncol Rep. 12:107–110. 2004.PubMed/NCBI
|
|
7
|
Zhan M and Han ZC: Hemangiopoietin
inhibits apoptosis of MO7e leukemia cells through
phosphatidylinositol 3-kinase-AKT pathway. Biochem Biophys Res
Commun. 317:198–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu ZS, Liu YJ, Lv LL, Han ZB, He R, Lu SH,
Wang T, Xu B, Chen ZZ and Han ZC: Bone marrow stromal cells
transduced with human hemangiopoietin gene support hematopoiesis in
vitro. Haematologica. 90:157–165. 2005.PubMed/NCBI
|
|
9
|
Guo NH, Krutzsch HC, Nègre E, Zabrenetzky
VS and Roberts DD: Heparin-binding peptides from the type I repeats
of thrombospondin. Structural requirements for heparin binding and
promotion of melanoma cell adhesion and chemotaxis. J Biol Chem.
267:19349–19355. 1992.PubMed/NCBI
|
|
10
|
Shimakura Y, Kawada H, Ando K, Sato T,
Nakamura Y, Tsuji T, Kato S and Hotta T: Murine stromal cell line
HESS-5 maintains reconstituting ability of ex vivo-generated
hematopoietic stem cells from human bone marrow and
cytokine-mobilized peripheral blood. Stem Cells. 18:183–189. 2000.
View Article : Google Scholar
|
|
11
|
Völker W, Hess S, Vischer P and Preissner
KT: Binding and processing of multimeric vitronectin by vascular
endothelial cells. J Histochem Cytochem. 41:1823–1832.
1993.PubMed/NCBI
|
|
12
|
Seiffert D: The glycosaminoglycan binding
site governs ligand binding to the somatomedin B domain of
vitronectin. J Biol Chem. 272:9971–9978. 1997.PubMed/NCBI
|
|
13
|
Deng G, Curriden SA, Hu G, Czekay RP and
Loskutoff DJ: Plasminogen activator inhibitor-1 regulates cell
adhesion by binding to the somatomedin B domain of vitronectin. J
Cell Physiol. 189:23–33. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zanetti A, Conforti G, Hess S,
Martìn-Padura I, Ghibaudi E, Preissner KT and Dejana E: Clustering
of vitronectin and RGD peptides on microspheres leads to engagement
of integrins on the luminal aspect of endothelial cell membrane.
Blood. 84:1116–11123. 1994.PubMed/NCBI
|
|
15
|
Burke C, Mayo KH, Skubitz AP and Furcht
LT: 1H NMR and CD secondary structure analysis of cell adhesion
promoting peptide F-9 from laminin. J Biol Chem. 266:19407–19412.
1991.PubMed/NCBI
|
|
16
|
Bittorf SV, Williams EC and Mosher DF:
Alteration of vitronectin. Characterization of changes induced by
treatment with urea. J Biol Chem. 268:24838–24846. 1993.PubMed/NCBI
|
|
17
|
Zhuang P, Chen AI and Peterson CB: Native
and multimeric vitronectin exhibit similar affinity for heparin.
Differences in heparin binding properties induced upon denaturation
are due to self-association into a multivalent form. J Biol Chem.
272:6858–6867. 1997. View Article : Google Scholar
|
|
18
|
Preissner KT and Jenne D: Structure of
vitronectin and its biological role in haemostasis. Thromb Haemost.
66:123–132. 1991.PubMed/NCBI
|
|
19
|
Zhuang P, Blackburn MN and Peterson CB:
Characterization of the denaturation and renaturation of human
plasma vitronectin. I Biophysical characterization of protein
unfolding and multimerization. J Biol Chem. 271:14323–14332. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guex N and Peitsch MC: SWISS-MODEL and the
Swiss-Pdb Viewer: an environment for comparative protein modeling.
Electrophoresis. 18:2714–2723. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Horn NA, Hurst GB, Mayasundari A,
Whittemore NA, Serpersu H and Peterson CB: Assignment of the four
disulfides in the N-terminal somatomedin B domain of native
vitronectin isolated from human plasma. J Biol Chem.
279:35867–35878. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kamikubo Y, Okumura Y and Loskutoff DJ:
Identification of the disulfide bonds in the recombinant
somatomedin B domain of human vitronectin. J Biol Chem.
277:27109–27119. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kamikubo Y, De Guzman R, Kroon G, Curriden
S, Neels JG, Churchill MJ, Dawson P, Ołdziej S, Jagielska A,
Scheraga HA, Loskutoff DJ and Dyson HJ: Disulfide bonding
arrangements in active forms of the somatomedin B domain of human
vitronectin. Biochemistry. 43:6519–6534. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gijsbers R, Ceulemans H and Bollen M:
Functional characterization of the non-catalytic ectodomains of the
nucleotide pyrophosphatase/phosphodiesterase NPP1. Biochem J.
371:321–330. 2003. View Article : Google Scholar
|