Introduction
Acute promyelocytic leukemia (APL), a particular
subtype of acute myeloid leukemia (AML) with a distinct cytological
morphology, is characterized by a chromosome reciprocal
translocation t(15;17), which results in the fusion between the
promyelocytic leukemia (PML) gene and the retinoic acid receptor
(RARα) gene (1,2). During the 1980s, the introduction of
all-trans retinoic acid (ATRA) created a precedent for tumor
molecular target therapy, enabling the complete remission of
>95% APL patients (3). However,
due to the wide application of ATRA, drug-resistant cases are
increasingly identified (4). As a
result, producing a more effective treatment for ATRA-resistant APL
patients has become a challenge in the clinic. Since the early
1990s, arsenic compounds have regained attention due to the
discovery of their clinical effects and unique mode of action on
refractory or relapsed patients, as well as patients newly
diagnosed with APL (1,5). Tetra-arsenic tetra-sulfide
(As4S4) is a type of traditional Chinese
medicine. In comparison with arsenic trioxide
(As2O3), As4S4 has
advantages including high remission rate, reduced side effects,
ease of use (may be taken orally) and cost-effectiveness. At
present, As4S4 has become one of the most
important therapies for APL (especially for recrudescent and
drug-resistant APL) (6).
In our previous study, we applied proteomics to
screen >20 differently expressed proteins before and after
treatment with As4S4 in the ATRA-resistant
APL cell line-NB4-R1. Among these proteins, SET expression was
observed to be markedly decreased following treatment with
As4S4 for 48 h, and the number of apoptotic
NB4-R1 cells increased significantly (7). However, the function, mechanism and
regulation pathways of SET in As4S4-induced
apoptosis requires further investigation.
RNA interference (RNAi) is a posttranscriptional
gene silencing mechanism which is mediated by double-stranded RNA
(dsRNA) molecules. RNAi is a new, powerful and widely used tool for
the analysis of gene function in mammals and plants (8,9).
Lentiviral vector-based RNAi is able to combine efficient infection
and integration of lentiviral DNA with suppression of specific
homologous gene expression of host RNA. Therefore, lentiviral
vector-based RNAi may provide a long-lasting knockdown influence
(10). In the present study, we
used powerful lentiviral-mediated RNAi eukaryotic vectors with
specific short hairpin RNA (shRNA) to block the expression of SET,
in order to lay the foundation for further investigation of its
mechanism in As4S4-induced APL cell
apoptosis, and provide new insight into the pharmacological actions
of traditional medicines.
Materials and methods
Cell culture
The NB4-R1 cell line was donated by Shanghai Jiao
Tong University School of Medicine. The cells were cultured in
RPMI-1640 (Gibco-BRL, Carlsbad, CA, USA) supplemented with 10%
heat-inactivated fetal bovine serum at 37°C in a humidified
incubator containing 5% CO2. Cell viability was
evaluated by the trypan blue dye exclusion assay and only cell
suspensions that presented >95% viability were used. The study
was approved by the ethics committee of Xi'an Jiaotong University,
Xi'an, China.
Design and clone of RNAi lentiviral
vectors
Four target shRNAs against human SET gene (Genbank
accession NM_003011) for RNAi were designed using the internet
application Ambion (Austin, TX, USA) (Table I). The sequence
5′-TTCTCCGAACGTGTCACGT-3′ which had no significant homology to any
known human genes was used as a negative control. Oligonucleotides
were synthesized according to the sequences, heated at 95°C for 5
min and then annealed at 37°C for 1 h. Annealed sequences were
ligated into the AgeI and EcoRI sites of pGCSIL-GFP
(containing human U6 promoter) to generate pGCSIL-GFP-SET (1–4)
vectors (Shanghai Genechem Co., Shanghai, China), which were
transformed into E. coli. Positive recombinant clones were
selected by PCR and DNA sequencing. The new recombinant lentiviral
vectors co-expressed the shRNA transcript and green fluorescent
protein (GFP).
 | Table IshRNA sequence. |
Table I
shRNA sequence.
| Marker | Target sequence |
|---|
| KD-1 |
GAGTTCACAGAAGCGGTGGAA |
| KD-2 |
GCTGCCGTCCATCACAACTGA |
| KD-3 |
CCGTGGGTACAGAAACCAATT |
| KD-4 |
CGTGGTGAACTCTGCCTTATA |
Lentiviral vector infection
Cells were divided into 3 groups: CON (normal NB4-R1
cells), NC (infection with negative control RNAi vector), and KD
(infection with pGCSIL-GFP-SET). According to different shRNA
sequences, the KD group was divided into 4 subgroups: KD1, KD2, KD3
and KD4.
The NB4-R1 cells were plated into 6-well plates at a
density of 1×105 cells/ml. The following day, cells were
infected with a mixture of 100 μl 2X polybrene and specific
negative control lentiviral vectors, at the multiplicity of
infection (MOI) 20, according to the preliminary results. The final
volume of culture medium was 1 ml/well. After centrifugation for 18
h, the supernatant was discarded and the precipitate was
resuspended in the culture medium with 10% FBS. The cells were
incubated, observed under fluorescence microscopy and harvested on
the 4th and 7th day after infection. The knockdown efficiency of
SET was analyzed by real-time quantitative PCR (RT-qPCR) and
western blotting.
RT-qPCR
After infection with lentiviruses for 4 days, the
total RNA from each group of cells was extracted in TRIzol
(Invitrogen, Carlsbad, CA, USA) and quantified by an ultraviolet
spectrophotometer at a wavelength of 260 nm. The reverse
transcription (RT) reaction was performed with a RevertAid First
Strand cDNA Synthesis kit (Fermentas, Burlington, ON, Canada)
according to manufacturer's instructions. The PCR conditions were
as follows: initial denaturation at 94°C for 5 min, the reaction
was repeated for 45 cycles, each cycle consisted of denaturing at
94°C for 30 sec, annealing at 61°C for 45 sec and synthesis at 72°C
for 45 sec. The primer sequences used to amplify SET and GAPDH are
listed in Table II. The cutoff
point (Ct) of each sample was plotted on the standard curve and the
mRNA copy numbers were calcuated. The GAPDH gene was used as an
endogenous control. The relative SET mRNA levels were expressed as
a ratio of SET to GAPDH.
 | Table IIForward (F) and reverse (R) primer
sequences for real-time quantitative PCR. |
Table II
Forward (F) and reverse (R) primer
sequences for real-time quantitative PCR.
| Target gene | Primer sequence | Amplification
length |
|---|
| SET-F |
5′-AAATATAACAAACTCCGCCAACC-3′ | 138 bp |
| SET-R |
5′-CAGTGCCTCTTCATCTTCCTC-3′ | |
| GAPDH-F |
5′-CACCCTGTTGCTGTAGCCAAA-3′ | 121 bp |
| GAPDH-R |
5′-CACCCTGTTGCTGTAGCCAAA-3′ | |
Western blotting
After infection with lentiviruses for 7 days, the
cells were lysed in RIPA buffer in the presence of proteinase
inhibitor cocktail (Sigma, Santa Clara, CA, USA). The protein
concentration was determined by BCA assay kit (Thermo, Waltham, MA,
USA). Protein (30 μg) was separated by 12% SDS-PAGE and transferred
to a nitrocellulose membrane. The membranes were blocked in 5%
skimmed milk for 2 h, incubated with primary antibodies against SET
(goat monoclonal, 1:1,000; Abcam, Cambridge, MA, USA) and GAPDH
(mouse monoclonal, 1:10,000; Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA) at room temperature for 2 h and 4°C overnight. The
membranes were washed with TBS containing 0.1% Tween-20 (TBST)
followed by an incubation of 1 h in goat anti-mouse and goat
anti-goat secondary antibody conjugated with HRP. After final
washing with TBST, the membranes were developed using
chemiluminescence and exposed to X-ray films. The immunoblots were
quantified with software Quantity One version 4.6.2. The expression
of SET in each sample was internally normalized to GAPDH and levels
were expressed relative to expression in control groups.
Statistical analysis
All the experiments were carried out in triplicate
and quantitative data in this study were expressed as mean ± SD.
Statistical differences between groups were compared using a
one-way analysis of variance (ANOVA). P<0.05 was considered to
indicate a statistically significant result.
Results
Construction and packaging of lentiviral
vectors
Four target shRNAs against human SET gene for RNAi
were constructed and ligated with lentiviral frame plasmids.
Positive recombinant clones were selected and identified with PCR.
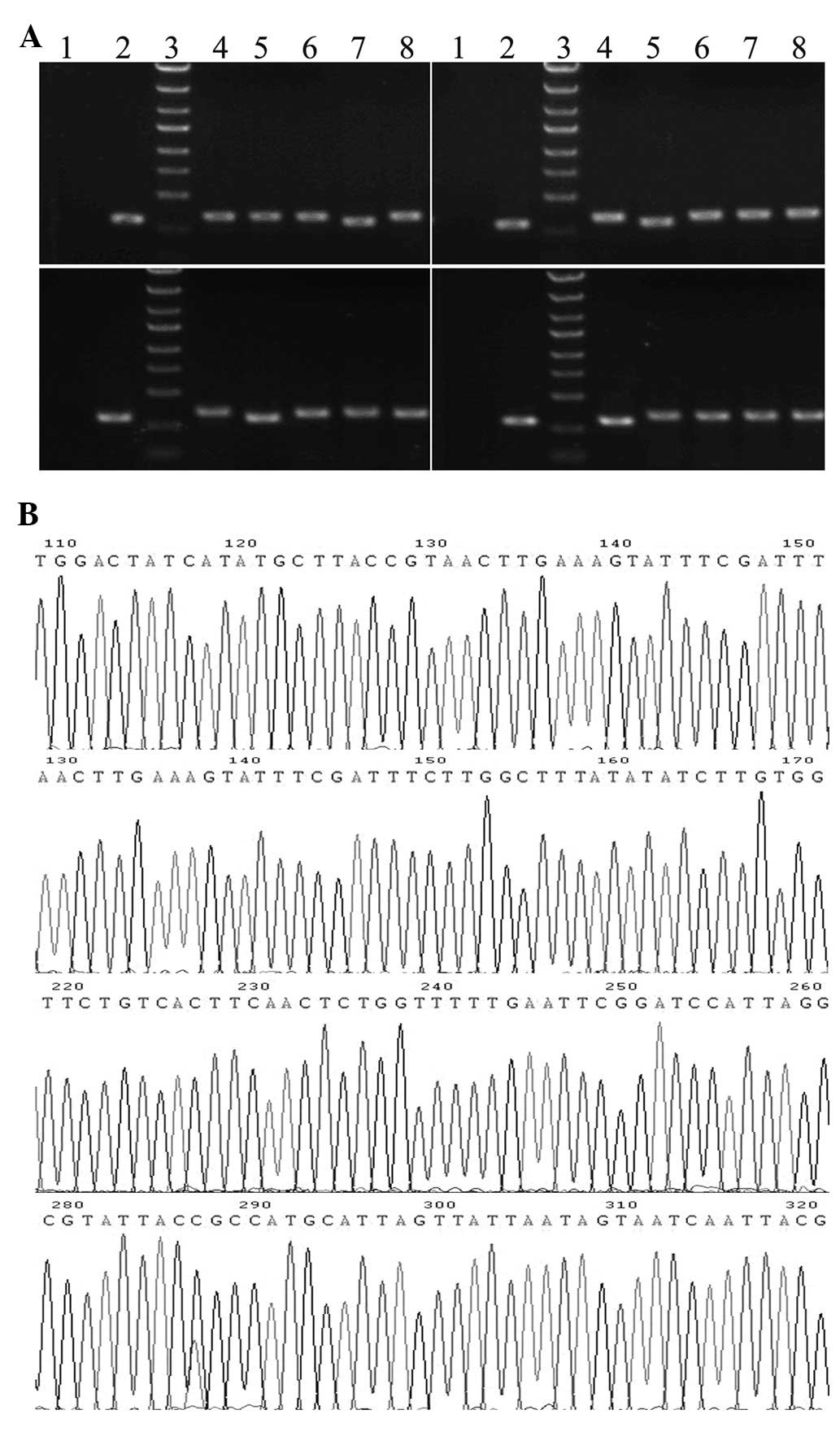
The sizes of PCR amplified fragments of recombinant clones and
unconnected shRNA empty vectors were 343 and 306 bp, respectively
(Fig. 1A). Sequence analysis
confirmed that the inserted SET shRNA sequences were correct
(Fig. 1B). The recombinant
lentiviral vectors were applied to infect NB4-R1 cells and virus
titers were determined with the hole by hole dilution method as
1×109 (KD1), 3×108 (KD2), 4×108
(KD3) and 6×108 (KD4) TU/ml, respectively.
Infection efficiency of NB4-R1 cells with
lentivirus
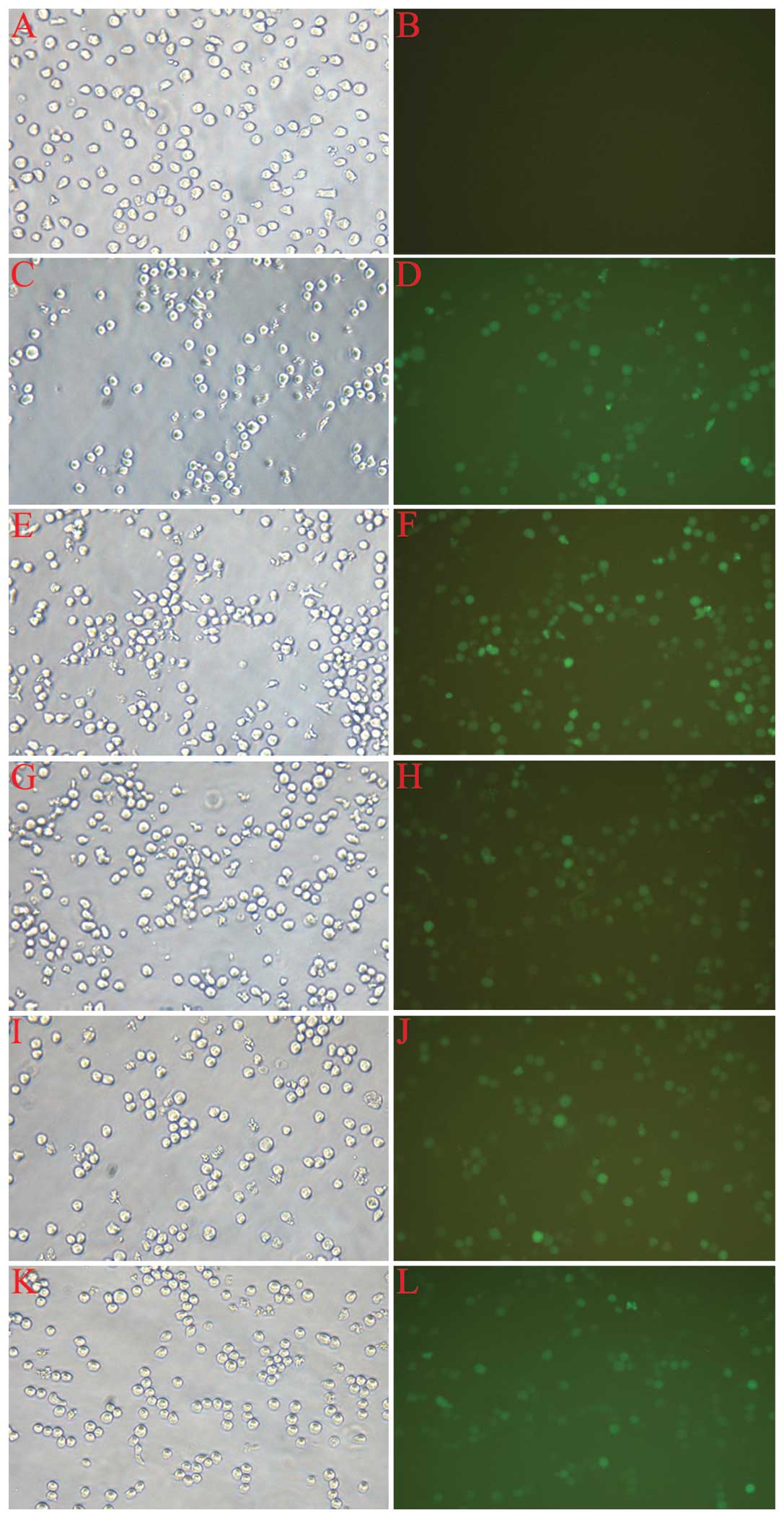
The assessment of infection rate was accomplished
with the determination of positive expression rate of GFP by
fluorescent microscopy 72 h after infection. As shown in Fig. 2, the cell growth state was good,
and the proportion of fluorescent cells ranged from 70–90% in each
group, with the exception of the normal control (CON). The cells
were then harvested for the following experiments.
Silencing efficacy of SET at mRNA
level
The silencing efficacy of SET shRNA lentivirus at
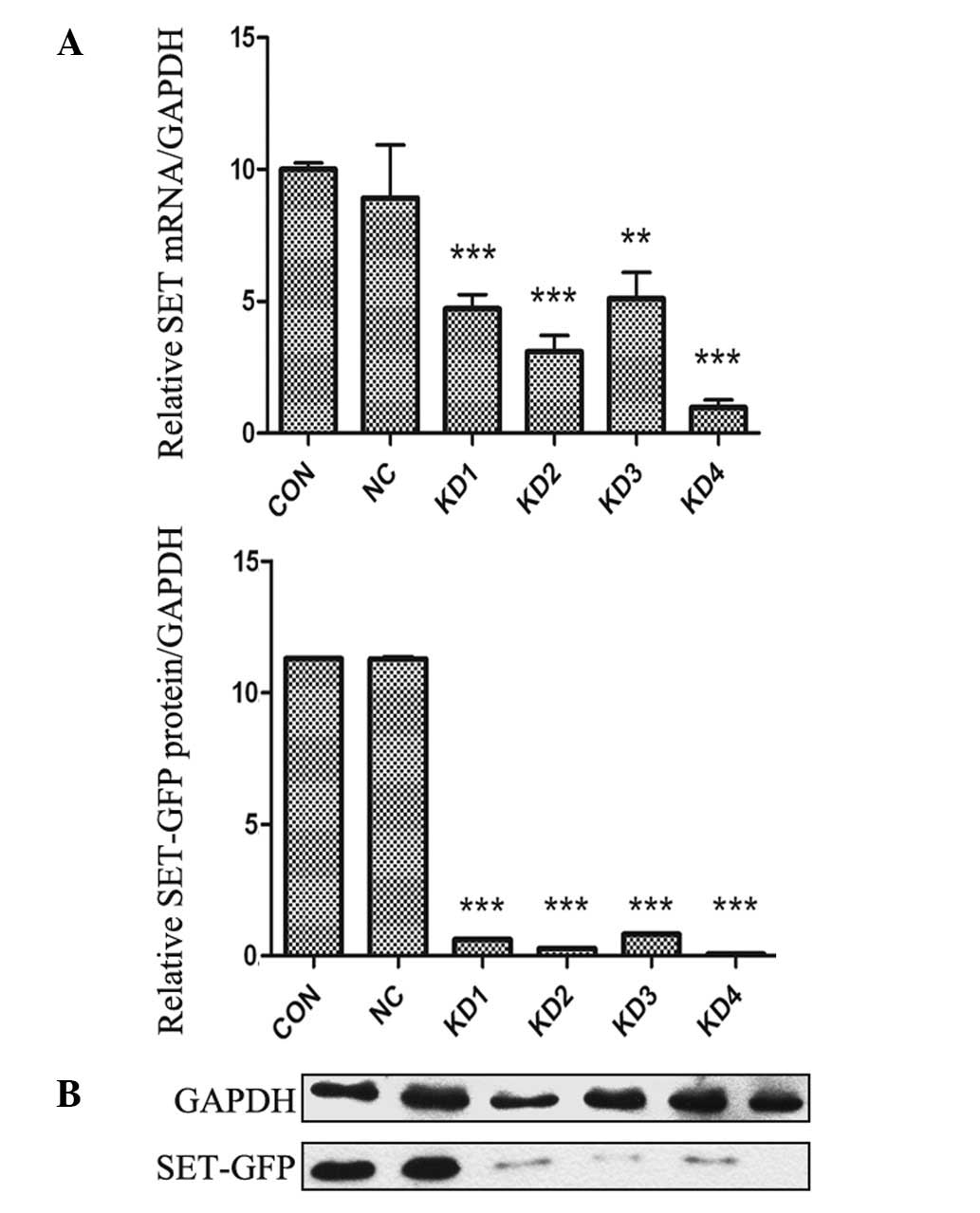
the mRNA level in NB4-R1 cells was determined by RT-qPCR. As shown
in Fig. 3A, infection of NB4-R1
cells with SET shRNA resulted in a significant decrease in mRNA
expression of SET. At the 4th day after infection, the amount of
SET mRNA in KD1-KD4 subgroups was notably reduced by 52.8, 69.1,
48.9 and 90.3%, respectively, compared with that of NC group.
However, there was no difference between the NC and CON groups.
Silencing efficacy of SET at protein
level
Western blot analysis of cell extracts was carried
out to determine whether decreased mRNA expression was correlated
with decreased translation of the gene product. As shown in
Fig. 3B, a similar trend was
observed in NB4-R1 cells. The protein expression levels of SET in
KD1-KD4 subgroups were decreased by 92.5, 96.3, 91.7 and 98.4%,
respectively, compared to the NC group. However, there was no
significant difference in the protein expression level between the
NC and CON groups. RT-qPCR and western blotting demonstrated that
lentiviral vectors were effective for SET silencing and KD4 was the
most effective construct. Stable infection was performed with KD4
construct.
Discussion
Acute promyelocytic leukemia (APL) is one of the
most common malignancies and has a high mortality rate. The
comprehensive treatments have been constantly improved; for
example, ATRA may enable the complete remission of >95% of APL
patients (3). However, an
increasing resistance trend restricted the long-term efficacy of
ATRA (11). In recent years,
arsenic compounds (mainly As2O3 and
As4S4) have gradually regained attention due
to their positive clinical effects and unique mode of action on APL
(12). However, a disadvantage of
As2O3 is that the daily intravenous infusion
required to achieve the ideal therapeutic result causes dermal and
gastrointestinal side effects (13). As4S4, another
arsenic compound, as a type of traditional Chinese medicine (also
named realgar or red orpiment), has been shown to be equally
effective in the treatment of APL, with less toxicity and side
effects, and As4S4 only requires oral
administration (14–16). Based on above advantages, this
orally administered agent would thus provide more benefits not only
for quality of life but also for easy access to consolidation and
maintenance therapy for leukemia.
In our previous study, SET was identified as one of
the differentially expressed proteins that correlated with
As4S4-induced NB4-R1 cell apoptosis. SET
(also known as TAF-1β, I2PP2A or IGAAD), localized to the nucleus
and cytoplasm, is a multifunctional protein. It is clear that SET
is a potent inhibitor of protein phosphatase 2A (PP2A), a
phosphatase with tumor suppressor activity (17). SET has also been described as an
inhibitor of the tumor suppressor NM23-H1 (a granzyme A
DNase-activated factor) (18) and
a negative regulator of histone acetylation (19).
PP2A is one of the major serine-threonine protein
phosphatase in all eukaryotic cells (20). PP2A is involved in almost all
cellular processes, including signal transduction (21), cell cycle progression (22) transcriptional and translational
regulation, DNA replication and chromosome depolymerization
(23,24). As a potential tumor suppressor,
inhibition of PP2A activity appears to be a common event in
different human neoplasia. For example, oncogenic viral proteins,
such as the SV40 virus small-t antigen, inhibits PP2A activity by
interacting with the subunits of PP2A (25). In addition, numerous genetic
alterations of the structure of PP2A subunit have been identified
in several types of human cancer. Consequently, exploring the
molecular targets inhibiting PP2A activity has an important
prevention significance for tumor treatment.
SET is the major cellular inhibitor of PP2A, and is
involved in the regulation of a variety of cellular processes and
signal transduction pathways (26). Although overexpression of SET has
been reported in tumor tissue in the uterus, stomach, colon and
rectum, as well as chronic myelogenous leukemia (27,28),
its role in the regulation of APL cells remains unclear. There is
evidence that SET may be involved in NB4-R1 cell apoptosis induced
by As4S4in vivo. Therefore, in this
study we designed 4 shRNA sequences targeting against SET and
constructed 4 recombinant lentiviral vectors. After infection with
NB4-R1 cells, RT-qPCR and western blotting were used to identify
silencing efficiency. Our results showed that the 4 RNAi vectors
all had high efficiency of infection; among these, the knockdown
efficiency of KD4 was the greatest, and was chosen to conduct the
follow-up research. This study will lay the foundation for further
study on the function and mechanism of SET. SET may be used in the
future as a potential target of gene therapy for ATRA-resistant APL
and other cancer.
Acknowledgements
This study was supported by the Natural Science
Foundation of China (nos. 30701133 and 81000218) and the
Fundamental Research Funds for the Central Universities and the
Shaanxi Province Science and Technology Development Fund, China
(nos. 2010K01-135 and 2012KTCL03-12). The authors express their
gratitude to Dr Xinyang Wang and Dr Wen Wen for their technological
assistance.
References
|
1
|
Wang ZY and Chen Z: Acute promyelocytic
leukemia: from highly fatal to highly curable. Blood.
111:2505–2515. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kogan SC: Curing APL: differentiation or
destruction? Cancer Cell. 15:7–8. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hoffman E and Mielicki WP: All-trans
retinoic acid (ATRA) in prevention and cancer therapy. Postepy Hig
Med Dosw (Online). 64:284–290. 2010.(In Polish).
|
|
4
|
Wang T, Ma X, Krausz KW, Idle JR and
Gonzalez FJ: Role of pregnane X receptor in control of all-trans
retinoic acid (ATRA) metabolism and its potential contribution to
ATRA resistance. J Pharmacol Exp Ther. 324:674–684. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yin T, Wu YL, Sun HP, et al: Combined
effects of As4S4 and imatinib on chronic
myeloid leukemia cells and BCR-ABL oncoprotein. Blood.
104:4219–4225. 2004.
|
|
6
|
Wu J, Shao Y, Liu J, Chen G and Ho PC: The
medicinal use of realgar (As4S4) and its
recent development as an anticancer agent. J Ethnopharmacol.
135:595–602. 2011.PubMed/NCBI
|
|
7
|
Qi J, He PC, Chen W, Wang H, Wang X and
Zhang M: Comparative proteome study of apoptosis induced by
As4S4 in retinoid acid resistant human acute
promyelocytic leukemia NB4-R1 cells. Leuk Res. 34:1506–1516. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pecot CV, Calin GA, Coleman RL,
Lopez-Berestein G and Sood AK: RNA interference in the clinic:
challenges and future directions. Nat Rev Cancer. 11:59–67. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hannon GJ: RNA interference. Nature.
418:244–251. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun BS, Dong QZ, Ye QH, et al:
Lentiviral-mediated miRNA against osteopontin suppresses tumor
growth and metastasis of human hepatocellular carcinoma.
Hepatology. 48:1834–1842. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gallagher R: Retinoic acid resistance in
acute promyelocytic leukemia. Leukemia. 16:1940–1958. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sanz MA, Grimwade D, Tallman MS, et al:
Management of acute promyelocytic leukemia: recommendations from an
expert panel on behalf of the European LeukemiaNet. Blood.
113:1875–1891. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin T, Wu YL, Sun HP, et al: Combined
effects of As4S4 and imatinib on chronic
myeloid leukemia cells and BCR-ABL oncoprotein. Blood.
104:4219–4225. 2004.
|
|
14
|
Wang L, Zhou GB, Liu P, et al: Dissection
of mechanisms of Chinese medicinal formula Realgar-Indigo naturalis
as an effective treatment for promyelocytic leukemia. Proc Natl
Acad Sci USA. 105:4826–4831. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang N, Wang LW, Gou BD, Zhang TL and Wang
K: Realgar-induced differentiation is associated with MAPK pathways
in HL-60 cells. Cell Biol Int. 32:1497–1505. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao W, Lu X, Yuan Y, et al: Effect of
size and processing method on the cytotoxicity of realgar
nanoparticles in cancer cell lines. Int J Nanomedicine.
6:1569–1577. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Janssens V and Rebollo A: The role and
therapeutic potential of Ser/Thr phosphatase PP2A in apoptotic
signalling networks in human cancer cells. Curr Mol Med.
12:268–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Switzer CH, Cheng RYS, Vitek TM,
Christensen DJ, Wink DA and Vitek MP: Targeting SET/I2PP2A
oncoprotein functions as a multi-pathway strategy for cancer
therapy. Oncogene. 30:2504–2513. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krajewski WA and Vassiliev OL: Histone
acetylation facilitates association of nucleosomes with SET domain
of ALL-1 methyltransferase in vitro. Biochem Biophys Res Commun.
397:112–116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu J, Kovach JS, Johnson F, et al:
Inhibition of serine/threonine phosphatase PP2A enhances cancer
chemotherapy by blocking DNA damage induced defense mechanisms.
Proc Natl Acad Sci USA. 106:11697–11702. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ratcliffe MJ, Itoh K and Sokol SY: A
positive role for the PP2A catalytic subunit in Wnt signal
transduction. J Biol Chem. 275:35680–35683. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sontag E, Nunbhakdi-Craig V, Bloom GS and
Mumby MC: A novel pool of protein phosphatase 2A is associated with
microtubules and is regulated during the cell cycle. J Cell Biol.
128:1131–1144. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kolupaeva V, Laplantine E and Basilico C:
PP2A-mediated dephosphorylation of p107 plays a critical role in
chondrocyte cell cycle arrest by FGF. PLoS One. 3:e34472008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aranda-Orgilles B, Rutschow D, Zeller R,
et al: Protein phosphatase 2A (PP2A)-specific ubiquitin ligase MID1
is a sequence-dependent regulator of translation efficiency
controlling 3-phosphoinositide-dependent protein kinase-1 (PDPK-1).
J Biol Chem. 286:39945–39957. 2011. View Article : Google Scholar
|
|
25
|
Sablina AA and Hahn WC: SV40 small T
antigen and PP2A phosphatase in cell transformation. Cancer
Metastasis Rev. 27:137–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gordon J, Hwang J, Carrier KJ, et al:
Protein phosphatase 2a (PP2A) binds within the oligomerization
domain of striatin and regulates the phosphorylation and activation
of the mammalian Ste20-Like kinase Mst3. BMC Biochem. 12:542011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guergnon J, Godet AN, Galioot A, et al:
PP2A targeting by viral proteins: a widespread biological strategy
from DNA/RNA tumor viruses to HIV-1. Biochim Biophys Acta.
1812:1498–1507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cristobal I, Blanco FJ, Garcia-Orti L, et
al: SETBP1 overexpression is a novel leukemogenic mechanism that
predicts adverse outcome in elderly patients with acute myeloid
leukemia. Blood. 115:615–625. 2010. View Article : Google Scholar
|