Introduction
Coronary heart disease (CHD) is one of the main
public health issues worldwide (1)
and also a major cause of morbidity and mortality in developed and
developing societies (2). As the
onset of 50% of all first coronary events is asymptomatic (3), the clinical task of finding the CHD
risk factors, which are able to accurately identify high-risk
individuals, is important in order to implement primary prevention
therapy and lifestyle changes. CHD is a complex disease.
Epidemiology studies have suggested that the etiology of CHD
involves interactions of genetic and environmental factors
(4). Genetic factors contribute
approximately half of the variability of the major risk factors in
the pathogenesis of CHD (5).
In the past decades, a number of linkages and
candidate-gene studies have been performed to identify the genes
characteristic of CHD. The Glu298Asp polymorphism of the human
endothelial nitric oxide synthase (eNOS) gene, which is located on
chromosome 7q35–q36 comprises 26 exons, spans 21 kb and encodes an
mRNA of 4052 nucleotides (6), is
thought to be one of the genes associated with CHD. A variant of
the eNOS gene has been identified within exon 7; a G to T
transversion at nucleotide position 894 of the eNOS cDNA, resulting
in a change of Glu 298 (GAG) to Asp (GAT) (7). In 1998, Shimasaki et
al(7) first reported that the
eNOS gene Glu298Asp polymorphism appeared to be an independent risk
factor for myocardial infarction (MI). Since then, numerous studies
focusing on the association between the eNOS gene Glu298Asp
polymorphism and CHD have been published (7,16–52).
Of these, certain studies further confirmed this association, while
others came to negative or even contrary conclusions.
In 2010, Li et al(8) performed a meta-analysis of 20 studies
involving a non-Asian population and 3 studies involving an Asian
population in order to detect the eNOS gene Glu298Asp polymorphism
and risk of CHD. A significant association was found between the
Asp (T) allele in the eNOS Glu298Asp (G894T) polymorphism and CHD
in the non-Asian population, but not in the Asian population. In
2012, Zhang et al(9)
performed a meta-analysis of 18 case-control studies to determine
whether the eNOS Glu298Asp polymorphism was associated with an
increased risk of CHD among Asian populations. The analysis
conluded that the eNOS Glu298Asp polymorphism may play a
significant role in the development of CHD among Asian populations.
However, the meta-analysis included certain irrelevant studies and
thus contained certain limitations. There have been 39 relevant
studies published thus far.
For these reasons, a comprehensive meta-analysis was
required to provide an updated result on the overall correlation
between the eNOS Glu298Asp polymorphism and CHD. A comprehensive
search was consequently conducted and the present meta-analysis was
performed according to the proposed PRISMA (preferred reporting
items for systematic reviews and meta-analyses) guidelines
(10), in order to clarify whether
an association existed between the eNOS gene Glu298Asp polymorphism
and the risk of CHD.
Subgroup analyses were also performed between the
Asian and non-Asian populations to investigate any
ethnicity-specific effects, to quantify heterogeneity between the
individual studies and to investigate the existence of any
potential publication bias.
Materials and methods
Literature search
A comprehensive electronic search covering the
period between 1st January, 1998 and 30th August, 2012 was
conducted in the PubMed database to search for any studies
attempting to detect an association between the eNOS Glu298Asp
polymorphism and CHD. Published and unpublished studies were
searched to ensure the analysis was as exhaustive as possible.
Keywords were used with the Boolean operators ‘OR’ and ‘AND’
combined with the following MeSH or text words: (‘endothelial
nitric oxide synthase’ OR ‘eNOS’) AND (‘polymorphism’ OR ‘genetic’
OR ‘mutation’ OR ‘variant’) AND (‘myocardial infarction’ OR
‘myocardial infarct’ OR ‘coronary artery disease’ OR ‘coronary
heart disease’ OR ‘CHD’ OR ‘ischemic heart disease’ OR ‘myocardial
ischemia’ OR ‘angina’ OR ‘CAD’). No restrictions were imposed and
the bibliographies of the included studies and previous
meta-analysis were checked for other relevant publications. As this
study ia a meta-analysis of primary studies, no specific ethical
approval is required.
Study selection
The eligibility of all studies retrieved from the
databases were independently evaluated by two authors according to
the following criteria: i) The study must have a case-control study
design; ii) the study must have investigated the association
between the eNOS Glu298Asp polymorphism and CHD susceptibility;
iii) only cases of CHD, without other diseases (e.g., diabetes
mellitus), were considered, the controls must have been from a
healthy population, and in the cases and controls, the diagnosis
must have been based on angiographic or clinical criteria with
clearly reported details; and iv) enough information must have been
provided to calculate the odds ratios (ORs) and the relevant 95%
confidence intervals (CIs). Disagreements between the two authors
were resolved by discussion.
In addition, the following exclusion criteria were
also used: i) no abstracts or reviews; ii) no studies in which the
genotype frequencies were not reported; and iii) no repeated or
overlapping publications. For studies with the same case series by
the same authors or the same institution in a same period, the most
recently published studies or the studies with the largest number
of subjects were included.
Data extraction
The following data were extracted by two authors
from all eligible publications: first author’s last name, year of
publication, ethnicity, age of included subjects, source of
controls, matching criteria, number of cases and controls, number
of different genotypes in cases and controls, Hardy-Weinberg
equilibrium (HWE) and minor allele frequency in controls. Any
disagreements were resolved by consensus. CHD was defined as
myocardial infarction (MI), angina pectoris or other ischemic heart
disease (IHD).
Statistical analysis
A pooled OR and 95% CI was computed for the risk
allele using the Comprehensive Meta Analysis software (version 2.2;
Biostat, Englewood, NJ, USA) (11)
to generate forest plots, in order to determine whether there was a
statistical association between cases and controls and to assess
the heterogeneity of the included studies. The Hardy-Weinberg
equilibrium (HWE) was tested by the Chi-square test at a
significance level of P<0.05. Heterogeneity was evaluated using
the Cochran’s Q (12) and
I2 statistics, the I2 statistic yielded
results ranging from 0 to 100% (0–25%, no heterogeneity; 25–50%,
moderate heterogeneity; 50–75%, large heterogeneity; and 75–100%,
extreme heterogeneity) (13). If
heterogeneity existed, the random effects model was used. If no
heterogeneity existed, the random effects model was considered to
have replaced the fixed effects model, therefore, all the analyses
of present study used the random effects model.
In addition, the effect of a single study on the
overall risk estimate was studied by the removal of each study in
turn. To test the robustness of the main results, subgroup analysis
was also conducted if significant heterogeneity was identified.
Potential publication bias was assessed by a visual inspection of
the funnel plots and by the Egger weighted regression (P<0.05
was considered to indicate a statistically significant difference)
(14) and ‘trim and fill’ methods
(15).
Results
Study identification
Of the 337 records initially identified, 38 articles
involving 39 case-control studies were included in the present
meta-analysis (7,16–52).
A detailed flowchart of the selection process is shown in Fig. 1.
Study characteristics
The major characteristics of the 39 case-control
studies are shown in Table I. All
the information from the studies was available. The number of
subjects ranged from 54 to 755 in the case group and from 43 to 624
in the control group, comprising a total of 7,489 cases and 7,051
controls. A total of 18 studies were conducted in the Asian
population (7,16,20,21,23,25,26,33–37,41,42,45,48,
51, 52) and 21 in the non-Asian population
(17–19,22,24,27–32,38–40,
43,44,46,47,49–50).
In terms of the source of the controls, 15 were from hosptial-based
(HB) studies (16,17,20,24,25,28,
30,34,35,37,41,43,45,47,51)
and 24 were from population-based (PB) studies (7,18,19,21–23,26,27,29,31–33,36,38–40,42,
44,46,48–50,52).
The genotyping methods included polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP) assays
(36 studies) and PCR-single strand conformation polymorphism
(PCR-SSCP) assays (3 studies) (18,19).
The genotype distributions in the controls of all studies were in
accordance with the HWE.
 | Table ICharacteristics of the studies
included in the meta-analysis. |
Table I
Characteristics of the studies
included in the meta-analysis.
| First author,
year | Ethnicity
(country) | Source of
controls | End points | Genotype
distribution | Genotyping
method | HWE P-value |
|---|
|
|---|
| Sample size | Case | Control |
|---|
|
|
|
|---|
| Case | Control | Glu/Glu | Glu/Asp | Asp/Asp | Glu/Glu | Glu/Asp | Asp/Asp |
|---|
| Yoshimura,
1998 | Asian (Japan) | HB | CHD | 113 | 100 | 89 | 23 | 1 | 91 | 9 | 0 | PCR-RFLP | 0.64 |
| Shimasaki,
1998 | Asian (Japan) | PB | MI | 285 | 607 | 225 | 59 | 1 | 526 | 80 | 1 | PCR-RFLP | 0.25 |
| Cai, 1999 | Non-Asian
(Australia) | HB | CHD | 605 | 158 | 286 | 249 | 70 | 66 | 70 | 22 | PCR-RFLP | 0.62 |
| Hingorani,
1999 | Non-Asian (UK) | PB | CHD | 298 | 138 | 120 | 71 | 107 | 66 | 58 | 14 | PCR-SSCP | 0.81 |
| Poirier, 1999 | Non-Asian
(France) | PB | MI | 368 | 421 | 163 | 156 | 49 | 148 | 219 | 54 | PCR-SSCP | 0.051 |
| Poirier, 1999 | Non-Asian
(Ireland) | PB | MI | 163 | 155 | 55 | 76 | 32 | 58 | 72 | 25 | PCR-SSCP | 0.74 |
| Yoon, 2000 | Asian (Korea) | HB | CHD | 110 | 128 | 94 | 15 | 1 | 110 | 18 | 0 | PCR-RFLP | 0.39 |
| Yoshimura,
2000 | Asian (Japan) | PB | CHD | 201 | 345 | 162 | 38 | 1 | 301 | 44 | 0 | PCR-RFLP | 0.21 |
| Granath, 2001 | Non-Asian
(Australia) | PB | CHD | 573 | 624 | 260 | 248 | 63 | 270 | 287 | 66 | PCR-RFLP | 0.42 |
| Wang, 2001 | Asian (China) | PB | CHD | 218 | 218 | 178 | 38 | 2 | 177 | 38 | 3 | PCR-RFLP | 0.56 |
| Colombo, 2002 | Non-Asian
(Italy) | HB | CHD | 201 | 114 | 91 | 78 | 32 | 48 | 59 | 7 | PCR-RFLP | 0.07 |
| Wei, 2002 | Asian (China) | HB | CHD | 106 | 108 | 84 | 19 | 3 | 98 | 10 | 0 | PCR-RFLP | 0.61 |
| Colombo, 2003 | Non-Asian
(Italy) | PB | CHD | 268 | 147 | 109 | 116 | 43 | 65 | 72 | 10 | PCR-RFLP | 0.09 |
| Chang, 2003 | Asian (Korea) | PB | CHD | 108 | 112 | 80 | 19 | 3 | 108 | 10 | 0 | PCR-RFLP | 0.63 |
| Agema, 2004 | Non-Asian (The
Netherlands) | PB | CHD | 755 | 574 | 343 | 333 | 79 | 216 | 270 | 88 | PCR-RFLP | 0.81 |
| Afrasyap, 2004 | Non-Asian
(Turkey) | HB | CHD | 250 | 150 | 114 | 103 | 33 | 74 | 62 | 14 | PCR-RFLP | 0.85 |
| Cam, 2005 | Non-Asian
(Turkey) | HB | CHD | 115 | 83 | 44 | 37 | 34 | 57 | 24 | 2 | PCR-RFLP | 0.78 |
| Kerkeni, 2006 | Non-Asian
(Tunisia) | PB | CHD | 100 | 120 | 45 | 44 | 11 | 72 | 43 | 5 | PCR-RFLP | 0.65 |
| Jaramillo,
2006 | Non-Asian
(Chile) | PB | CHD | 112 | 72 | 73 | 31 | 8 | 51 | 20 | 1 | PCR-RFLP | 0.54 |
| Ji, 2007 | Asian (China) | PB | CHD | 165 | 190 | 125 | 39 | 1 | 157 | 32 | 1 | PCR-RFLP | 0.64 |
| Wang, 2007 | Asian (China) | HB | CHD | 58 | 43 | 25 | 21 | 12 | 21 | 15 | 7 | PCR-RFLP | 0.15 |
| Kim, 2007 | Asian (Korea) | HB | CHD | 147 | 222 | 119 | 28 | 0 | 181 | 38 | 0 | PCR-RFLP | 0.16 |
| Vasilakou,
2008 | Non-Asian
(Greece) | PB | CHD | 209 | 161 | 109 | 85 | 15 | 76 | 74 | 11 | PCR-RFLP | 0.21 |
| Mathew, 2008 | Asian (India) | PB | CHD | 100 | 100 | 72 | 26 | 2 | 79 | 18 | 3 | PCR-RFLP | 0.14 |
|
Zakrzewski-Jakubiak, 2008 | Non-Asian
(Canada) | PB | CHD | 58 | 111 | 22 | 23 | 12 | 40 | 57 | 14 | PCR-RFLP | 0.36 |
| Tamemoto, 2008 | Asian (Japan) | HB | CHD | 54 | 283 | 38 | 13 | 3 | 249 | 32 | 2 | PCR-RFLP | 0.39 |
| Alp, 2009 | Non-Asian
(Turkey) | PB | CHD | 146 | 122 | 76 | 59 | 11 | 71 | 40 | 11 | PCR-RFLP | 0.14 |
| Angeline, 2010 | Asian (India) | PB | MI | 100 | 100 | 56 | 30 | 14 | 67 | 31 | 2 | PCR-RFLP | 0.46 |
| Alkharfy, 2010 | Asian (Saudi
Arabia) | HB | CHD | 142 | 145 | 65 | 67 | 10 | 98 | 40 | 7 | PCR-RFLP | 0.28 |
| Bor-Kucukatay,
2010 | Non-Asian
(Turkey) | HB | CHD | 83 | 74 | 48 | 28 | 7 | 50 | 21 | 3 | PCR-RFLP | 0.68 |
| Velloso, 2010 | Non-Asian
(Brazil) | PB | CHD | 100 | 103 | 49 | 47 | 4 | 36 | 51 | 16 | PCR-RFLP | 0.77 |
| Isordia-Salas,
2010 | Non-Asian
(Mexico) | PB | MI | 180 | 180 | 104 | 62 | 14 | 134 | 42 | 4 | PCR-RFLP | 0.74 |
| Salimi, 2010 | Asian (Iran) | HB | CHD | 241 | 261 | 112 | 103 | 26 | 160 | 84 | 17 | PCR-RFLP | 0.20 |
| Saini, 2011 | Asian (India) | PB | CHD | 60 | 50 | 45 | 15 | 0 | 44 | 6 | 0 | PCR-RFLP | 0.65 |
| Motawi, 2011 | Non-Asian
(Egypt) | HB | CHD | 100 | 50 | 46 | 34 | 20 | 19 | 28 | 3 | PCR-RFLP | 0.80 |
| Rahimi, 2012 | Asian (Iran) | HB | CHD | 105 | 92 | 61 | 33 | 11 | 67 | 24 | 1 | PCR-RFLP | 0.47 |
| da Costa Escobar
Piccoli, 2012 | Non-Asian
(Brazil) | PB | CHD | 135 | 115 | 58 | 52 | 18 | 62 | 44 | 7 | PCR-RFLP | 0.83 |
| Rai, 2012 | Asian (India) | PB | CHD | 253 | 174 | 159 | 84 | 10 | 119 | 50 | 5 | PCR-RFLP | 0.93 |
| Gad, 2012 | Non-Asian
(Egypt) | PB | MI | 104 | 101 | 52 | 47 | 5 | 59 | 34 | 8 | PCR-RFLP | 0.33 |
Meta-analyses
Table II shows the
main results and the results of the heterogeneity test on the
meta-analyses. Overall, substantial heterogeneity existed that all
the genetic models used the random-effects models. A significant
association was identified between the eNOS Glu298Asp polymorphism
and CHD susceptibility for all the genetic models, with a lower Asp
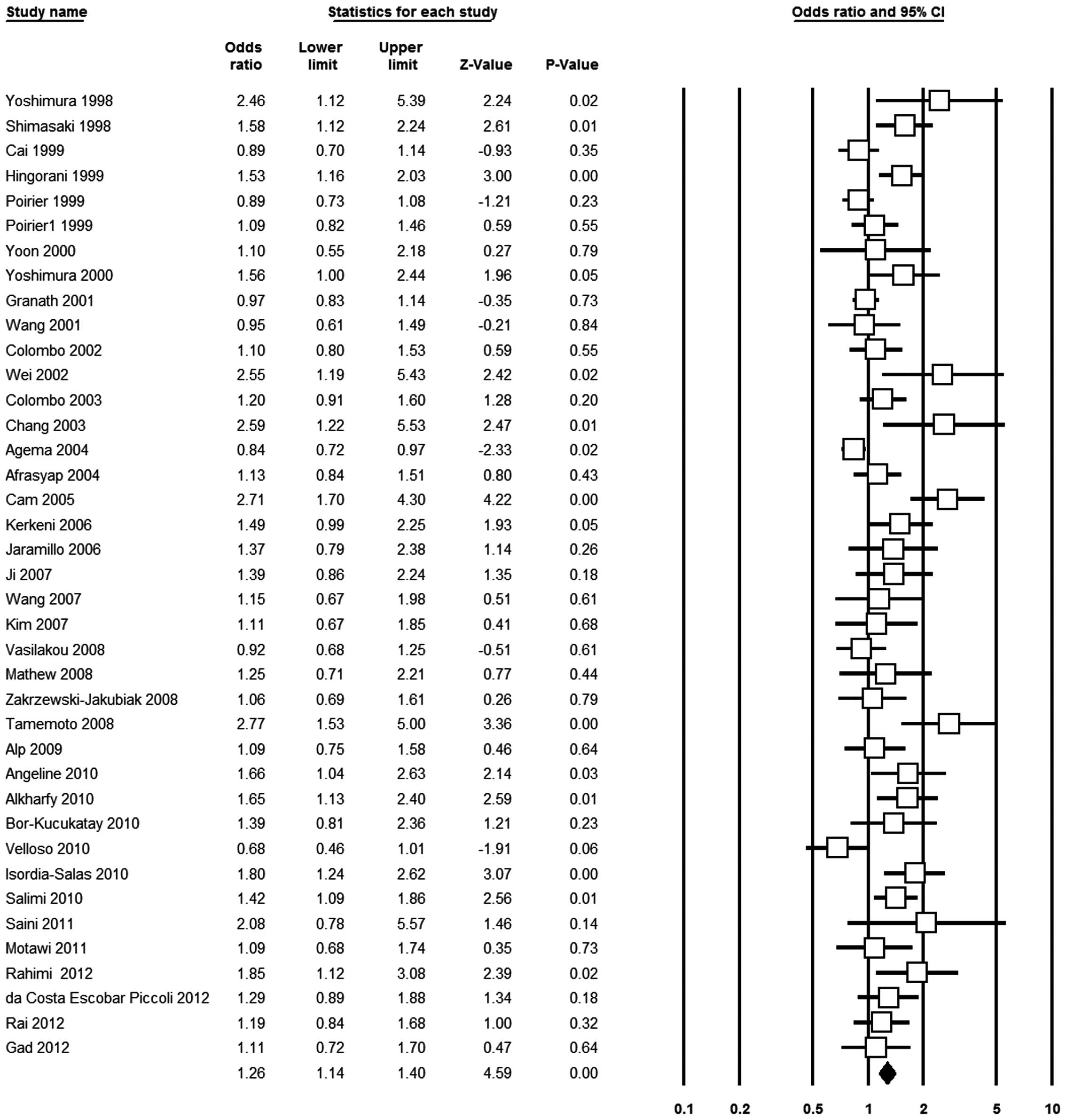
allele frequency shown in the cases than in the controls [Asp vs.
Glu, OR 1.26, 95% CI 1.14–1.40, P<0.001, Fig. 2; Asp/Asp vs. Glu/Glu, OR 1.58, 95%
CI 1.23–2.02, P<0.001; Glu/Asp vs. Glu/Glu, OR 1.12, 95% CI
1.03–1.22, P=0.001; (Glu/Asp+Asp/Asp) vs. Glu/Glu, OR 1.17, 95% CI
1.07–1.27, P<0.001; Asp/Asp vs. (Glu/Glu+Glu/Asp), OR 1.59, 95%
CI 1.25–2.03, P<0.001].
 | Table IIPooled ORs and 95% CIs of the
association between eNOS Glu298Asp polymorphism and CHD. |
Table II
Pooled ORs and 95% CIs of the
association between eNOS Glu298Asp polymorphism and CHD.
| Total and
subgroups | Studies | Asp vs. Glu | Asp/Asp vs.
Glu/Glu | Glu/Asp vs.
Glu/Glu | (Glu/Asp+Asp/Asp)
vs. Glu/Glu | Asp/Asp vs.
(Glu/Glu+Glu/Asp) |
|---|
|
|
|
|
|
|---|
| OR (95% CI) | P-value | I2
(%) | OR (95% CI) | P-value | I2
(%) | OR (95% CI) | P-value | I2
(%) | OR (95% CI) | P-value | I2
(%) | OR (95% CI) | P-value | I2
(%) |
|---|
| Total | 39 | 1.26
(1.14–1.40) | <0.001 | 64.73 | 1.58
(1.23–2.02) | <0.001 | 56.78 | 1.12
(1.03–1.22) | 0.001 | 29.38 | 1.17
(1.07–1.27) | <0.001 | 37.77 | 1.59
(1.25–2.03) | <0.001 | 57.75 |
| Ethnicity |
| Asian | 18 | 1.48
(1.32–1.65) | <0.001 | 16.74 | 2.15
(1.49–3.10) | <0.001 | 0 | 1.40
(1.24–1.59) | <0.001 | 0 | 1.43
(1.27–1.61) | <0.001 | 0 | 1.88
(1.31–2.69) | <0.001 | 0 |
| Non-Asian | 21 | 1.13
(1.00–1.26) | 0.04 | 65.63 | 1.38
(1.04–1.84) | 0.03 | 66.29 | 0.96
(0.89–1.05) | 0.38 | 0 | 1.02
(0.94–1.11) | 0.67 | 13.86 | 1.47
(1.10–1.97) | 0.01 | 69.77 |
| Source of
control |
| HB | 15 | 1.43
(1.19–1.72) | <0.001 | 63 | 2.14
(1.40–3.27) | <0.001 | 46.34 | 1.40
(1.24–1.59) | 0.02 | 31.5 | 1.29
(1.09–1.53) | <0.001 | 40.91 | 2.09
(1.38–3.16) | <0.001 | 45.96 |
| PB | 24 | 1.18
(1.06–1.32) | <0.001 | 61.84 | 1.34
(1.00–1.80) | 0.05 | 57.75 | 0.96
(0.89–1.05) | 0.38 | 24.88 | 1.11
(1.01–1.22) | 0.04 | 29.73 | 1.38
(1.03–1.86) | 0.03 | 60.82 |
| End point |
| CHD | 33 | 1.26
(1.13–1.41) | <0.001 | 64.22 | 1.60
(1.21–2.12) | <0.001 | 57.47 | 1.11
(1.01–1.21) | 0.03 | 23.74 | 1.16
(1.06–1.27) | <0.001 | 34.03 | 1.63
(1.23–2.16) | <0.001 | 59.75 |
| MI | 6 | 1.28
(0.99–1.65) | 0.06 | 72.65 | 1.55
(0.83–2.88) | 0.17 | 60.54 | 1.08
(0.91–1.52) | 0.21 | 57.25 | 1.21
(0.95–1.55) | 0.12 | 59.91 | 1.48
(0.85–2.57) | 0.16 | 53.06 |
| Genotyping
type |
| PCR-RFLP | 36 | 1.28
(1.15–1.43) | <0.001 | 63.85 | 1.63
(1.24–2.15) | <0.001 | 55.67 | 1.15
(1.05–1.26) | <0.001 | 24.83 | 1.20
(1.09–1.31) | <0.001 | 37.47 | 1.60
(1.22–2.08) | <0.001 | 54.51 |
| PCR-SSCP | 3 | 1.13
(0.82–1.56) | 0.46 | 79.99 | 1.38
(0.71–2.68) | 0.34 | 77.36 | 0.86
(0.71–1.04) | 0.12 | 0 | 0.97
(0.81–1.16) | 0.75 | 10.28 | 1.61
(0.78–3.33) | 0.19 | 82.78 |
In the stratified analysis, a marked and exact
association was identified between the eNOS Glu298Asp polymorphism
and CHD susceptibility in the Asian population, hospital-based, CHD
and PCR-RFLP subgroups of all the genetic models; however, the
results of the non-Asian population and population-based subgroups
were inconsistent. In contrast, the MI and PCR-SSCP subgroups
showed no association with the eNOS Glu298Asp polymorphism.
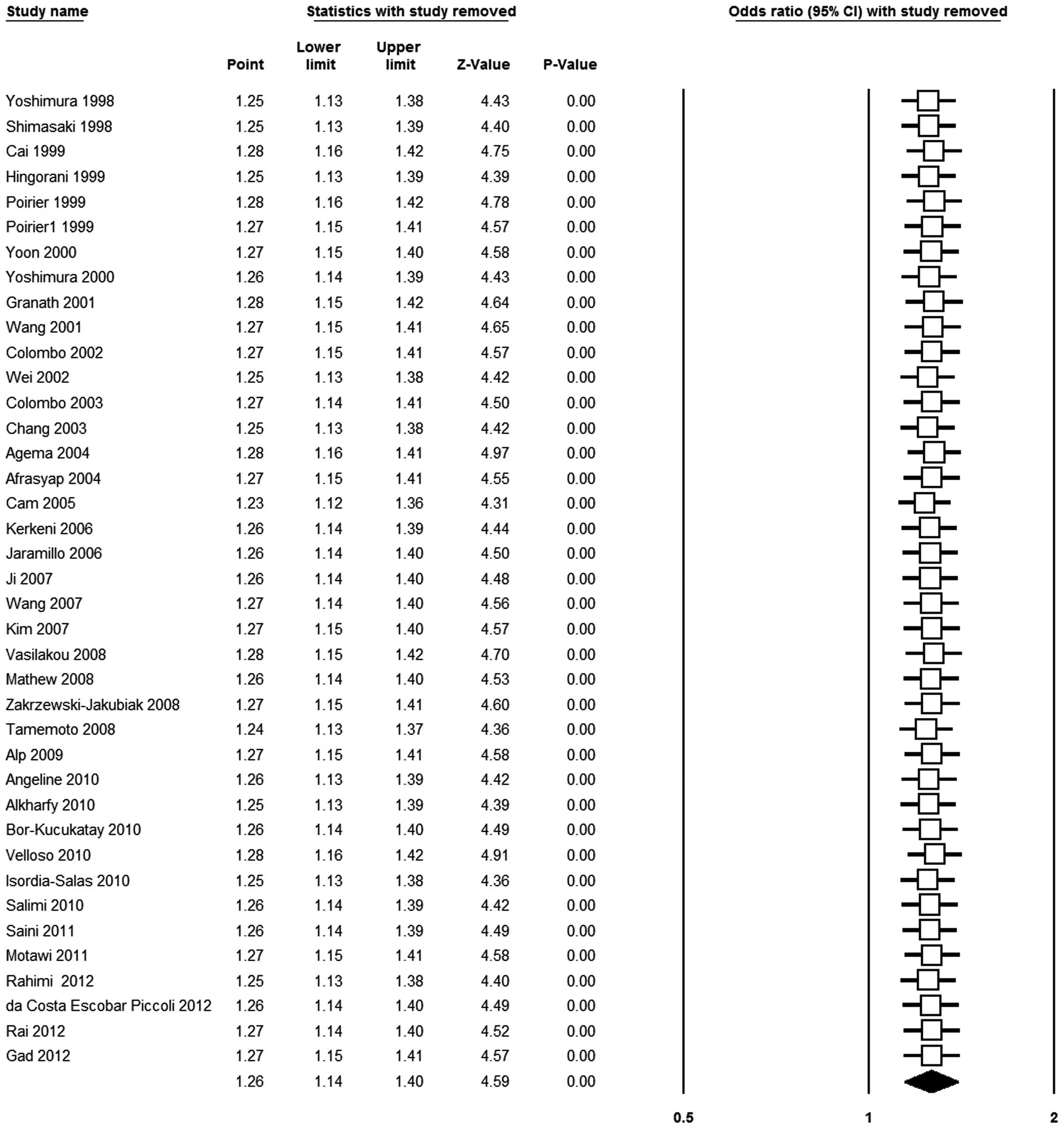
Sensitivity analysis
A sensitivity analysis was carried out by omitting
each study included in the overall meta-analysis in turn. The
results in any of the genetic models were not materially altered,
which showed that the results were robust (Asp vs. Glu model,
Fig. 3).
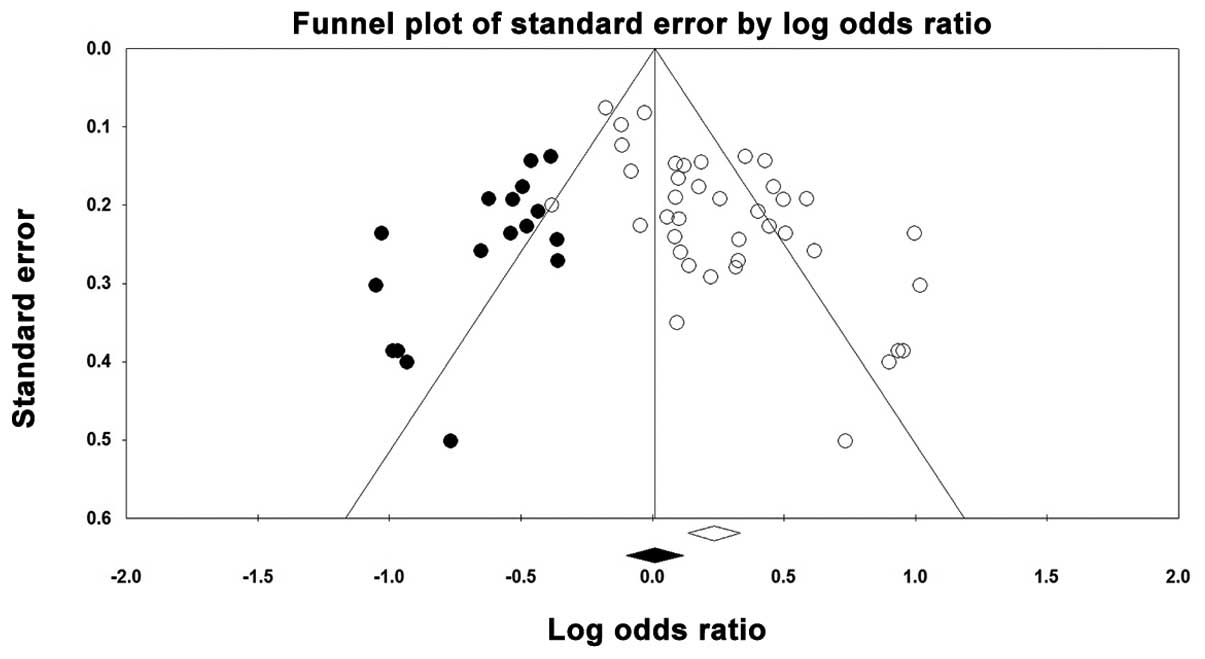
Publication bias
A funnel plot was created and the Egger’s test was
performed to assess any possible publication bias. The funnel plot
for the genetic Asp vs. Glu model showed that a weak publication
bias was detected (blank circles in Fig. 4) and the ‘trim and fill’ method
estimated that there were 17 possible missing studies (black spots
in Fig. 4). This finding was also
confirmed by the results of the Egger’s test [Asp vs. Glu, Asp/Asp
vs. Glu/Glu, Glu/Asp vs. Glu/Glu and (Glu/Asp+Asp/Asp) vs. Glu/Glu,
P<0.001; Asp/Asp vs. (Glu/Glu+Glu/Asp), P=0.001].
Discussion
CHD often presents suddenly with little warning and
easily results in mortality. Therefore, the early identification of
asymptomatic but susceptible individuals is extremely important.
However, traditional risk factors are inadequate for identifying
these asymptomatic high-risk individuals. At present, genetic
testing is a well-accepted method for the early identification of
patients with subclinical CHD, as it is non-invasive, relatively
simple and inexpensive when compared with non-invasive coronary
imaging tools. Additionally, the genetic markers are fixed and
require measuring only once in an individual’s lifetime, they are
able to guide therapy selection and they may be of use in family
counseling (53). To identify the
genetic markers of an increased CHD risk is a key aim and the eNOS
gene Glu298Asp polymorphism is a novel solution.
The human eNOS gene is located on chromosome
7q35–q36 (6). A polymorphic
variant of this gene in exon 7 characterized by a Glu298Asp
(894G→T) polymorphism has been identified. Although the mechanism
responsible for the eNOS Glu298Asp mutation that is related to
endothelial dysfunction remains unknown, various studies (8,9,54–57)
have determined the correlation of its genotypes and numerous
associated diseases. A number of studies have identified a lower
Asp allele frequency in CHD cases than in healthy controls, as the
two alleles are considered to have codominant effects on eNOS
levels. However, certain studies have generated results that
disagreed with each other. This may be due to the studies including
varying populations (e.g., varied race or country) sampling
strategies or genotyping procedures or small sample sizes. The
present meta-analysis was performed in all the populations and
subgroup analyses were conducted according to these factors.
The present meta-analysis of 39 case-control studies
revealed that the eNOS Glu298Asp polymorphic variant was associated
with CHD susceptibility. The results also revealed that a greater
Asp allele frequency gave rise to an increased susceptibility to
CHD, with the random effects ORs ranging from 1.12 to 1.59 for the
five genetic models under investigation. The results remained
statistically significant following sensitivity analyses that
omitted one study at a time and confirmed the result was robust. In
the subgroups analyses, the association depended on four variables:
ethnicity, source of control, end point and genotying method. A
strong association was identified between the eNOS Glu298Asp
polymorphism and CHD in the Asian population, hospital-based, and
CHD subgroups. The ORs and relevant 95% CIs of each subgroup were
similar, which also confirmed that an association existed between
the eNOS Glu298Asp polymorphism and CHD. However, the results of
the non-Asian population and population-based analyses were
inconsistent. In contrast, the the MI and PCR-SSCP subgroup showed
no association with the eNOS Glu298Asp polymorphism.
Compared with a previous meta-analysis (8), the strength of the present
meta-analysis was based on a more comprehensive search and the
identification of a greater number of published studies, thereby
providing more statistical power to detect the genetic effect
estimates. A further sensitivity analysis confirmed that there was
a significant association between the eNOS Glu298Asp polymorphism
and the CHD risk among the Asian population. The results of the
present study were also consistent with another previous
meta-analysis and externally confirmed the validity of its results,
which indicated that the association between the eNOS Glu298Asp
polymorphism and CHD risk was significant among Asian populations
(9).
There are certain limations to the present study.
First, heterogeneity was detected in the present meta-analysis and
although heterogeneity between studies is extremely common in the
meta-analysis of genetic association studies, this should not be
ignored. Heterogenity may be due to a number of reasons, including
differences in the recruitment procedures of the study population,
environmental backgrounds, genotyping methods or diagnostic
criteria. Subgroup analyses were performed to assess these
criteria, but the heterogeneity remained. Secondly, the sample
sizes in the majority of the studies were relatively small.
Compared with those studies with a large sample size, the studies
with a small sample size may overestimate the true association,
whereas a study with a large sample size may reflect a true
association as it has greater statistical power. Thirdly, the
present study performed a more comprehensive search, but a weak
reporting bias was detected. Reporting bias is a significant threat
to any literature-based review or meta-analysis. Lastly, the
present results were based on unadjusted estimates and the lack of
original data from the eligible studies limited the evaluation of
the effects of the gene-gene and gene-environment interactions in
CHD development. Undoubtedly, all these limitations may have
affected the final conclusions.
The present meta-analysis may also provide certain
suggestions for clinical practice and further research. Further
research should clarify the association of other polymorphisms and
their interactions with the eNOS Glu298Asp polymorphism and CHD.
Clinically, the eNOS Glu298Asp polymorphism may be used as a basis
to judge whether an Asian individual has a susceptibility to CHD or
not and to provide more attention to those who are susceptible.
In conclusion, the results of the present study
support the significant association of the eNOS Glu298Asp
polymorphism and CHD susceptibility. The Asp allele was associated
with an increased CHD susceptibility, while the Glu allele was
associated with a decreased CHD susceptibility. However,
large-scale studies with consideration of the gene-gene and
gene-environment interactions should be conducted in the future to
investigate this association.
Acknowledgements
This study was partly supported by grants from the
Beijing Medicine Research and Development Fund (SF-2009-II-18) and
the Public Health Application Research and Vaccine preventable
diseases Research Funds of the Chinese Preventive Medicine
Association (20103345), without commercial or not-for-profit
sectors.
References
|
1
|
Negi S and Anand A: Atherosclerotic
coronary heart disease-epidemiology, classification and management.
Cardiovasc Hematol Disord Drug Targets. 10:257–261. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Murray CJ and Lopez AD: Mortality by cause
for eight regions of the world: Global burden of disease study.
Lancet. 349:1269–1276. 1997. View Article : Google Scholar
|
|
3
|
Rosamond W, Flegal K, Friday G, et al;
American Heart Association Statistics Committee and Stroke
Statistics Subcommittee. Heart disease and stroke statistics - 2007
update: a report from the American Heart Association Statistics
Committee and Stroke Statistics Subcommittee. Circulation.
115:e69–e171. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mi X, Eskridge KM, George V and Wang D:
Structural equation modeling of gene-environment interactions in
coronary heart disease. Ann Hum Genet. 75:255–265. 2011.PubMed/NCBI
|
|
5
|
Kullo IJ and Ding K: Mechanisms of
disease: The genetic basis of coronary heart disease. Nat Clin
Pract Cardiovasc Med. 4:558–569. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marsden PA, Heng HH, Scherer SW, et al:
Structure and chromosomal localization of the human constitutive
endothelial nitric oxide synthase gene. J Biol Chem.
268:17478–17488. 1993.PubMed/NCBI
|
|
7
|
Shimasaki Y, Yasue H, Yoshimura M, et al:
Association of the missense Glu298Asp variant of the endothelial
nitric oxide synthase gene with myocardial infarction. J Am Coll
Cardiol. 31:1506–1510. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Wu X, Li X, Feng G, He L and Shi Y:
The endothelial nitric oxide synthase gene is associated with
coronary artery disease: a meta-analysis. Cardiology. 116:271–278.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang K, Bai P, Shi S, et al: The G894T
polymorphism on endothelial nitric oxide synthase gene is
associated with increased coronary heart disease among Asia
population: evidence from a Meta analysis. Thromb Res. 130:192–197.
2012. View Article : Google Scholar
|
|
10
|
Moher D, Liberati A, Tetzlaff J and Altman
DG; PRISM Group. Preferred reporting items for systematic reviews
and meta-analyses: the PRISMA statement. PLoS Med. 6:e10000972009.
View Article : Google Scholar
|
|
11
|
Borenstein M, Hedges L, Higgins J,
Rothstein H and Englewood NJ: Comprehensive meta-analysis: A
computer program for meta-analysis. Biostat Inc; 2007, [Computer
software].
|
|
12
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duval S and Tweedie R: Trim and fill: A
simple funnel-plot-based method of testing and adjusting for
publication bias in meta-analysis. Biometrics. 56:455–463. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoshimura M, Yasue H, Nakayama M, et al: A
missense Glu298Asp variant in the endothelial nitric oxide synthase
gene is associated with coronary spasm in the Japanese. Hum Genet.
103:65–69. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cai H, Wilcken DE and Wang XL: The
Glu-298→Asp (894G→T) mutation at exon 7 of the endothelial nitric
oxide synthase gene and coronary artery disease. J Mol Med (Berl).
77:511–514. 1999.
|
|
18
|
Hingorani AD, Liang CF, Fatibene J, et al:
A common variant of the endothelial nitric oxide synthase
(Glu298→Asp) is a major risk factor for coronary artery disease in
the UK. Circulation. 100:1515–1520. 1999.
|
|
19
|
Poirier O, Mao C, Mallet C, et al:
Polymorphisms of the endothelial nitric oxide synthase gene - no
consistent association with myocardial infarction in the ECTIM
study. Eur J Clin Invest. 29:284–290. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yoon Y, Song J, Hong SH and Kim JQ: Plasma
nitric oxide concentrations and nitric oxide synthase gene
polymorphisms in coronary artery disease. Clin Chem. 46:1626–1630.
2000.PubMed/NCBI
|
|
21
|
Yoshimura M, Yasue H, Nakayama M, et al:
Genetic risk factors for coronary artery spasm: significance of
endothelial nitric oxide synthase gene T-786-->C and missense
Glu298Asp variants. J Investig Med. 48:367–374. 2000.PubMed/NCBI
|
|
22
|
Granath B, Taylor RR, van Bockxmeer FM and
Mamotte CD: Lack of evidence for association between endothelial
nitric oxide synthase gene polymorphisms and coronary artery
disease in the Australian Caucasian population. J Cardiovasc Risk.
8:235–241. 2001. View Article : Google Scholar
|
|
23
|
Wang CL, Hsu LA, Ko YS, Ko YL and Lee YH:
Lack of association between the Glu298Asp variant of the
endothelial nitric oxide synthase gene and the risk of coronary
artery disease among Taiwanese. J Formos Med Assoc. 100:736–740.
2001.PubMed/NCBI
|
|
24
|
Colombo MG, Andreassi MG, Paradossi U, et
al: Evidence for association of a common variant of the endothelial
nitric oxide synthase gene (Glu298→Asp polymorphism) to the
presence, extent, and severity of coronary artery disease. Heart.
87:525–528. 2002.PubMed/NCBI
|
|
25
|
Wei D, Shan J, Chen Z and Shi Y: The G894T
mutation of the endothelial nitric oxide synthase gene is
associated with coronary atherosclerotic heart disease in Chinese.
Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 19:471–474. 2002.(In
Chinese).
|
|
26
|
Chang K, Baek SH, Seung KB, et al: The
Glu298Asp polymorphism in the endothelial nitric oxide synthase
gene is strongly associated with coronary spasm. Coron Artery Dis.
14:293–299. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Colombo MG, Paradossi U, Andreassi MG, et
al: Endothelial nitric oxide synthase gene polymorphisms and risk
of coronary artery disease. Clin Chem. 49:389–395. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Afrasyap L and Ozturk G: NO level and
endothelial NO synthase gene polymorphism (Glu298Asp) in the
patients with coronary artery disease from the Turkish population.
Acta Biochim Biophys Sin (Shanghai). 36:661–666. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Agema WR, de Maat MP, Zwinderman AH, et
al: An integrated evaluation of endothelial constitutive nitric
oxide synthase polymorphisms and coronary artery disease in men.
Clin Sci (Lond). 107:255–261. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cam SF, Sekuri C, Tengiz I, et al: The
G894T polymorphism on endothelial nitric oxide synthase gene is
associated with premature coronary artery disease in a Turkish
population. Thromb Res. 116:287–292. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jaramillo PC, Muñoz MA, Lanas MC, Lanas ZF
and Salazar LA: Endothelial nitric oxide synthase G894T gene
polymorphism in Chilean subjects with coronary artery disease and
controls. Clin Chim Acta. 371:102–106. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kerkeni M, Addad F, Chauffert M, et al:
Hyperhomocysteinemia, endothelial nitric oxide synthase
polymorphism, and risk of coronary artery disease. Clin Chem.
52:53–58. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ji XW, Zhang AY and Guan LX: Association
between angiotensin-converting enzyme and endothelial nitric oxide
synthase gene polymorphism and risk of coronary artery disease.
Zhonghua Xin Xue Guan Bing Za Zhi. 35:1024–1028. 2007.(In
Chinese).
|
|
34
|
Kim IJ, Bae J, Lim SW, et al: Influence of
endothelial nitric oxide synthase gene polymorphisms (−786T>C,
4a4b, 894G>T) in Korean patients with coronary artery disease.
Thromb Res. 119:579–585. 2007.
|
|
35
|
Wang ZL, Zhao YQ, Liao JD, et al:
Association of endothelial nitric oxide synthase gene polymorphism
with level of uric acid in serum for acute coronary syndrome.
Zhongguo Wei Zhong Bing Ji Jiu Yi Xue. 19:652–656. 2007.(In
Chinese).
|
|
36
|
Mathew J, Narayanan P, Sundaram R, et al:
Lack of association between Glu(298) asp polymorphism of
endothelial nitric oxide synthase (eNOS) gene and coronary artery
disease in Tamilian population. Indian Heart J. 60:223–227.
2008.PubMed/NCBI
|
|
37
|
Tamemoto H, Ishikawa SE and Kawakami M:
Association of the Glu298Asp polymorphism of the eNOS gene with
ischemic heart disease in Japanese diabetic subjects. Diabetes Res
Clin Pract. 80:275–279. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vasilakou M, Votteas V, Kasparian C, et
al: Lack of association between endothelial nitric oxide synthase
gene polymorphisms and risk of premature coronary artery disease in
the Greek population. Acta Cardiol. 63:609–614. 2008. View Article : Google Scholar
|
|
39
|
Zakrzewski-Jakubiak M, de Denus S, Dubé
MP, Bélanger F, White M and Turgeon J: Ten renin-angiotensin
system-related gene polymorphisms in maximally treated Canadian
Caucasian patients with heart failure. Br J Clin Pharmacol.
65:742–751. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Alp E, Menevse S, Tulmac M, et al: Lack of
association between matrix metalloproteinase-9 and endothelial
nitric oxide synthase gene polymorphisms and coronary artery
disease in Turkish population. DNA Cell Biol. 28:343–350. 2009.
View Article : Google Scholar
|
|
41
|
Alkharfy KM, Al-Daghri NM, Al-Attas OS,
Alokail MS, Draz HM and Hussain T: Endothelial nitric oxide
synthase gene polymorphisms (894G > T and −786T > C) and risk
of coronary artery disease in a Saudi population. Arch Med Res.
41:134–141. 2010.
|
|
42
|
Angeline T, Isabel W and Tsongalis GJ:
Endothelial nitric oxide gene polymorphisms, nitric oxide
production and coronary artery disease risk in a South Indian
population. Exp Mol Pathol. 89:205–208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bor-Kucukatay M, Demir S, Akbay R,
Dursunoglu D, Akdag B and Semiz E: Relationship between
hemorheology and Glu(298)Asp polymorphism of endothelial nitric
oxide synthase gene in patients with coronary artery disease. Mol
Biol Rep. 37:171–178. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Isordia-Salas I, Leaños-Miranda A and
Borrayo-Sánchez G: The Glu298ASP polymorphism of the endothelial
nitric oxide synthase gene is associated with premature ST
elevation myocardial infarction in Mexican population. Clin Chim
Acta. 411:553–557. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Salimi S, Firoozrai M, Zand H, et al:
Endothelial nitric oxide synthase gene Glu298Asp polymorphism in
patients with coronary artery disease. Ann Saudi Med. 30:33–37.
2010.PubMed/NCBI
|
|
46
|
Velloso MW, Pereira SB, Gouveia L, et al:
Endothelial nitric oxide synthase Glu298Asp gene polymorphism in a
multi-ethnical population with heart failure and controls. Nitric
Oxide. 22:220–225. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Motawi T, Shaker O, Taha M, Sedrak H and
Nabil M: Endothelial nitric oxide synthase and angiotensinogen gene
polymorphism in coronary artery diseases in Egypt. Angiology.
62:191–197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Saini V, Bhatnagar MK and Bhattacharjee J:
Association of endothelial dysfunction with endothelin, nitric
oxide and eNOS Glu298Asp gene polymorphism in coronary artery
disease. Dis Markers. 31:215–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
da Costa Escobar Piccoli J, Manfredini V,
Hamester FI, et al: Interaction between endothelial nitric oxide
synthase gene polymorphisms (−786T>C, 894G>T and intron 4
a/b) and cardiovascular risk factors in acute coronary syndromes.
Arch Med Res. 43:205–211. 2012.
|
|
50
|
Gad MZ, Abdel Rahman MF, Hashad IM,
Abdel-Maksoud SM, Farag NM and Abou-Aisha K: Endothelial nitric
oxide synthase (G894T) gene polymorphism in a random sample of the
Egyptian population: comparison with myocardial infarction
patients. Genet Test Mol Biomarkers. 16:695–700. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rahimi Z and Nourozi-Rad R: Association of
endothelial nitric oxide synthase gene variant (G894T) with
coronary artery disease in Western Iran. Angiology. 63:131–137.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Rai H, Fitt J, Sharma AK, et al: Lack of
association between Glu298Asp polymorphism and coronary artery
disease in North Indians. Mol Biol Rep. 39:5995–6000. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Superko HR, Roberts R, Agatston A, et al:
Genetic testing for early detection of individuals at risk of
coronary heart disease and monitoring response to therapy:
challenges and promises. Curr Atheroscler Rep. 13:396–404. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Devuyst O: Variable renal disease
progression in autosomal dominant polycystic kidney disease: a role
for nitric oxide? J Nephrol. 16:449–452. 2003.PubMed/NCBI
|
|
55
|
Takaoka M: NOS gene polymorphism. Nihon
Rinsho. 62:103–109. 2004.(In Japanese).
|
|
56
|
León-Velarde F and Mejía O: Gene
expression in chronic high altitude diseases. High Alt Med Biol.
9:130–139. 2008.
|
|
57
|
Su MT, Lin SH and Chen YC: Genetic
association studies of angiogenesis- and vasoconstriction-related
genes in women with recurrent pregnancy loss: a systematic review
and meta-analysis. Hum Reprod Update. 17:803–812. 2011. View Article : Google Scholar : PubMed/NCBI
|